Abstract
Objective:
To demonstrate the sensitivity of a recently developed whole-brain magnetic resonance spectroscopic imaging (MRSI) sequence to cerebral pathology and disability in amyotrophic lateral sclerosis (ALS), and compare with measures derived from diffusion tensor imaging.
Methods:
Whole-brain MRSI and diffusion tensor imaging were undertaken in 13 patients and 14 age-similar healthy controls. Mean N-acetylaspartate (NAA), fractional anisotropy, and mean diffusivity were extracted from the corticospinal tract, compared between groups, and then in relation to disability in the patient group.
Results:
Significant reductions in NAA were found along the course of the corticospinal tracts on whole-brain MRSI. There were also significant changes in fractional anisotropy (decreased) and mean diffusivity (increased) in the patient group, but only NAA showed a significant relationship with disability (r = 0.65, p = 0.01).
Conclusion:
Whole-brain MRSI has potential as a quantifiable neuroimaging marker of disability in ALS. It offers renewed hope for a neuroimaging outcome measure with the potential for harmonization across multiple sites in the context of a therapeutic trial.
Biomarkers for amyotrophic lateral sclerosis (ALS) are a research priority.1 Although median survival from symptom onset is 3 years, there is a longer “tail” to the survival curve, extending past 10 years for a minority. Diagnosis requires a history of progressive weakness and the demonstration of pertinent upper motor neuron and lower motor neuron (LMN) signs on examination.2 Therapeutic trials rely on survival or change in rate of decline of functional measures.
Degeneration of white matter (WM) tracts detected using diffusion tensor imaging (DTI)3 and primary motor cortical thinning detected by surface-based morphometry4 are 2 promising structural measures, but sensitivity to disability has been variable.5 Progress is being made toward defining a functional signature of cortical damage in ALS.6,7
Magnetic resonance spectroscopic imaging (MRSI) is a noninvasive method of tissue metabolite profiling, demonstrating widespread cortical involvement in ALS.8 The most consistently assessed metabolite is N-acetylaspartate (NAA), which reflects neuronal mitochondrial function and is therefore widely expressed within cerebral neuronal tissues, including cortical pyramidal neurons.9
The Metabolite Imaging and Data Analysis System (MIDAS) is an integrative processing platform for MRSI,10 recently developed to allow whole-brain imaging.11 Data acquired have high intrasubject reliability12 and robust quantification, meaning that accurate concentrations of neurochemicals can be obtained.10 Whole-brain MRSI was applied to a group of patients with ALS (including 2 with primary lateral sclerosis [PLS]) and compared with healthy controls, with a targeted region-of-interest analysis to compare MRSI- with DTI-derived measures of structural damage in relation to disability.
METHODS
Participants.
Patients were diagnosed according to standard criteria for ALS13 and PLS,14 assessed by 2 neurologists working in a tertiary referral center (M.R.T. and K.T.). Participation in the study was offered sequentially to all patients attending the clinic, including new referrals and those in active follow-up without assisted ventilation, able to lie flat for at least 1 hour, and with the capacity to provide informed consent. A minimum sample size of 11 provided 95% power to detect a 10% decrease in NAA with an SD of 10%.
All except 1 patient were apparently sporadic (i.e., did not report a family history of ALS or frontotemporal dementia, although participants were not systematically screened for genetic mutations linked to ALS). All were examined on the day of study (M.R.T.). Duration of disease was calculated as the difference in months between date of initial scan and date of symptom onset. Disability was scored using the ALS Functional Rating Scale–revised (ALSFRS-R, score 0–48, with lower scores representing higher disability).15 Disease progression was calculated as 48 minus the ALSFRS-R score, divided by the disease duration in months. Controls were age-similar healthy individuals.
Standard protocol approvals, registrations, and patient consents.
The study was approved by the South Central Oxford Research Ethics Committee B (08/H0605/85), and all participants provided written informed consent.
Data acquisition.
