Abstract
Arabinosylation of hydroxyproline (Hyp) is a post-translational modification often found in secreted peptide signals in plants. The physiological importance of this modification was highlighted by the finding that CLAVATA3 (CLV3), a key peptide signal for regulating the fate of stem cells in the shoot apical meristem in Arabidopsis, contains three l-arabinose residues linked via linear β-1,2-linkages. However, understanding the functions and properties of arabinosylated peptides has been hindered by difficulties in synthesizing the complex arabinose chain. Here we report the stereoselective total synthesis of β-1,2-linked triarabinosylated CLV3 peptide ([Ara3]CLV3). Chemically synthesized [Ara3]CLV3 restricted stem cell activity more effectively than did unmodified CLV3 peptide. Comparison of mono-, di- and triarabinosylated CLV3 glycopeptides revealed that the biological activity increased progressively as the arabinose chain length increased. Thus, the arabinose chain length of CLV3 is important for its biological activity. Nuclear magnetic resonance spectroscopy and nuclear Overhauser effect-based structure calculations further revealed the structural impact of the arabinose chain on peptide conformation. The arabinose chain of [Ara3]CLV3 extends toward the C-terminal end of the peptide, and its non-reducing end is positioned proximal to the peptide backbone. Consequently, the arabinose chain causes distinct distortion in the C-terminal half of the peptide in a highly directional manner. The established synthetic route of [Ara3]CLV3 will greatly contribute to our understanding of the biology and biochemistry of arabinosylated peptide signals in plants.
Keywords: Arabinose, Meristem, Peptide hormone, Post-translational modification
Introduction
Hydroxyproline (Hyp) residues in small secreted peptide signals in plants are often post-translationally modified with pentose sugars. This modification was first identified in the defense-related peptides TobHypSys (tobacco hydroxyproline-rich systemin) I and II, which are small secreted peptides isolated from wounded tobacco leaf extracts (Pearce et al. 2001). TobHypSys I and II exhibit proteinase inhibitor-inducing activities at subnanomolar concentrations. The pentose sugars in TobHypSys peptides are physiologically critical, since chemically synthesized TobHypSys analogs devoid of these sugar chains are 10,000 times less active than the native glycopeptides. Pentose modification was subsequently found in the PSY1 peptide, which regulates cellular proliferation and expansion in Arabidopsis (Amano et al. 2007). Using sugar composition analysis, all three pentose residues of PSY1 have been identified to be l-arabinose.
The physiological importance of Hyp arabinosylation was further highlighted by the identification of the CLAVATA3 (CLV3) glycopeptide in Arabidopsis (Ohyama et al. 2009). Plants continuously produce organs from the self-renewing shoot apical meristem (SAM). A receptor kinase gene, CLAVATA1 (CLV1) (Clark et al. 1997), and a secreted peptide gene, CLV3 (Fletcher et al. 1999), are both expressed in adjacent regions in the SAM. These two peptides are key components of the regulatory network that controls stem cell renewal and differentiation in Arabidopsis (Fletcher and Meyerowitz 2000). Mutations in the CLV genes cause the accumulation of undifferentiated stem cells due to the overactivation of the WUSCHEL (WUS) transcription factor pathway (Schoof et al. 2000), leading to enlargement of the meristem. In contrast, overexpression of CLV3 results in a premature loss of stem cells that leads to developmental arrest. CLV3 belongs to the CLV3/ESR (CLE) family of peptides that contain a short conserved domain (the CLE domain) at or near the C-terminus (Cock and McCormick 2001). We previously identified that the mature CLV3 peptide present in the apoplast of CLV3-overexpressing Arabidopsis plants was a 13 amino acid glycopeptide in which the Hyp7 residue was modified with three l-arabinose residues linked via linear β-1,2-linkages (Ohyama et al. 2009). This arabinosylated CLV3 peptide exhibited considerably higher activity in restricting stem cell proliferation within the SAM, and interacted with its receptor CLV1 more strongly than non-arabinosylated forms (Ogawa et al. 2008, Ohyama et al. 2009).
Despite increasing interest in arabinosylated peptides, the mechanism by which linear β-1,2-linked triarabinoside contributes biological activity and peptide conformation is not well understood. The main difficulty in understanding the functions and properties of a glycopeptide is that chemical synthesis of glycopeptides remains a difficult task due to the absolute stereoselective nature of glycosidic linkages. The installation of a 1,2-cis-glycosidic bond such as β-1,2-linked triarabinoside is more challenging as compared with other sugar linkages due to steric and electronic factors.
