Abstract
Shoot apical meristems (SAMs), which are maintained at the tips of stems, are indeterminate structures and sources of stem cells from which all aerial organs are ultimately derived. Although mechanisms that regulate the homeostasis of the stem cells have been extensively investigated, identification of further unknown regulators should provide better understanding of the regulation. Here, we report that members of the Arabidopsis ERECTA (ER) receptor kinase family redundantly play a significant role in the regulation of stem cell homeostasis. In wild-type seedlings, the expression of WUSCHEL (WUS), a central regulator of the stem cell population, is stimulated by cytokinin. Interestingly, however, the SAM morphology and the expression of CLAVATA3 (CLV3), which is expressed in stem cells and therefore serves as a stem cell marker, are relatively stable against cytokinin treatment regardless of increased WUS expression. These findings indicate the presence of a mechanism to buffer stem cell homeostasis against an increase in cytokinin. Mutant seedlings lacking all ER-family members, which are expressed in the SAM, show an increase in the stem cell population and also the up-regulation of a cytokinin-responsive gene in the SAM. In this mutant, WUS expression is stimulated by cytokinin treatment as efficiently as in wild-type plants. However, in contrast to wild-type plants, SAM morphology and CLV3 expression respond drastically to cytokinin treatment, suggesting that the buffering mechanism to maintain stem cell homeostasis against an increase in cytokinin is severely impaired in this mutant. We suggest that the ER family regulates stem cell homeostasis via buffering its cytokinin responsiveness in the SAM.
Keywords: Arabidopsis thaliana, Cytokinin, ERECTA family, Receptor kinases, Shoot apical meristem, Stem cells
Introduction
The shoot apical meristem (SAM) plays significant roles in establishment of the plant architecture. The SAM, which is maintained at the tips of stems, is an indeterminate structure and serves as a source of stem cells from which all post-embryonic aerial organs are ultimately derived. Mechanisms that regulate the homeostasis of the stem cells in the SAM have been investigated and some key regulators have been reported (Besnard et al. 2011). Among them, WUSCHEL (WUS) is expressed at the organizing center just below the stem cell region of the SAM and is required for the maintenance of stem cells (Laux et al. 1996, Mayer et al. 1998). WUS activates the expression of CLAVATA3 (CLV3) in stem cells (Fletcher et al. 1999), which encodes a secreted small peptide hormone (Kondo et al. 2006, Ohyama et al. 2009) and also serves as a useful stem cell marker gene (Yadav et al. 2009). The CLV3 peptides in turn repress WUS expression (Kondo et al. 2006, Ohyama et al. 2009). This local WUS–CLV3 feedback loop ensures a constant number of stem cells in the SAM (Brand et al. 2000, Schoof et al. 2000, Betsuyaku et al. 2011). Interestingly, in the inflorescence SAM, this WUS–CLV3 regulatory circuit was shown to behave relatively stably against fluctuations of the amounts of each component, suggesting the presence of a failsafe mechanism to guarantee stem cell maintenance (Muller et al. 2006, Gordon et al. 2009, Chickarmane et al. 2012). The plant hormone cytokinin is also known to participate in the regulation of stem cell homeostasis by the WUS–CLV3 circuit (Leibfried et al. 2005, Gordon et al. 2009, Chickarmane et al. 2012). Although cytokinin signaling activates WUS expression, cytokinin treatment did not cause the drastic enhancement of SAM size in wild-type inflorescences, suggesting the presence of a mechanism buffering stem cell homeostasis against an increasing amount of cytokinin in the inflorescence SAM (Gordon et al. 2009, Chickarmane et al. 2012). However, although some genes involved in cytokinin signaling pathways have been shown to function in this buffering mechanism (Gordon et al. 2009, Chickarmane et al. 2012), information about further unknown participants is still missing. Moreover, it is unknown whether such a buffering mechanism for stem cell homeostasis also exists in the vegetative SAM at the seedling stage.
The Arabidopsis ERECTA (ER) family consists of ER, ERECTA-LIKE 1 (ERL1) and ERECTA-LIKE 2 (ERL2) (Shpak et al. 2004), and encodes receptor kinases (Torii et al. 1996, Shpak et al. 2004). The ER-family members are broadly expressed in various tissues (Yokoyama et al. 1998, Shpak et al. 2004, Uchida et al. 2012) and play roles in diverse aspects of plant development by mediating cell–cell signals that sense and coordinate organ development (Torii et al. 1996, Shpak et al. 2004, Uchida et al. 2012). Although loss-of-function mutations in the ER gene do not show obvious defects in SAM regulation (Torii et al. 1996), er mutations have been reported to modify meristem defects caused by other mutations (Xu et al. 2003, Qi et al. 2004, Uchida et al. 2011), suggesting that ER might play a significant but unknown role in SAM regulation. Also, simultaneous loss of all ER-family members confers extreme dwarfism (Shpak et al. 2004), implying the possibility that the ER-family members are redundantly involved in SAM function, although this possibility has not been investigated so far.