All data were acquired on a Siemens 3T Trio MR System at the Oxford Centre for Magnetic Resonance Research. The MRSI data were acquired using a 32-channel head coil as described previously.10,16 DTI data (b = 1,000 smm−2, 60 directions, 2 × 2 ×2 mm resolution) and T1-weighted images were acquired using the 12-channel head coil, as described previously.3
MRSI analysis.
MRSI data were analyzed using the MIDAS software, with a fully automated approach using the standard defaults10 (appendix e-1 on the Neurology® Web site at www.neurology.org). Briefly, B0 and eddy corrections were applied to the data, the anatomical extent of which was masked by calculating water and lipid masks. Spectral fitting was performed using a Gaussian lineshape model.17 MRSI data were then signal-intensity normalized by referencing to the simultaneously acquired nonsuppressed water data, taking into account spatially dependent variations in signal intensity. The MIDAS approach includes algorithms for rejecting poor-quality data on a voxel-by-voxel basis. Resulting NAA maps were registered to standard space using registration tools within FSL (www.fmrib.ox.ac.uk/fsl) with inhouse modifications.18,19 To test for differences between patients and controls we used permutation-based testing as implemented in “randomize” within FSL.18 Statistical thresholding was performed by cluster-based analysis with corrected cluster p < 0.05 using threshold-free cluster enhancement.
DTI analysis.
The Diffusion Toolbox of the FMRIB (Centre for Functional Magnetic Resonance of the Brain) was used to fit a tensor model to the DTI data at each voxel. Voxel-wise values of fractional anisotropy (FA; a measure of the directional dependence of water dependence, where higher values reflect greater tissue integrity) and mean diffusivity (MD; a measure of total water diffusion, where lower values reflect greater tissue integrity) were then calculated. Tract-based spatial statistics (TBSS), a method for voxel-wise statistical comparison of DTI metrics, was used to test for differences in FA and MD between patients and controls across the whole-brain WM.20 First, FA data from all subjects were aligned into a common space using the nonlinear registration tool FNIRT,21 which uses a b-spline representation of the registration warp field.22 Next, the mean FA image was created and thinned to produce a mean FA skeleton, representing the centers of all tracts common to the group. Individual subject's FA values were warped onto this group skeleton for statistical comparisons by searching perpendicularly from the skeleton for maximum FA values, and the resulting data were fed into voxel-wise cross-subject statistics. MD values from the same voxels were also projected onto the skeleton for statistical analysis. To test for statistical differences between patients and controls, permutation-based testing was used, as implemented in “randomise” within FSL. As with the MIDAS data, statistical thresholding was determined using a threshold-free cluster enhancement approach with a fully corrected cluster threshold of p < 0.05. For ease of visualization, the results of the TBSS analysis were thickened using “tbss_fill” within FSL.
Region-of-interest analyses.
A corticospinal tract (CST) region of interest was derived from a previously acquired healthy control dataset.23 To do this, an M1 mask was drawn on the standard Montreal Neurological Institute template high-resolution structural image, registered into each subject's native space, and a probabilistic tractography approach was used to reconstruct the CST.24,25 Pathways in each subject were then registered into standard space, binarized, and overlaid to produce population probability maps for each pathway in which voxel values reflected the proportion of subjects in whom a pathway was present. These were then thresholded so that only voxels present in at least 40% of the subjects were included in the mask. To ensure that this CST mask, derived from previously acquired data, was unbiased by potential atrophy in patients in this study, a global WM map was derived by segmenting the T1-weighted scan for each subject within this current study using “FAST” (within FSL); the resulting WM maps were then summed so that only voxels that were WM in every subject were included. The CST mask was then limited to the WM using this WM mask.
The mean NAA, FA, and MD within the CST was then calculated bilaterally for each subject. To directly compare the strength of the correlation between CST NAA and disability and the CST DTI metrics and disability, we performed a Fisher r-to-z comparison (http://faculty.vassar.edu/lowry/rdiff.html).