We here describe the stereoselective total synthesis of β-1,2-linked triarabinosylated CLV3 peptide using an intramolecular aglycon delivery approach. We also analyzed the solution structure of arabinosylated CLV3 by nuclear magnetic resonance (NMR) spectroscopy to elucidate how the arabinose chain contributes to the conformation of the CLV3 peptide backbone.
Results
Total synthesis of triarabinosylated CLV3 with linear β-1,2-linkages
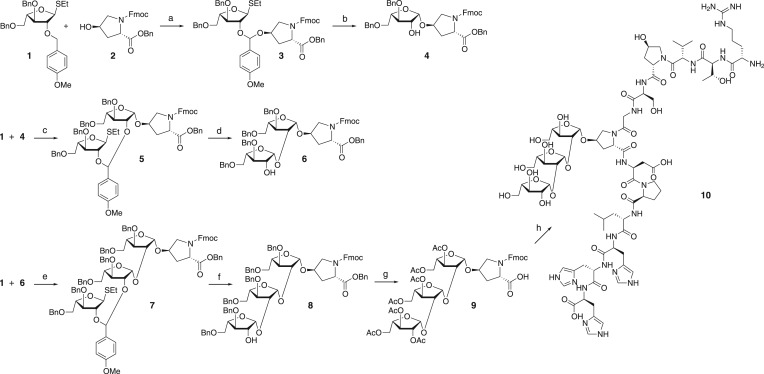
Total synthesis of β-1,2-linked triarabinosylated CLV3 ([Ara3]CLV3) started from the synthesis of the Hyp triarabinoside building block. From the various methods for stereoselective 1,2-cis glycosylation, we chose the intramolecular aglycon delivery approach, which uses the p-methoxybenzyl group as a temporal linker (Ito and Ogawa 1994, Desire and Prandi 1999, Marotte et al. 2003). In this methodology, the glycosyl acceptor is temporarily appended to the C-2-O-protecting p-methoxybenzyl group of the glycosyl donor and is transferred to C-1 from the same face as C-2-O upon activation. This results in the formation of a 1,2-cis-glycosyl bond in an absolutely stereoselective manner.
An arabinosyl donor, ethyl thioarabinoside 1, and an acceptor, Fmoc-Hyp-OBn 2, were treated with 2,3-dichloro-5,6-dicyanobenzoquinone (DDQ) to give the intermediate mixed acetal 3 in 74% yield (Fig. 1). Intramolecular aglycon delivery initiated by iodonium dicollidine perchlorate (IDCP) gave the β-linked Hyp arabinofuranoside 4 in 82% yield. The β configuration of 4 was confirmed by the 1H- and 13C-NMR data (δH-1 4.99 p.p.m., JH-1,H-2 4.7 Hz, δC-1 101.4 p.p.m.). Hyp arabinofuranoside with a free 2-OH group 4 was further reacted with arabinosyl donor 1 and DDQ to give the intermediate acetal 5 in 46% yield. IDCP-promoted intramolecular aglycon delivery afforded the β-1,2-linked Hyp diarabinofuranoside 6 in 75% yield. Finally, reaction of 6 with arabinosyl donor 1 and DDQ gave the intermediate acetal 7 in 54% yield, and intramolecular aglycon delivery as above gave β-1,2-linked Hyp triarabinofuranoside 8 in 65% yield. Because the side chain O-benzyl protecting group of 8 was found to be resistant to deprotection within the peptide chain, the benzyl protecting group was replaced by an acetyl group to give 9 in 31% yield. This triarabinosylated Hyp building block 9 was incorporated into the peptide resin at Hyp7 by solid-phase peptide synthesis. After acidolytic release from the resin, the crude glycopeptide was deacetylated to yield the [Ara3]CLV3 glycopeptide 10.
Fig. 1.
Synthesis of [Ara3]CLV3 glycopeptide. (a) DDQ, CH2Cl2, 74%; (b) IDCP, CH2Cl2, 82%; (c) DDQ, CH2Cl2, 46%; (d) IDCP, CH2Cl2, 75%; (e) DDQ, CH2Cl2, 54%; (f) IDCP, CH2Cl2, 65%; (g) H2, Pd(OH)2/C, CH2Cl2/MeOH/AcOH, then Fmoc-OSu, CH3CN/H2O, NaHCO3, then Ac2O, pyridine, 31%, three steps; (h) solid-phase peptide synthesis, then NaOMe, MeOH.