In this study we report that in seedling plants lacking all ER-family members, which are expressed in the SAM, defects in stem cell homeostasis are observed and the regulation of the cytokinin responsiveness in the SAM is severely impaired. We suggest that the ER family regulates stem cell homeostasis via buffering of its cytokinin responsiveness in the vegetative SAM.
Results
All ER-family members are redundantly involved in SAM regulation
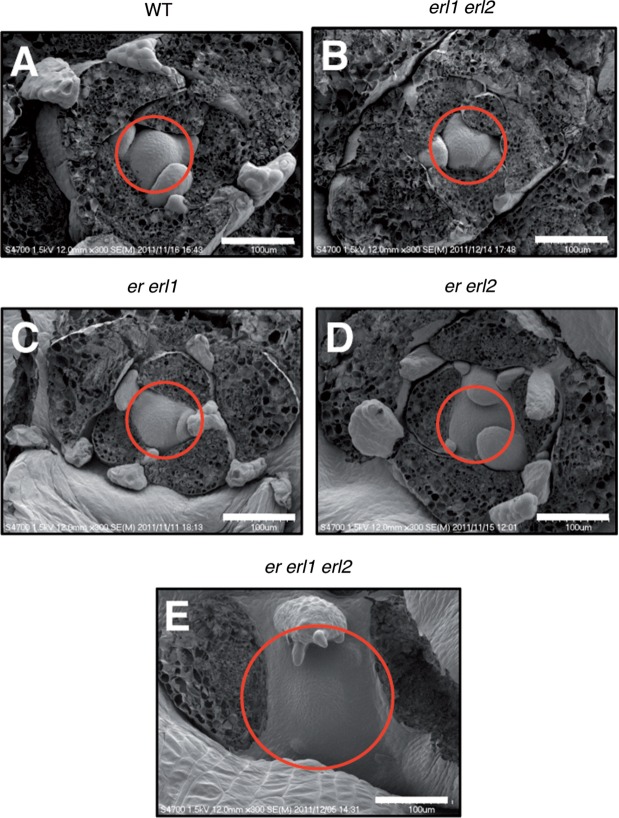
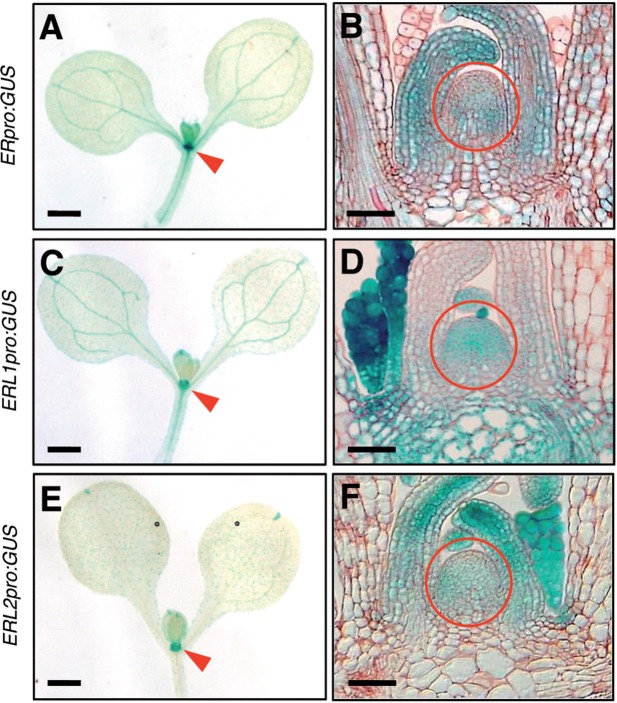
Although er mutations do not show obvious defects of SAM regulation, they affect meristem phenotypes caused by other mutations (Xu et al. 2003, Qi et al. 2004, Uchida et al. 2011), suggesting that ER might play a significant but unknown role in SAM regulation. We assumed that its two paralogous genes, ERL1 and ERL2, might mask the appearance of potential defects in the SAM of er mutants. Therefore, to investigate the involvement of ER-family members in SAM regulation, we compared SAM morphologies of multiple mutants of various combinations among er-family members (Fig. 1). Among the multiple mutants we analyzed, erl1 erl2, er erl1 and er erl2 double mutants did not exhibit apparent changes in SAM morphology compared with wild-type seedlings (Fig. 1A–D). On the other hand, er erl1 erl2 triple mutant seedlings clearly showed the abnormal SAM morphology (Fig. 1E). The SAM of er erl1 erl2 seedlings appeared to be flat and broadened compared with the wild-type SAM (Fig. 1A, E). This observation was supported by examination of longitudinal sections of the SAM (see later Figs. 3B, D, F, 5G). Thus, all ER-family members are redundantly involved in the regulation of the vegetative SAM. Although the promoter activities of the ER-family members were roughly detected at regions including the shoot apex at the seedling stage in a previous study (Shpak et al. 2004), the authors did not examine whether they were active inside the SAM at that stage. When we examined the promoter activities of ER, ERL1 and ERL2 using β-glucuronidase (GUS) reporter genes at the seedling stage, all promoters showed strong GUS signals at regions including the shoot apex (Fig. 2A, C, E) as reported (Shpak et al. 2004). Our observation of thin plastic sections of the shoot apex regions clearly indicated that GUS signals were detected inside the SAM in all cases (Fig. 2B, D, F). These expression patterns of ER-family members within the SAM appear consistent with the fact that all ER-family members are redundantly involved in SAM regulation (Fig. 1E).