To make an informal assessment of the anatomical specificity of NAA decreases, a mask of the optic radiations (ORs)—as a tract not typically involved in ALS—was derived in a manner similar to the CST mask. The distribution of the t statistic for the comparison between patients and controls within the CST and OR masks was calculated for NAA, and within the TBSS skeleton of these masks for FA and MD.
RESULTS
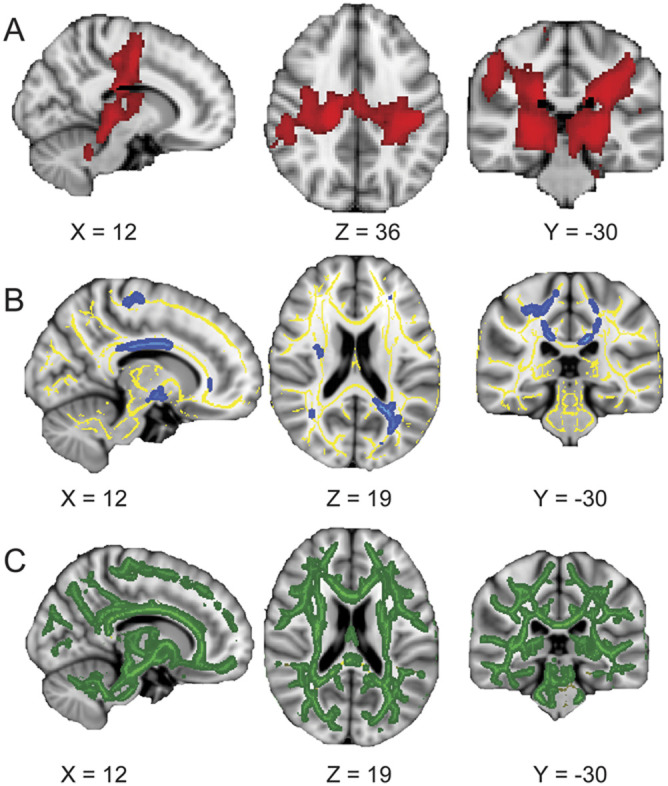
Thirteen patients (8 with “classic” ALS, 3 with LMN-predominant “flail arm” ALS, and 2 with PLS; mean age 62 ± 13 years) and 14 age-similar controls (mean age 58 ± 14 years) were recruited. Clinical details for the patients are shown in the table.
Table.
Patient characteristicsa
Whole-brain MRSI and DTI changes in patients vs controls.
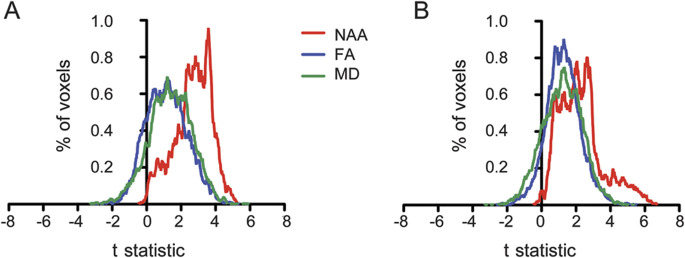
A voxel-wise, whole-brain analysis revealed significantly lower NAA in patients than controls throughout the course of the cerebral CSTs bilaterally (figure 1A). This result remained unchanged in an additional analysis excluding the patients with PLS, and subsequently also the patients with flail arm ALS (appendix e-1). Standard analysis of FA and MD within the WM tracts showed decreases in FA and increases in MD throughout the WM in patients compared with healthy controls (figure 1, B and C).
Figure 1. Regional changes in whole-brain magnetic resonance spectroscopic imaging diffusion tensor imaging in a group of 11 patients with amyotrophic lateral sclerosis and 2 patients with primary lateral sclerosis compared with age-similar healthy controls.