Biological activity of chemically synthesized [Ara3]CLV3
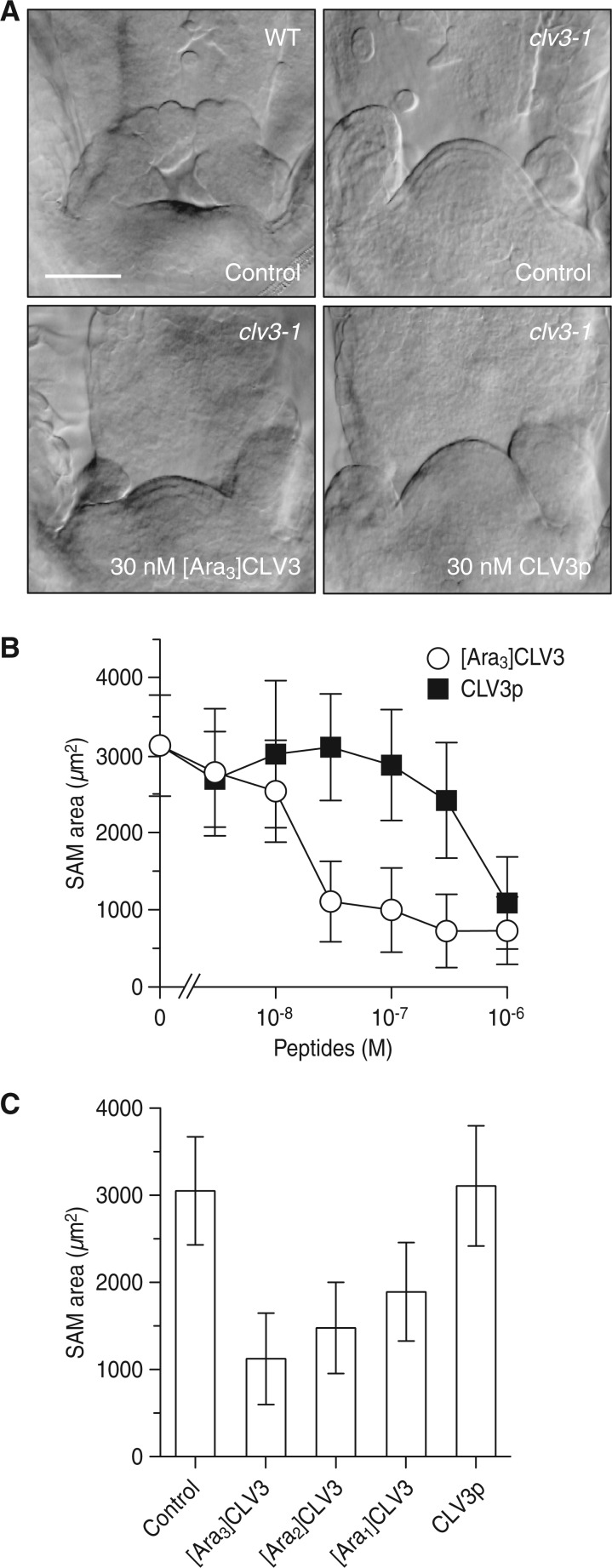
In the Arabidopsis SAM, the CLV3–CLV1 pathway negatively regulates stem cell accumulation by repressing the expression of WUS, a key transcription factor that promotes stem cell identity. Because a loss-of-function mutation in CLV3 causes the overactivation of WUS signaling, seedlings of the clv3 mutant display a large conical SAM due to accumulation of stem cells (Schoof et al. 2000). To test whether chemically synthesized [Ara3]CLV3 rescues the clv3 mutation, we treated clv3-1 mutant seedlings with synthesized [Ara3]CLV3. The SAM of clv3-1 seedlings treated with [Ara3]CLV3 at 30 nM were substantially reduced in size, comparable with the wild type (Fig. 2A, B). In contrast, synthetic peptide devoid of arabinoside (CLV3p) showed weak activity even at 300 nM.
Fig. 2.