Fig. 1.
SAM morphologies of multiple mutants of various combinations among ER-family members. The SAMs of 9-day-old seedlings were observed by scanning electron microscopy. Circles indicate the SAMs. Bars = 100 µm.
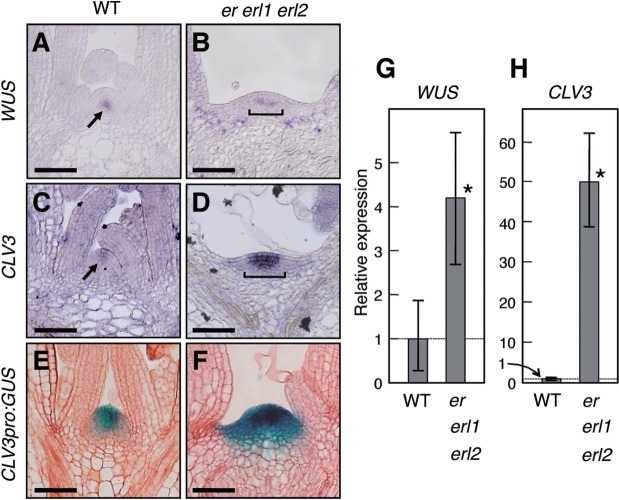
Fig. 3.
Expression patterns and levels of WUS and CLV3. Nine-day-old seedlings were analyzed. (A–D) In situ RNA hybridization. Arrows and brackets indicate signals. Bars = 50 µm. (E and F) CLV3pro:GUS. Bars = 50 µm. (G and H) Aerial parts were analyzed by qRT-PCR. The expression levels were normalized with respect to that of β-TUBULIN and the normalized values in the wild type were set at 1. Data shown are means of three independent samples (10 individuals were collected as a pool for each sample) with error bars representing the SD. Asterisks indicate significant differences by Student’s t-test (P < 0.05).
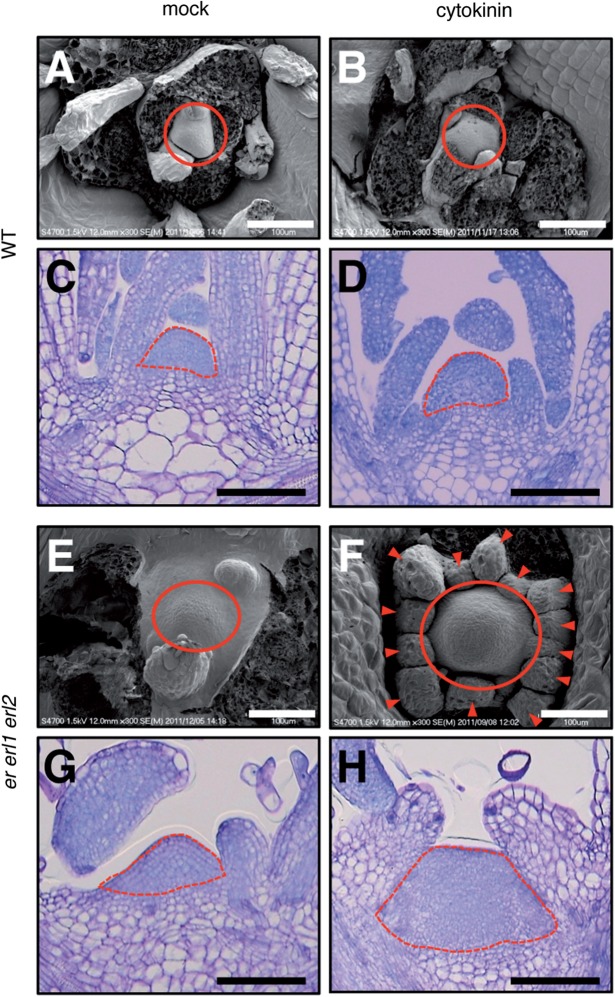
Fig. 5.
SAM morphologies with/without cytokinin treatment. The SAMs of 10-day-old seedlings grown on normal or cytokinin-containing media were observed. (A, B, E and F) Observation by scanning electron microscopy. Circles indicate the SAMs. Arrowheads indicate leaf primordium-like structures. Bars = 100 µm. (C, D, G and H) Thin plastic sections. Dotted lines show the SAMs. Bars = 100 µm.