Row (A) shows the results of the whole-brain N-acetylaspartate (NAA) analysis, with significant changes in red; (B) shows the results of the fractional anisotropy (FA) analysis on the white-matter skeleton, with skeleton in yellow and significant changes in blue; whereas (C) shows mean diffusivity (MD) results on the white-matter skeleton, with significant changes in green. Decreased NAA is seen throughout the length of the corticospinal tract (CST) (A), reduced FA particularly within the superior CSTs and corpus callosum (B), and increased MD throughout the white matter tracts (C).
CST MRSI- and DTI-derived measures.
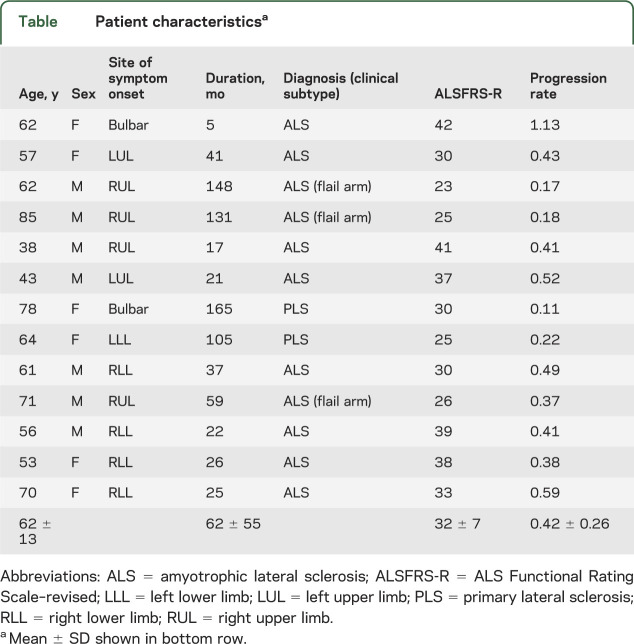
Patients had a significantly lower mean CST NAA than healthy controls (patient NAA = 2,205 ± 49; control NAA = 2479 ± 41; t(25) = 4.336, p = 0.0002, figure 2A). A significantly lower CST FA and higher CST MD was found in the patient group compared with the healthy controls (patient FA = 0.48 ± 0.006; control FA = 0.51 ± 0.005; t(25) = 3.69, p = 0.001, figure 2B; patient MD = 7.47 ± 0.19*104, control MD = 7.22 ± 0.20*104; t(25) = 2.96, p = 0.0005, figure 2C).
Figure 2. Comparison of CST NAA, FA, and MD between patients and controls (A–C), and in relation to disability in the patient group (D–F, 2 patients with PLS are shown as unfilled triangles in each).
Although all 3 measures are significantly different in the patient group, only NAA has a significant relationship with disability. (D) NAA: r = 0.65, p = 0.01; (E) FA: r = −0.03, p = 0.91; (F) MD: r = −0.29, p = 0.32. ALSFRS-R = Amyotrophic Lateral Sclerosis Functional Rating Scale–revised; CST = corticospinal tract; FA = fractional anisotropy; MD = mean diffusivity; NAA = N-acetylaspartate; PLS = primary lateral sclerosis.
Comparison of MRSI- and DTI-derived measures in relation to disability.
Patients with less disability (higher ALSFRS-R score) had higher mean CST NAA concentration (r = 0.65, p = 0.01, figure 2D), and this relationship persisted after correction for age as a potential confound (r = 0.775, p = 0.003). There was no significant relationship between CST DTI measures and disability (FA: r = −0.03, p = 0.91, figure 2E; MD: r = −0.29, p = 0.32, figure 2F; even after correction for age). Using Fisher r-to-z comparison, CST NAA was significantly more correlated with disability than either of the DTI metrics (compared with FA: z = 2.87, p = 0.004; compared with MD: z = 2.4, p = 0.01).