Biological activities of synthetic [Ara3]CLV3 glycopeptide. (A) Nomarski micrographs of the SAM of wild-type (ecotype Ler) and clv3-1 seedlings treated with the indicated concentration of peptide for 5 d. The upper panel shows the SAM of wild-type and clv3-1 seedlings without peptide treatment (control). The lower panel shows the SAM of clv3-1 seedlings treated with [Ara3]CLV3 and CLV3p at 30 nM. Scale bar = 50 µm. (B) SAM area of clv3-1 treated with various concentrations of peptide for 5 d. The area of the SAM was measured on a median plane by calculating the area above the straight line between the basal edges of two opposite leaf primordia. Data represent mean values ± SD (n = 18–24). (C) Comparison of the activities of [Ara1]CLV3, [Ara2]CLV3 and [Ara3]CLV3 glycopeptides. The clv3-1 seedlings were treated with each peptide at 30 nM for 5 d. Data represent mean values ± SD (n = 19–40).
We further synthesized mono- and diarabinosylated CLV3 ([Ara1]CLV3 and [Ara2]CLV3) by using mono- and diarabinosylated Hyp building blocks prepared from the synthetic intermediates 4 and 6, respectively. Comparison of the activities of mono-, di- and triarabinosylated CLV3 glycopeptides revealed that the biological activity increased progressively as arabinose chain length increased (Fig. 2C). These results indicate that the arabinose chain of CLV3 is important for full biological activity.
Conformational analysis of [Ara3]CLV3 glycopeptide by NMR
To investigate from a structural aspect how the arabinose chain linked to the Hyp7 residue contributes to the biological functions of [Ara3]CLV3, we compared the solution structures of non-glycosylated CLV3p and [Ara3]CLV3 glycopeptide by NMR spectroscopy. All the proton signals of CLV3p and [Ara3]CLV3 were assigned using a standard procedure based on two-dimensional total correlation spectroscopy (TOCSY) and nuclear Overhauser effect spectroscopy (NOESY) spectra in 90% H2O/10% D2O (Supplementary Tables S1, S2).
A summary of the sequential nuclear Overhauser effect (NOE) connectivities and three-bond coupling constants 3JNα are shown in Supplementary Fig. S1A. Abundant consecutive dαN(i,i+1) connectivities accompanied by intermediate values of 3JNα coupling constants (5.7–8.1 Hz) indicated a predominance of extended backbone conformation in both CLV3p and [Ara3]CLV3. Relatively high temperature coefficients (8.2–8.9 p.p.b. K–1) of the amide protons indicate the absence of shielded hydrogen bonds (Supplementary Tables S1, S2). Strong dαδ(i,i+1) connectivities indicate that Hyp4, Hyp7 and Pro9 are in a trans conformation. However, a comparison of the amide proton chemical shifts between CLV3p and [Ara3]CLV3 revealed that arabinosylation of Hyp7 caused a considerable shift in the δ values for multiple amide protons within the Gly6–His13 region, suggesting a conformational change within the C-terminal half of the peptide upon arabinosylation (Supplementary Fig. S1B). Indeed, a dNN(i,i+1) NOE, which is indicative of turn structure in the peptide backbone, appeared in the Leu10–His11 segment of [Ara3]CLV3. Supporting these results, a sugar–peptide NOE was observed between the δ protons of Hyp7 and the anomeric proton of Ara1, which suggests that the β-face (bottom face) of the Ara1 residue to which the arabinose chain is further linked is turned towards the C-terminal side of the peptide (Supplementary Fig. S1C).
Molecular modeling
In order to gain more insights into the structural changes induced by Hyp arabinosylation, we applied NOE-constrained simulated annealing protocols to calculate possible conformations of CLV3p and [Ara3]CLV3 using CNS software (Brunger et al. 1998). Simulated annealing in the presence of NOE constraints has often been used to obtain optimized structures that are fully consistent with the observed NMR data. A family of 100 structures was generated from an initial extended structure on the basis of interproton distance restraints derived from the observed NOE data. The five structures with the lowest energies were selected for analysis after refinement using conjugate gradient energy minimization. We observed characteristic distortion of the peptide backbone around the Pro-Leu-His sequence within the C-terminal region of [Ara3]CLV3 (Fig. 3A, B). The linear arabinose chain adopts a helical conformation and faces the C-terminal side of the peptide, thus positioning the non-reducing end of the arabinose chain close to the peptide main chain. This bulky arabinose chain appears to cause steric repulsion with the peptide main chain at the C-terminal portion of [Ara3]CLV3 and leads to a characteristic distortion in the peptide. Collectively, the data suggest that Hyp-bound triarabinoside induces a conformational alteration in the peptide backbone toward the C-terminus and contributes to full biological activity.