Fig. 2.
GUS reporter lines to show promoter activities of ER-family members. Nine-day-old seedlings were analyzed. (A, C and E) Whole seedlings. Bars = 1 mm. (B, D and F) Thin plastic sections of the SAMs. Bars = 50 µm. Arrows and circles indicate the shoot apex and the SAMs, respectively.
Stem cell homeostasis is severely impaired in er erl1 erl2 seedlings
To analyze the SAM defects of er erl1 erl2 seedlings further, we examined the expression pattern and level of WUS, a central regulator of the stem cell population, and also CLV3 as a stem cell marker gene. WUS expression was observed only in a limited region in the SAM of wild-type plants (Fig. 3A) as reported (Mayer et al. 1998), while er erl1 erl2 plants displayed a slightly broadened expression pattern (Fig. 3B) probably because of the broadened shape of the SAM, but the signal intensities of WUS transcripts between wild-type and er erl1 erl2 plants appeared almost comparable (Fig. 3A, B). This observation was supported by measurement of the WUS transcripts by quantitative real-time PCR (qRT-PCR), which indicated only about a 4-fold increase in the expression in er erl1 erl2 (Fig. 3G). On the other hand, when we examined CLV3 expression, drastic changes in both the expression pattern and level were observed between wild-type and er erl1 erl2 seedlings. In wild-type seedlings, CLV3 was expressed only in several cells at the tip of the SAM (Fig. 3C), while in er erl1 erl2 seedling CLV3 expression was strongly detected in an increased number of cells (Fig. 3D). qRT-PCR indicated a dramatic increase in CLV3 expression in er erl1 erl2 seedlings (Fig. 3H). This observation was further supported by the examination of the CLV3 promoter activity using a GUS reporter gene (Fig. 3H). Thus, stem cell homeostasis is severely impaired in the SAM of er erl1 erl2 seedlings.
The expression of the cytokinin-responsive ARR15 is expanded in the SAM of er erl1 erl2 seedlings
The plant hormone cytokinin is one of the key factors affecting stem cell homeostasis (Leibfried et al. 2005, Gordon et al. 2009, Chickarmane et al. 2012). To examine the involvement of the action of cytokinin in the defects observed in the SAM of er erl1 erl2 seedlings, we checked the expression pattern of the type-A ARABIDOPSIS RESPONSE REGULATOR 15 (ARR15), which has been reported to be one of the representative cytokinin-responsive genes in the SAM (Leibfried et al. 2005, Zhao et al. 2010). The promoter activity of ARR15 using a GUS reporter gene was strongly detected in the central region of the SAM, but rarely or very weakly in the peripheral regions of the SAM (Fig. 4A). On the other hand, the ARR15 promoter was active in the entire region of the SAM of er erl1 erl2 seedlings (Fig. 4B). This raised the possibility that the SAM of er erl1 erl2 seedlings might possess the potential to respond to cytokinin more than the wild-type SAM.
Fig. 4.
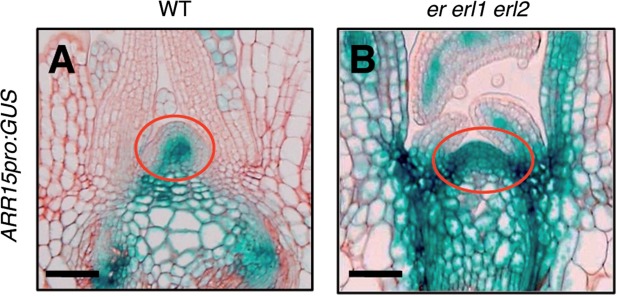
Expression patterns of ARR15pro:GUS. Nine-day-old seedlings were analyzed. Circles indicate the SAMs. Bars = 50 µm.
The SAM responds drastically to cytokinin when the activity of the whole ER family is attenuated
To examine the above possibility, we compared responses to cytokinin treatment between wild-type and er erl1 erl2 plants. When wild-type seedlings were treated with cytokinin, the SAM morphology did not obviously change (Fig. 5A, B). This observation was supported by the examination of longitudinal sections of the SAM (Fig. 5C, D), suggesting that, from the viewpoint of morphology, the vegetative SAM is resistant to an increase in cytokinin in wild-type seedlings. On the other hand, when er erl1 erl2 seedlings were treated with cytokinin, the SAM morphology was altered dramatically (Fig. 5E, F). The originally flat SAM of er erl1 erl2 seedlings (Fig. 5E) rose into an enlarged mound-like structure (Fig. 5F) with formation of multiple bumps (Fig. 5F, arrowheads), which had the appearance of leaf primordia, at the SAM periphery. Furthermore, longitudinal sectioning (Fig. 5G, H) clearly indicated that the mound-like SAM of the cytokinin-treated er erl1 erl2 seedlings was filled with a very large number of small cells (Fig. 5H). These show that the mechanism to buffer the cytokinin responsiveness, which is functional in the wild-type SAM, is severely impaired in the SAM of er erl1 erl2 seedlings.