There was no statistically significant correlation between mean CST FA levels and mean CST NAA levels (r = −0.35, p = 0.2), nor between mean CST MD levels and mean CST NAA levels (r = −0.18, p = 0.5). The relationship between NAA and disability remained significant when a partial correlation was performed between CST NAA and ALSFRS-R correcting for CST FA (r = 0.69, p = 0.01), CST MD (r = 0.62, p = 0.02), or both (r = 0.86, p = 0.02).
There was no significant relationship between duration of disease and any MRI metrics within the CST (NAA: r = −0.43, p = 0.14; FA: r = 0.07, p = 0.81; MD: r = 0.227, p = 0.46). There was no significant relationship between the rate of disease progression and any MRI metrics within the CST (NAA: r = 0.36, p = 0.22; FA: r = −0.05, p = 0.87; MD: r = −0.38, p = 0.20).
Anatomical specificity of MRSI- and DTI-derived measures.
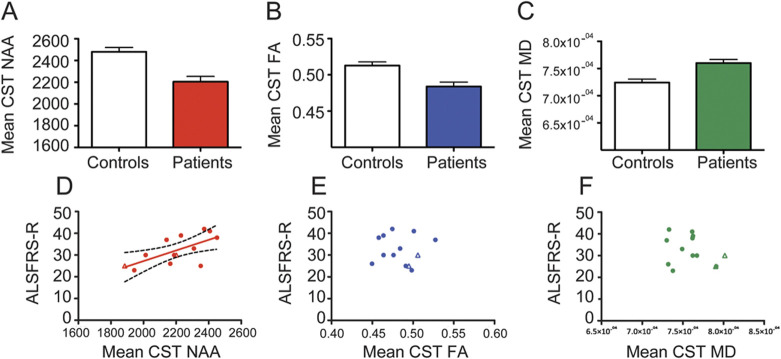
Comparison of the 3 approaches showed greater differences between patients and controls in NAA within the CST compared with FA and MD, as demonstrated by higher t-statistic values within the CST. By comparison, in the OR, the 3 distributions were largely overlapping (figure 3), supporting the greater anatomical specificity of the NAA results.
Figure 3. t Statistic distributions within the corticospinal tract (CST) (left panel) and optic radiation (right panel) showing increased sensitivity of N-acetylaspartate (NAA) compared with fractional anisotropy (FA) and mean diffusivity (MD) within the CST.
DISCUSSION
This whole-brain MRSI study in ALS has demonstrated:
Reduced NAA within the CST as a consistent finding across a group of patients of varying clinical subtype and disease duration.
A significant relationship between CST NAA levels and disability, in contrast to DTI-derived measures.
It has not been possible until recently to acquire MRSI measurements across the entire brain during a single scan. Typically, discrete larger voxels of interest (typically in the order of 2 × 2 × 2 cm) were manually placed instead, relying therefore on subjective (and so potentially inconsistent) judgments about anatomical landmarks, making standardization across subjects highly challenging, and precluding an unbiased assessment of regional metabolite changes. Furthermore, high intrasubject variability in measurements have led to coefficients of variation for the major metabolites, even in healthy controls in optimal conditions, of 10% to 22.6%,26 meaning that measures of NAA have been limited in their utility.
One other recently published whole-brain MIDAS study in ALS extracted metabolite spectra from several points along the CST, and confirmed reduced NAA in the patient group.11 The authors did not report regional findings from whole-brain analysis, nor compare with DTI-derived measures. Their study reported correlations between reduced CST NAA and finger-tapping speed (a measure linked to upper motor neuron dysfunction). It did not note any correlation with disability as measured by ALSFRS-R, which is the current “gold standard” outcome measure in therapeutic trials. Historically, single-voxel MRSI studies have reported a relationship between motor cortex or CST NAA and ALSFRS scores,27,28 as well as finger-tapping speed.29 We did not find a relationship between disease duration or rate of progression and CST NAA. This may be attributable in part to the relatively long disease duration of some of the participants, in whom subjective recall about the timing of initial symptom onset may also be less reliable, and also to the small study size.