Fig. 3.
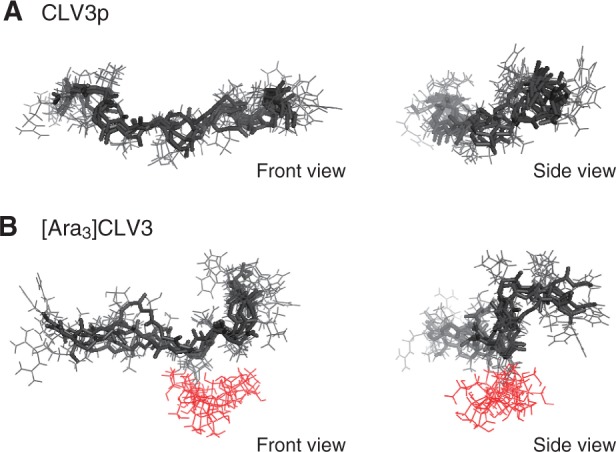
Energy-minimized structures of CLV3p and [Ara3]CLV3 resulting from a simulated annealing protocol that incorporated NOE-derived distance restraints. (A) Superimposition of the five best conformers obtained from simulated annealing calculations on CLV3p (left panel, front view; right panel, side view from the C-terminus). The peptide main chain is depicted by thick lines. (B) Superimposition of the five best conformers obtained from simulated annealing calculations on [Ara3]CLV3. The arabinose chain is represented in red.
Discussion
Secreted peptide signals often undergo post-translational modifications that can affect specific receptor interactions and biological functions (Matsubayashi 2011). One such modification is glycosylation. However, a detailed understanding of the functions of a glycopeptide is often hindered by the difficulty in chemically synthesizing these molecules due to the absolute stereoselective nature of glycosidic linkages. The plant-specific O-glycosylation, Hyp arabinosylation, is one such challenging target for chemical synthesis.
Arabidopsis CLV3, a glycopeptide that regulates stem cell fate in the SAM, is a secreted peptide signal in which Hyp7 is post-translationally modified with three l-arabinose residues connected by linear β-1,2-linkages (Ohyama et al. 2009). The installation of a 1,2-cis-glycosidic bond is more challenging than other linkages due to steric and electronic factors. In the current work, we employed a p-methoxybenzyl-assisted intramolecular aglycon delivery approach for stereoselective synthesis of the Hyp triarabinoside building block and succeeded in the total synthesis of arabinosylated CLV3 glycopeptide. This Hyp triarabinoside building block can be incorporated into any other peptide, and thus will greatly contribute to our understanding of the biology and biochemistry of arabinosylated peptide signals in plants. Chemically synthesized [Ara3]CLV3 exhibited higher stem cell restricting activity than did unmodified CLV3 peptide, indicating the physiological importance of arabinosylation in CLV3 signaling.
In small glycopeptides with high conformational flexibility, glycosylation is generally thought to affect the local secondary structure of the peptide at the residues proximal to the glycosylation site by reducing the number of conformations through steric interactions between the peptide backbone and the sugar chains (Imperiali and Rickert 1995). Sugar chains are highly flexible; therefore, the peptide backbone demonstrates a tendency to bend away from the sugar residue in a rather non-specific way. In contrast, our conformational study of [Ara3]CLV3 revealed that the arabinose chain on the Hyp7 residue specifically faces toward the C-terminal side of the peptide backbone and induces a conformational change at residues somewhat distal from the arabinosylation site. This glycopeptide conformation results from the characteristic triarabinoside branching structure of the vicinal β-1,2-linkages. This branching imparts helical character to the sugar chain, in common with other sugar chains with cis 1,2-linkages (Nitz et al. 2002). The rotational freedom of the arabinose chain is limited by steric constraints between the helical triarabinoside and the peptide N-terminal backbone because of the trans conformation of Hyp7. The arabinose chain, therefore, faces toward the C-terminal side and its non-reducing end extends proximal to the peptide backbone, thereby causing distinct distortion of the C-terminal half of the peptide. We propose that Hyp-bound β-1,2-linked triarabinoside defines a novel class of post-translational glycosylation that induces conformational changes within the peptide backbone in a highly directional manner.