The above observation raised a question as to whether the loss of function of the entire ER family might confer enhanced cytokinin responsiveness of the whole plant body. To examine this, we checked expression levels of several cytokinin-responsive type-A ARR genes (ARR4, ARR5, ARR6, ARR7, ARR8, ARR9, ARR15 and ARR16) in the whole plant bodies by qRT-PCR. All genes we examined were up-regulated by cytokinin treatment to a similar extent between wild-type and er erl1 erl2 seedlings (Supplementary Fig. S1), indicating that the global cytokinin responsiveness was not obviously altered between wild-type and er erl1 erl2 plants. Given that the SAM morphology of er erl1 erl2 seedlings was drastically altered by cytokinin treatment (Fig. 5F, H) in contrast to that of wild-type plants, the SAM appears to be a tissue that displays enhanced responsiveness to cytokinin when the activity of the whole ER family is attenuated.
Also, as judged by excessive formation of leaf primordium-like structures (Fig. 5F, arrowheads) in cytokinin-treated er erl1 erl2 seedlings and the appearance of cells in the structures (Fig. 5F, arrowheads, and 5H), it was implied that, in addition to homeostasis of the SAM, leaf development might also be affected by cytokinin in er erl1 erl2 plants. Actually, as shown in Supplementary Fig. S2, cytokinin treatment induced excessive formation of apparently abnormal leaves with a squat shape in er erl1 erl2 plants (Supplementary Fig. S2C, D), while cytokinin treatment of wild-type seedlings did not exert significant effects on leaf morphology (Supplementary Fig. S2A, B). In cytokinin-treated er erl1 erl2 seedlings, trichomes were still observed on the malformed leaves (Supplementary Fig. S2D), suggesting that their fundamental leaf structure seemed to be maintained. It remains to be investigated whether the SAM of cytokinin-treated er erl1 erl2 seedlings generates abnormal leaf primordia that secondarily result in malformed leaves or if er erl1 erl2 leaves show enhanced cytokinin responsiveness during their development.
The mechanism to buffer CLV3 expression against the increased expression of WUS by cytokinin treatment is impaired in er erl1 erl2 seedlings
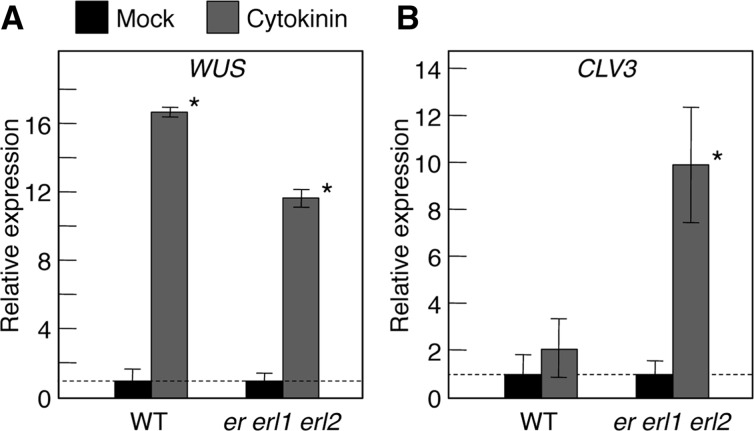
Previously it was reported that, although cytokinin treatment greatly activated WUS expression in the inflorescence SAM, CLV3 expression was relatively stable against the treatment, indicating that the inflorescence SAM possesses a mechanism to maintain CLV3 homeostasis regardless of the increased expression of WUS by cytokinin treatment (Gordon et al. 2009). Because the morphology of the vegetative SAM of wild-type seedlings displayed a significant resistance to cytokinin treatment (Fig. 5A–D), we examined whether a similar buffering system to maintain the CLV3 homeostasis is functional in the vegetative SAM as well as in the inflorescence SAM. When wild-type seedlings were treated by cytokinin, WUS expression was greatly up-regulated (Fig. 6A), while CLV3 expression was not significantly altered (Fig. 6B), indicating that the vegetative SAM also possesses the functional buffering mechanism to maintain CLV3 homeostasis regardless of an increase in WUS induced by cytokinin treatment. Next, we examined whether this buffering mechanism is still functional in er erl1 erl2. When er erl1 erl2 seedlings were treated with cytokinin, WUS expression was up-regulated similarly to the case of wild-type seedlings (Fig. 6A). On the other hand, CLV3 expression was also greatly up-regulated in er erl1 erl2 (Fig. 6B) in contrast to the case of wild-type plants where CLV3 expression was relatively stable (Fig. 6B). This indicates that the mechanism to buffer CLV3 homeostasis against an increase of cytokinin is impaired in er erl1 erl2.
Fig. 6.