In many MRS studies, accurate absolute quantification of NAA has not been possible, meaning that is often quoted as a ratio to other metabolites such as creatine and choline. The former is considered indicative of cellular energy status and the latter to reflect membrane structural integrity. Levels of both increase with neuronal dysfunction, and so giving NAA as a ratio may reduce the specificity of MRS findings. The MIDAS processing has the advantage of allowing calculation of intensity-normalized metabolite values, which enables comparison of results for individual metabolites between subjects.
This study made a direct comparison with DTI-derived biomarker candidates. DTI is currently a leading contender for a noninvasive biomarker of pathology in ALS, with more than 10 years of development since the first observations.30 The WM tract degeneration that DTI is sensitive to is a well-recognized feature of even clinically LMN-only cases of ALS.31 The DTI changes observed in the superior CSTs and corpus callosum mirror closely what has been observed in histologic postmortem ALS studies.32 However, it is perhaps surprising that DTI studies have not consistently demonstrated a relationship between FA (or MD) and disability (studies reviewed in reference 5). An apparent paradox has also emerged whereby higher FA values (thought to reflect greater WM tract integrity) have been noted in patients with the longest disease duration,3,33 despite evidence of decreasing FA values in some, but not all longitudinal studies.34,35 There are also data specifically supporting a role for baseline FA in prognostication.36,37 This raises the possibility that DTI measures might not be as simplistically related to WM tract degeneration in ALS as previously thought.
As well as showing sensitivity to changes over time in patients with ALS,38 MRS has, uniquely, also demonstrated sensitivity to changes associated with the administration of drug therapy.39 Our study is small and cross-sectional, involving a heterogeneous group of patients, and requires validation in other patient cohorts. The performance of longitudinal whole-brain MIDAS will be important to evaluate its potential as a therapeutic efficacy marker. In the longer term, the aspiration of the “Neuroimaging Symposium in ALS” (NISALS) is the standardization of image-acquisition parameters and analysis, with harmonization across multiple sites to translate advanced MRI measures as a source of outcome measures in therapeutic trials.40 The MIDAS platform, as a more standardized methodology, may mark the re-emergence of MRSI as a leading modality in this aspiration.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Matthew Taylor for invaluable technical assistance and David Flitney for providing IT support. The authors are indebted to Care Centre Research Assistant Melanie Lord and Centre Coordinator & Specialist Nurse Rachael Marsden, as well as to the ALS patients and their carers who make inspirational personal effort participating in nontherapeutic research.
Glossary
- ALS
amyotrophic lateral sclerosis
- ALSFRS-R
Amyotrophic Lateral Sclerosis Functional Rating Scale–revised
- CST
corticospinal tract
- DTI
diffusion tensor imaging
- FA
fractional anisotropy
- LMN
lower motor neuron
- MD
mean diffusivity
- MIDAS
Metabolite Imaging and Data Analysis System
- MRSI
magnetic resonance spectroscopic imaging
- NAA
N-acetylaspartate
- OR
optic radiation
- PLS
primary lateral sclerosis
- TBSS
tract-based spatial statistics
- WM
white matter
Footnotes
Supplemental data at www.neurology.org
Editorial, page 606
AUTHOR CONTRIBUTIONS
C.J.S.: analysis and interpretation, critical revision of the manuscript for important intellectual content. S.K.: acquisition of data, critical revision of the manuscript for important intellectual content. K.T.: critical revision of the manuscript for important intellectual content. M.J.: analysis and interpretation, critical revision of the manuscript for important intellectual content. A.A.M.: critical revision of the manuscript for important intellectual content. M.R.T.: study concept and design, acquisition of data, analysis and interpretation, critical revision of the manuscript for important intellectual content, study supervision.