Hyp triarabinosylation has been conserved in land plants including mosses and ferns, suggesting the physiological importance of this modification in plants (Lamport and Miller 1971, Bollig et al. 2007). Plants largely rely on passive diffusion through the cell wall for local cell to cell signaling in the apoplast, but molecular size and diffusion efficiency are inversely proportional: the smaller the molecular weight, the faster the rate of diffusion. Peptide size and structural diversity are, however, also inversely proportional: the shorter the sequence length, the lower the structural diversity. To balance these requirements, plants might have evolved unique post-translational modifications that afford specific structural alterations to small peptides composed of a limited set of amino acids. The plant-specific post-translational modification, Hyp arabinosylation, might be a result of adaptation to such plant-specific endogenous environmental factors.
Materials and Methods
Synthesis of Fmoc-[AcAra3]Hyp-OH
The detailed synthetic procedures are described in the Supplementary methods. In brief, an arabinosyl donor 1 (Desire and Prandi 1999) and an acceptor 2 were treated with DDQ in CH2Cl2 to give the intermediate mixed acetal 3. Intramolecular aglycon delivery initiated by IDCP (Marotte et al. 2003) in CH2Cl2 gave the β-linked Hyp arabinofuranoside 4. Compound 4 was further reacted with arabinosyl donor 1 in the presence of DDQ in CH2Cl2 to provide the intermediate acetal 5. IDCP-promoted intramolecular aglycon delivery in CH2Cl2 as above afforded the β-1,2-linked Hyp diarabinofuranoside 6. Reaction of 6 with arabinosyl donor 1 in the presence of DDQ gave the intermediate acetal 7, and intramolecular aglycon delivery as above gave Hyp triarabinofuranoside 8. All the protecting groups of 8 were deprotected by hydrogenolysis on Pd(OH)2/C and re-protected by treatment with Fmoc-OSu, followed by Ac2O/pyridine, to provide the β-1,2-linked Hyp triarabinofuranoside building block 9.
1H-NMR (500 MHz, CDCl3) (a mixture of Hyp cis/trans rotational conformers): δ = 7.77–7.65 (m, 2 H), 7.61–7.53 (m, 2 H), 7.42–7.28 (m, 4 H), 5.32–4.92 (m, 7 H), 4.72–3.57 (m, 18 H), 2.83–2.28 (m, 2 H), 2.11–1.82 (m, 21 H). ESI-MS: m/z calculated for [C49H57NO24+Na]+: 1,066.3; found: 1,066.4 ([M+Na]+).
Synthesis of [Ara3]CLV3
Building block 9 was incorporated into the peptide at Hyp7 by solid-phase peptide synthesis. The peptides were cleaved from the resin by trifluoroacetic acid and lyophilized. Finally, the acetyl groups on the arabinoses were deprotected by sodium methoxide in dry methanol. HPLC purification using an amide column (TSK-gel Amide-80, TOSOH) gave analytically pure [Ara3]CLV3 10.
ESI-MS: m/z calculated for [C78H120N22O32+2H]2+: 939.4; found: 939.4 ([M+2H]2+).
Bioassay
The clv3-1 loss-of-function mutant has been described previously (Fletcher et al. 1999). Surface-sterilized Arabidopsis seeds (∼40 seeds) were directly sown into 1.0 ml of B5 medium containing 1.0% sucrose in the absence or presence of the indicated concentrations of peptide in 24-well microplates, then incubated without shaking under continuous light at 22°C. After 5 d, the seedlings were fixed with acetic acid–ethanol (1 : 9), cleared in a mixture of chloral hydrate, glycerol and water (8 : 1 : 2, w/v/v), and observed under a microscope equipped with Nomarski optics. The area of the shoot apical meristem was measured as previously described (Ohyama et al. 2009).
NMR spectroscopy
NMR spectroscopy for the conformation analysis was carried out using a JEOL JNM-ECA600 instrument equipped with a HCN triple resonance probe. NMR samples were 1.6 mM ([Ara3]CLV3) and 6.4 mM (CLV3p) in 90% H2O/10% D2O. The pH of the aqueous samples was adjusted to 4.8 with a small amount of NaOH. Spectra were collected at 5°C using 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as an internal standard. TOCSY spectra were recorded with an isotropic mixing time of 70 ms. NOESY spectra were acquired with mixing times of 200 and 400 ms. The temperature coefficients of the amide protons were studied by collecting one-dimensional 1H spectra at seven different temperatures between 5 and 35°C in 5°C increments. NMR spectroscopy for the structural assignment was carried out using a JEOL JNM-LA500 instrument.