Expression levels of WUS and CLV3 with/without cytokinin treatment. Aerial parts of 10-day-old seedlings grown on normal or cytokinin-containing media were analyzed by qRT-PCR. The expression levels were normalized with respect to that of β-TUBULIN and the normalized values without cytokinin treatment were set at 1. Data shown are means of four independent samples (10 individuals were collected as a pool for each sample) with error bars representing the SD. Asterisks indicate significant differences by Student’s t-test (P < 0.05).
Discussion
ER-family receptor kinases redundantly play a significant role in regulation of the SAM
The simultaneous loss of functions of the all ER-family members, which were all expressed in the SAM at the seedling stage (Fig. 2), caused defects in SAM regulation (Figs. 1, 3, 4), clearly indicating that ER-family members redundantly play a significant role in this regulation. The ER family encodes leucine-rich repeat receptor-like kinases in a subfamily of transmembrane-type signaling receptors (Torii et al. 1996, Shpak et al. 2004), and it has been inferred that ER-family proteins function in receptor complexes through associations with multiple partners (Shpak et al. 2003). Although such partners are assumed for regulation of stomatal development (Nadeau and Sack 2002, Lee et al. 2012) in which the ER family has also been reported to be involved (Shpak et al. 2005), information about interacting partners of the ER family in SAM regulation is completely lacking. Moreover, although some ligands for ER-family receptors in other types of developmental regulation have been reported (Hara et al. 2007, Hara et al. 2009, Hunt and Gray 2009, Abrash and Bergmann 2010, Kondo et al. 2010, Sugano et al. 2010, Abrash et al. 2011, Uchida et al. 2012), such ligands for SAM regulation are still unknown. Thus, identification of interacting partners for the receptor complexes and the ligands would be important for further understanding of SAM regulation by the ER-family receptor kinases. Also analysis of intracellular signaling pathways downstream of the ER family would be an interesting future issue.
The ER family regulates stem cell homeostasis via buffering of its cytokinin responsiveness in the vegetative SAM
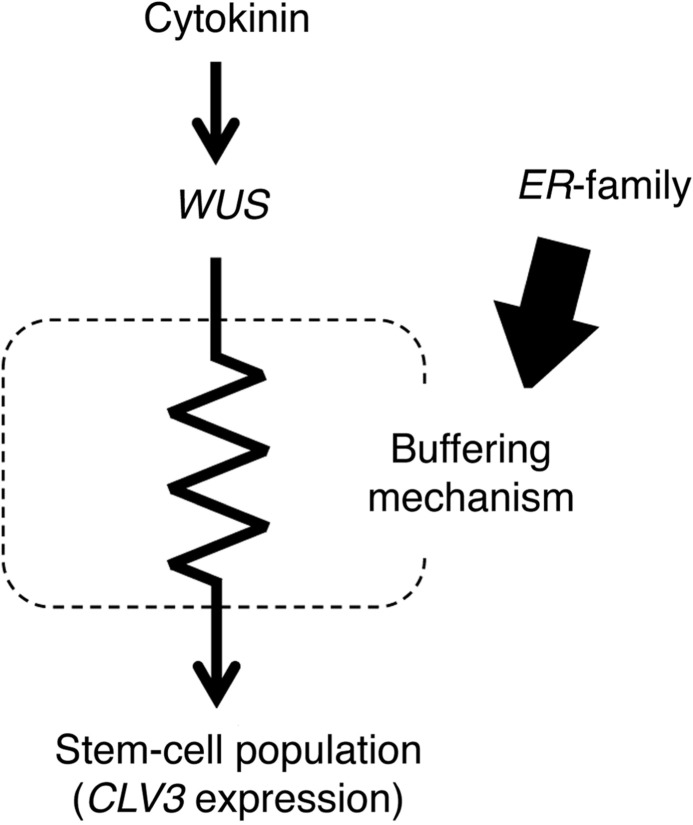
Our results indicate that when the activity of the whole ER family is attenuated, the SAM can no longer buffer cytokinin responsiveness to maintain SAM homeostasis (Figs. 5, 6). WUS expression was greatly stimulated by cytokinin treatment (Fig. 6A) both in the wild-type and in er erl1 erl2 plants, while responses of the stem cell marker CLV3 differed greatly between them (Fig. 6B); wild-type seedlings did not show a significant increase in CLV3, while er erl1 erl2 seedlings did. These findings suggest that the buffering mechanism to maintain SAM homeostasis could be located downstream of the WUS function and upstream of the stem cell regulation marked by CLV3 expression, and that the ER family is involved in the buffering mechanism (Fig. 7). Further investigation of this mechanism would be an important future interest. To that end, we will discuss a few points in the next section.
Fig. 7.
Brief illustration of the proposed system to buffer stem cell homeostasis against an increase of cytokinin. See text for the explanation.