STUDY FUNDING
C.J.S. is funded by the Oxford NIHR Biomedical Research Centre. M.R.T. is funded by the Medical Research Council & Motor Neurone Disease Association UK Lady Edith Wolfson Fellowship. The Oxford Motor Neuron Disease Care & Research Centre (M.R.T. and K.T.) receives funding from the Motor Neurone Disease Association UK Care Centre Program. The MIDAS development (A.A.M.) is supported by NIH grant R01EB000822.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.Turner MR, Kiernan MC, Leigh PN, Talbot K. Biomarkers in amyotrophic lateral sclerosis. Lancet Neurol 2009;8:94–109. [DOI] [PubMed] [Google Scholar]
- 2.Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet 2011;377:942–955. [DOI] [PubMed] [Google Scholar]
- 3.Filippini N, Douaud G, Mackay CE, Knight S, Talbot K, Turner MR. Corpus callosum involvement is a consistent feature of amyotrophic lateral sclerosis. Neurology 2010;75:1645–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verstraete E, Veldink JH, Hendrikse J, Schelhaas HJ, van den Heuvel MP, van den Berg LH. Structural MRI reveals cortical thinning in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2012;83:383–388. [DOI] [PubMed] [Google Scholar]
- 5.Turner MR, Agosta F, Bede P, Govind V, Lule D, Verstraete E. Neuroimaging in amyotrophic lateral sclerosis. Biomark Med 2012;6:319–337. [DOI] [PubMed] [Google Scholar]
- 6.Douaud G, Filippini N, Knight S, Talbot K, Turner MR. Integration of structural and functional magnetic resonance imaging in amyotrophic lateral sclerosis. Brain 2011;134:3470–3479. [DOI] [PubMed] [Google Scholar]
- 7.Agosta F, Canu E, Valsasina P, et al. Divergent brain network connectivity in amyotrophic lateral sclerosis. Neurobiol Aging 2013;34:419–427. [DOI] [PubMed] [Google Scholar]
- 8.Pioro EP, Antel JP, Cashman NR, Arnold DL. Detection of cortical neuron loss in motor neuron disease by proton magnetic resonance spectroscopic imaging in vivo. Neurology 1994;44:1933–1938. [DOI] [PubMed] [Google Scholar]
- 9.Simmons ML, Frondoza CG, Coyle JT. Immunocytochemical localization of N-acetyl-aspartate with monoclonal antibodies. Neuroscience 1991;45:37–45. [DOI] [PubMed] [Google Scholar]
- 10.Maudsley AA, Darkazanli A, Alger JR, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed 2006;19:492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govind V, Sharma KR, Maudsley AA, Arheart KL, Saigal G, Sheriff S. Comprehensive evaluation of corticospinal tract metabolites in amyotrophic lateral sclerosis using whole-brain (1)H MR spectroscopy. PLoS One 2012;7:e35607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR Biomed 2010;23:251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000;1:293–299. [DOI] [PubMed] [Google Scholar]
- 14.Gordon PH, Cheng B, Katz IB, et al. The natural history of primary lateral sclerosis. Neurology 2006;66:647–653. [DOI] [PubMed] [Google Scholar]
- 15.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci 1999;169:13–21. [DOI] [PubMed] [Google Scholar]
- 16.Ebel A, Maudsley AA. Improved spectral quality for 3D MR spectroscopic imaging using a high spatial resolution acquisition strategy. Magn Reson Imaging 2003;21:113–120. [DOI] [PubMed] [Google Scholar]
- 17.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med 1998;40:822–831. [DOI] [PubMed] [Google Scholar]
- 18.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23(suppl 1):S208–S219. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001;5:143–156. [DOI] [PubMed] [Google Scholar]
- 20.Smith SM, Jenkinson M, Johansen-Berg H, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 2006;31:1487–1505. [DOI] [PubMed] [Google Scholar]
- 21.Andersson J, Smith S, Jenkinson M. FNIRT: FMRIB's non-linear image registration tool. In: Fourteenth Annual Meeting of the Organization for Human Brain Mapping; 2008; Melbourne.