Molecular modeling and structure calculations
The 400 ms NOESY experiments were used to generate interproton distance constraints. Peaks were classified as strong, medium or weak. These classifications corresponded to upper bounds of 2.5, 3.5 and 5.0 Å, respectively. The simulated annealing protocol of the CNS 1.21 program (Brunger 2007) was employed to generate a set of 100 structures, starting from extended template structures, using Cartesian dynamics at 1,000 K followed by cooling to 0 K. The five lowest energy structures were further energy minimized using the conjugate gradient minimization algorithm. Image rendering was achieved using PyMOL (http://www.pymol.org/). Parameter and topology files for l-arabinose and Hyp were obtained by submitting their structural file in Mol-format to the PRODRG2 server (http://davapc1.bioch.dundee.ac.uk/prodrg/) (Schuttelkopf and van Aalten 2004). This program generated parameter and topology files for various structure calculation programs. Minor alterations were made to the nomenclature within these files to incorporate them into CNS format.
Supplementary data
Supplementary data are available at PCP online.
Funding
This research was supported by the Ministry of Education, Culture, Sports, Science, and Technology [Grant-in-Aid for Scientific Research for Priority Areas (No. 19060010 to Y.M. and No. 23012020 to H.S.)]; the Japan Society for the Promotion of Science [Funding Program for Next Generation World-Leading Researchers (No. GS025 to Y.M.)].
Supplementary Material
Acknowledgments
We thank Dr. Yoshinori Uekusa (Institute for Molecular Science, Japan) for helpful discussions on molecular modeling by CNS, and Dr. Michiko Nakano (Institute for Molecular Science) for NMR measurements.
Glossary
Abbreviations
- CLV3
CLAVATA3
- DDQ
2,3-dichloro-5,6-dicyanobenzoquinone
- Hyp
hydroxyproline
- IDCP
iodonium dicollidine perchlorate
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- SAM
shoot apical meristem
- TobHypSys
tobacco hydroxyproline-rich systemin
- TOCSY
total correlation spectroscopy
- WUS
WUSCHEL.
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig K, Lamshoft M, Schweimer K, Marner FJ, Budzikiewicz H, Waffenschmidt S. Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii—conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 2007;342:2557–2566. doi: 10.1016/j.carres.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Cock JM, McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desire J, Prandi J. Synthesis of methyl beta-d-arabinofuranoside 5-[1d (and l)-myo-inositol 1-phosphate], the capping motif of the lipoarabinomannan of Mycobacterium smegmatis. Carbohydr. Res. 1999;317:110–118. doi: 10.1016/s0008-6215(99)00078-6. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Meyerowitz EM. Cell signaling within the shoot meristem. Curr. Opin. Plant Biol. 2000;3:23–30. doi: 10.1016/s1369-5266(99)00033-3. [DOI] [PubMed] [Google Scholar]
- Imperiali B, Rickert KW. Conformational implications of asparagine-linked glycosylation. Proc. Natl Acad. Sci. USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Ogawa T. A novel approach to the stereoselective synthesis of β-mannosides. Angew. Chem. Int. Ed. Engl. 1994;33:1765–1766. [Google Scholar]
- Lamport DT, Miller DH. Hydroxyproline arabinosides in the plant kingdom. Plant Physiol. 1971;48:454–456. doi: 10.1104/pp.48.4.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotte K, Sanchez S, Bamhaoud T, Prandi J. Synthesis of oligoarabinofuranosides from the mycobacterial cell wall. Eur. J. Org. Chem. 2003;2003:3587–3598. [Google Scholar]
- Matsubayashi Y. Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 2011;52:5–13. doi: 10.1093/pcp/pcq169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz M, Ling CC, Otter A, Cutler JE, Bundle DR. The unique solution structure and immunochemistry of the Candida albicans β-1,2-mannopyranan cell wall antigens. J. Biol. Chem. 2002;277:3440–3446. doi: 10.1074/jbc.M109274200. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science. 2008;319:294. doi: 10.1126/science.1150083. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA. Production of multiple plant hormones from a single polyprotein precursor. Nature. 2001;411:817–820. doi: 10.1038/35081107. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.