Candidate factors that could be integrated into the mechanism to buffer SAM homeostasis involving the ER family
We observed that the activity of the ARR15 promoter was expanded in the SAM of er erl1 erl2 seedlings (Fig. 4B). ARR15 is considered to be a cytokinin-responsive gene (D’Agostino et al. 2000), while its gene products, ARR15 proteins, are known to serve as a negative factor of cytokinin signaling for negative feedback regulation (Kiba et al. 2003). Therefore, although the SAM of er erl1 erl2 seedlings responds drastically to cytokinin (Figs. 5, 6), it is likely that the negative regulation of cytokinin signaling by ARR15 proteins is still functional even in er erl1 erl2 plants. Because this negative effect of ARR15 proteins is reported to play a significant role in stem cell maintenance (Leibfried et al. 2005), it could serve as one of modifiers of the buffering ability of the system in which the ER family is involved.
Another important aspect of negative feedback regulation of cytokinin signaling is down-regulation of cytokinin biosynthesis enzyme genes, which is induced by activation of cytokinin signaling (Miyawaki et al. 2004). The LONELY GUY (LOG) family encodes enzymes to catalyze the final activation steps to produce active cytokinins (Kurakawa et al. 2007, Kamada-Nobusada and Sakakibara 2009, Kuroha et al. 2009, Tokunaga et al. 2012). Interestingly, a recent paper reported that, among Arabidopsis LOG-family members, regulation of LOG4 expression is integrated into the WUS–CLV3 regulatory circuit (Chickarmane et al. 2012). Actually, when we examined the LOG4 expression levels by qRT-PCR, they were decreased in er erl1 erl2 (Supplementary Fig. S3). Thus, it would be an attractive scenario that the ER family regulates stem cell homeostasis via the regulation of LOG4 expression. In addition, it was reported that LOG1 and LOG7 are also expressed in the SAM (Kuroha et al. 2009, Yadav et al. 2009) and that LOG7 in particular plays a pivotal role in the maintenance of the SAM (Tokunaga et al. 2012). It might be possible that LOG1 and/or LOG7 are also integrated into the WUS–CLV3 regulatory circuit and involved in the buffering system where ER-family members exert their functions.
Recently it has been reported that a cross-talk between cytokinin signaling and auxin signaling plays a significant role in maintenance of the stem cell population (Zhao et al. 2010). Interestingly, the pathways triggered by both hormones directly converge on the promoters of ARR7 and ARR15 (Zhao et al. 2010), which negatively regulate cytokinin signaling; auxin represses their expression, while cytokinin activates it. Although it remains to be shown fully how this cytokinin–auxin cross-talk is integrated into the WUS–CLV3 regulatory circuit, it is possible that auxin signaling is also a candidate component of the buffering system in which the ER family is involved.
Although the detailed molecular mechanisms as to how the ER family contributes to the buffering system to maintain stem cell homeostasis are still largely unknown, we believe that our findings provide a cue to investigate a novel aspect of the regulation of the SAM.
Materials and Methods
Plant materials and growth conditions
The wild-type accession of Arabidopsis thaliana used in this study was Columbia (Col). er-105, erl1-2 and erl2-1 mutants were reported previously (Torii et al. 1996, Shpak et al. 2004). Promoter–GUS reporter lines of the ER family (Shpak et al. 2004), CLV3 (Brand et al. 2002) and ARR15 (Zhao et al. 2010) were also described previously.
Surface-sterilized seeds were plated on Murashige and Skoog medium and then transferred to a growth room at 22°C under continuous white light. For cytokinin treatment, plants were grown on media containing 5 µM 6-benzylaminopurine (BAP).
Quantitative RT-PCR
RNA isolation, synthesis of first-strand cDNA and real-time PCR were performed as previously described (Uchida et al. 2011). Primers used for real-time PCR were as follows; WUS, gcgatgcttatctggaacat and cttccagatggcaccactac; CLV3, aaagtgaatgggttggagca and tcatgtagtcctaaacccttcgt; ARR4, gtcatcgagagattgcttcgt and acgccatccactatctaccg; ARR5, tcagagaacatcttgcctcgt and atttcacaggcttcaataagaaatc; ARR6, gaacattttgcctcgtattgatag and cgagagttttaccggcttca; ARR7, tcatctgagaacatcttacctcgt and ttcaccggtttcaacaagaat; ARR8, acgttcctgcaagaatctcc and ggtttcaacttggtaagatcagc; ARR9, acgttcctgcaagaatcagc and cagccaatcttactggtttcaa; ARR15, gagaacatacaacctcgtatagaacaa and gctaatttcaccggttttagca; ARR16, atggatgtggtggcttatga and tctaagcgcattctctgctg; and LOG4, gtttgatgggtttggtttcg and caccggtcaactctctaggc.
Histology
The procedures of GUS staining were previously described (Uchida et al. 2007). To make plastic sections, materials were fixed in formalin/acetic acid/alcohol (FAA) and then embedded in Technovit 7100 resin as previously described (Uchida et al. 2012). Sections of 4 µm were stained with 0.02% toluidine blue or 0.04% neutral red.