- 22.Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 1999;18:712–721. [DOI] [PubMed] [Google Scholar]
- 23.Stepens A, Stagg CJ, Platkajis A, Boudrias MH, Johansen-Berg H, Donaghy M. White matter abnormalities in methcathinone abusers with an extrapyramidal syndrome. Brain 2010;133:3676–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrens TE, Woolrich MW, Jenkinson M, et al. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med 2003;50:1077–1088. [DOI] [PubMed] [Google Scholar]
- 25.Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 2007;34:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall I, Wardlaw J, Cannon J, Slattery J, Sellar RJ. Reproducibility of metabolite peak areas in 1H MRS of brain. Magn Reson Imaging 1996;14:281–292. [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Poptani H, Woo JH, et al. Amyotrophic lateral sclerosis: diffusion-tensor and chemical shift MR imaging at 3.0 T. Radiology 2006;239:831–838. [DOI] [PubMed] [Google Scholar]
- 28.Sivak S, Bittsansky M, Kurca E, et al. Proton magnetic resonance spectroscopy in patients with early stages of amyotrophic lateral sclerosis. Neuroradiology 2010;52:1079–1085. [DOI] [PubMed] [Google Scholar]
- 29.Rooney WD, Miller RG, Gelinas D, Schuff N, Maudsley AA, Weiner MW. Decreased N-acetylaspartate in motor cortex and corticospinal tract in ALS. Neurology 1998;50:1800–1805. [DOI] [PubMed] [Google Scholar]
- 30.Ellis CM, Simmons A, Jones DK, et al. Diffusion tensor MRI assesses corticospinal tract damage in ALS. Neurology 1999;53:1051–1058. [DOI] [PubMed] [Google Scholar]
- 31.Ince PG, Evans J, Knopp M, et al. Corticospinal tract degeneration in the progressive muscular atrophy variant of ALS. Neurology 2003;60:1252–1258. [DOI] [PubMed] [Google Scholar]
- 32.Smith MC. Nerve fibre degeneration in the brain in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 1960;23:269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwata NK, Kwan JY, Danielian LE, et al. White matter alterations differ in primary lateral sclerosis and amyotrophic lateral sclerosis. Brain 2011;134:2642–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Graaff MM, Sage CA, Caan MW, et al. Upper and extra-motoneuron involvement in early motoneuron disease: a diffusion tensor imaging study. Brain 2011;134:1211–1228. [DOI] [PubMed] [Google Scholar]
- 35.Blain CR, Williams VC, Johnston C, et al. A longitudinal study of diffusion tensor MRI in ALS. Amyotroph Lateral Scler 2007;8:348–355. [DOI] [PubMed] [Google Scholar]
- 36.Agosta F, Pagani E, Petrolini M, et al. MRI predictors of long-term evolution in amyotrophic lateral sclerosis. Eur J Neurosci 2010;32:1490–1496. [DOI] [PubMed] [Google Scholar]
- 37.Menke RA, Abraham I, Thiel CS, et al. Fractional anisotropy in the posterior limb of the internal capsule and prognosis in amyotrophic lateral sclerosis. Arch Neurol Epub 2012 Aug 20. [DOI] [PubMed]
- 38.Pohl C, Block W, Karitzky J, et al. Proton magnetic resonance spectroscopy of the motor cortex in 70 patients with amyotrophic lateral sclerosis. Arch Neurol 2001;58:729–735. [DOI] [PubMed] [Google Scholar]
- 39.Kalra S, Tai P, Genge A, Arnold DL. Rapid improvement in cortical neuronal integrity in amyotrophic lateral sclerosis detected by proton magnetic resonance spectroscopic imaging. J Neurol 2006;253:1060–1063. [DOI] [PubMed] [Google Scholar]
- 40.Turner MR, Grosskreutz J, Kassubek J, et al. Towards a neuroimaging biomarker for amyotrophic lateral sclerosis. Lancet Neurol 2011;10:400–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.