In situ RNA hybridization
In situ hybridization was performed according to Nahar et al. (2001). Templates for transcription of WUS and CLV3 antisense probes were described in Hamada et al. (2000) and Kinoshita et al. (2010), respectively.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology [Grant-in-Aid for Young Scientists (B) (22770038), for Scientific Research (C) (24570050) and for Scientific Research on Innovative Areas (24113513) to N.U., and Grant-in-Aid for Scientific Research (B) (22370019), for Scientific Research on Priority Areas (19060007) and for Exploratory Research (22657015) to M.T.].
Supplementary Material
Acknowledgments
We thank Drs. Keiko U. Torii, Rüdiger Simon, Shinichiro Sawa and Jan U. Lohmann for providing materials.
Glossary
Abbreviations
- ARR
ARABIDOPSIS RESPONSE REGULATOR
- CLV3
CLAVATA3
- ER
ERECTA
- ERL1
ERECTA-LIKE 1
- ERL2
ERECTA-LIKE 2
- GUS
β-glucuronidase
- qRT-PCR
quantitative real-time PCR
- SAM
shoot apical meristem
- WUS
WUSCHEL.
References
- Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development. 2010;137:447–455. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- Abrash EB, Davies KA, Bergmann DC. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand–receptor interactions. Plant Cell. 2011;23:2864–2879. doi: 10.1105/tpc.111.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Vernoux T, Hamant O. Organogenesis from stem cells in planta: multiple feedback loops integrating molecular and mechanical signals. Cell. Mol. Life Sci. 2011;68:2885–2906. doi: 10.1007/s00018-011-0732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, et al. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011;52:14–29. doi: 10.1093/pcp/pcq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Brand U, Grunewald M, Hobe M, Simon R. Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 2002;129:565–575. doi: 10.1104/pp.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. Cytokinin signaling as a positional cue for patterning the apical–basal axis of the growing Arabidopsis shoot meristem. Proc. Natl Acad. Sci. USA. 2012;109:4002–4007. doi: 10.1073/pnas.1200636109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino IB, Deruere J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl Acad. Sci. USA. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Onouchi H, Tanaka H, Kudo M, Liu YG, Shibata D, et al. Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 2000;24:91–101. doi: 10.1046/j.1365-313x.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, Peterson KM, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- Kamada-Nobusada T, Sakakibara H. Molecular basis for cytokinin biosynthesis. Phytochemistry. 2009;70:444–449. doi: 10.1016/j.phytochem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Kiba T, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, et al. The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:868–874. doi: 10.1093/pcp/pcg108. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- Kondo T, Kajita R, Miyazaki A, Hokoyama M, Nakamura-Miura T, Mizuno S, et al. Stomatal density is controlled by a mesophyll-derived signaling molecule. Plant Cell Physiol. 2010;51:1–8. doi: 10.1093/pcp/pcp180. [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–848. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, et al. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes Dev. 2012;26:126–136. doi: 10.1101/gad.179895.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Matsumoto-Kitano M, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37:128–138. doi: 10.1046/j.1365-313x.2003.01945.x. [DOI] [PubMed] [Google Scholar]
- Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar MA, Ishida T, Smyth DR, Tasaka M, Aida M. Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 2011;53:1134–1143. doi: 10.1093/pcp/pcs057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Qi Y, Sun Y, Xu L, Xu Y, Huang H. ERECTA is required for protection against heat-stress in the AS1/AS2 pathway to regulate adaxial–abaxial leaf polarity in Arabidopsis. Planta. 2004;219:270–276. doi: 10.1007/s00425-004-1248-z. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- Shpak ED, Lakeman MB, Torii KU. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell. 2003;15:1095–1110. doi: 10.1105/tpc.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, Mori M, et al. Stomagen positively regulates stomatal density in Arabidopsis. Nature. 2010;463:241–244. doi: 10.1038/nature08682. [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, et al. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69:355–365. doi: 10.1111/j.1365-313X.2011.04795.x. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Igari K, Bogenschutz NL, Torii KU, Tasaka M. Arabidopsis ERECTA-family receptor kinases mediate morphological alterations stimulated by activation of NB-LRR-type UNI proteins. Plant Cell Physiol. 2011;52:804–814. doi: 10.1093/pcp/pcr032. [DOI] [PubMed] [Google Scholar]
- Uchida N, Lee JS, Horst RJ, Lai HH, Kajita R, Kakimoto T, et al. Regulation of inflorescence architecture by intertissue layer ligand–receptor communication between endodermis and phloem. Proc. Natl Acad. Sci. USA. 2012;109:6337–6342. doi: 10.1073/pnas.1117537109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Townsley B, Chung KH, Sinha N. Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc. Natl Acad. Sci. USA. 2007;104:15953–15958. doi: 10.1073/pnas.0707577104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, et al. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl Acad. Sci. USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J. 1998;15:301–310. doi: 10.1046/j.1365-313x.1998.00203.x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, Schultheiss SJ, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.