Abstract
RNA helicases are compact, machine-like proteins that can harness the energy of nucleoside triphosphate binding and hydrolysis to dynamically remodel RNA structures and protein-RNA complexes. Through such activities, helicases participate in virtually every process associated with the expression of genetic information. Often found as components of multi-enzyme assemblies, RNA helicases facilitate the processivity of RNA degradation, the remodeling of protein interactions during maturation of structured RNA precursors, and fidelity checks of RNA quality. In turn, the assemblies modulate and guide the activities of the helicases. We describe the roles of RNA helicases with a conserved “DExD/H box” sequence motif in representative examples of such machineries from bacteria, archaea and eukaryotes. The recurrent occurrence of such helicases in complex assemblies throughout the course of evolution suggests a common requirement for their activities to meet cellular demands for the dynamic control of RNA metabolism.
Keywords: DExD/H box helicase, RNA degradosome, RNA helicase, RNA surveillance, RNA turnover, RNA-protein interactions, exosome, non-coding RNA, post-transcriptional regulation, ribonuclease
Introduction
Life and its myriad biochemical processes are seldom at standstill. A salient illustration is the dynamic nature of the control of gene expression; consider for instance the cellular RNA transcript pool, which is in continuous flux and responds sensitively to changes in the balance of synthesis and degradation rates. In all kingdoms of life the regulation of transcript degradation is a crucial mechanism of cellular development and response to varying environmental conditions. Key to these processes are numerous enzymes, such as RNA helicases, which manipulate RNA structure and remodel ribonucleoprotein complexes, and a variety of ribonucleases (RNases). The RNases encompass different catalytic mechanisms and substrate preferences, and they often play dual roles in RNA metabolism through their capacity not only to degrade RNA, but also to specifically process precursors of structured RNA into their mature forms.1,2 RNases may also function in conjunction with trans-acting small RNAs to rapidly modulate the expression of specific transcripts, often as part of stress responses.3 While some RNases are capable of working autonomously, many require protein partners, such as the RNA helicases, for full functionality.
This review will explore the interplay of RNases and RNA helicases (particularly the members of the DExD/H box family) in RNA degradation and processing. This interplay is best understood currently in the context of multi-component assemblies, and we discuss how these assemblies affect the functional and allosteric regulation of the helicases, and how the helicases modulate RNA processing and decay.
Nucleic Acid Helicases in Broad Perspective
DNA and RNA helicases are ubiquitous enzymes involved in the manipulation of nucleic acids and their complexes with proteins. Since their identification in the 1980s, it has become increasingly apparent that helicases participate in virtually every cellular process involving nucleic acids, including DNA replication, transcription, RNA-folding and ribosome genesis.4-7 Helicases have been widely understood to be nucleoside triphosphate-dependent enzymes that can interact with nucleic acids and unwind duplex substrates. Unexpectedly, they were also found to displace bound proteins to remodel nucleic acid-protein complexes.5,8
Helicases may be classified into six superfamilies (SF1 to SF6) based on conserved sequence and structural elements,7,9 with all family members possessing Walker A and B motifs that are common to many NTPase enzymes.10 Broadly, helicases can be grouped into two structural categories based on whether they form oligomeric (mostly hexameric) rings or not (http://www.rnahelicase.org/index.html). The ring-forming helicases comprise SFs 3 to 6, and are represented by proteins such as the bacterial Rho transcription termination factor and bacteriophage P4.11
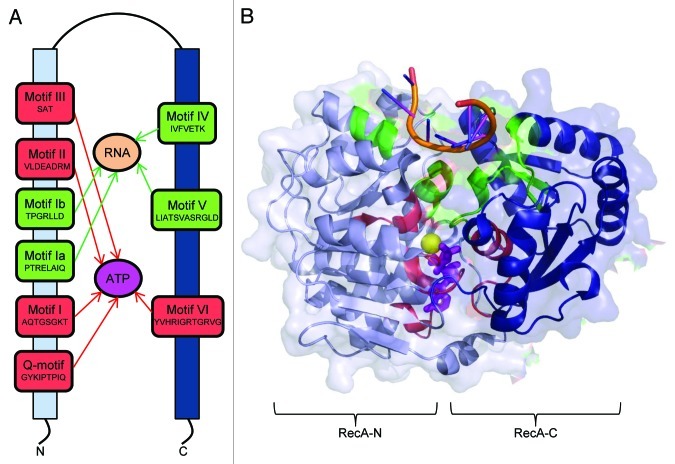
The largest family, SF2, is formed by the DEAD-box and related DEAH, DExH and DExD RNA helicases. These proteins share at least 12 characteristic sequence motifs, some of which are conserved across the SF2 family, while others vary between the sub groups.12 DEAD family members are defined at the simplest level by the presence of an Asp-Glu-Ala-Asp (DEAD) motif (and DExD/H variations thereof) within the ATPase motif II. The conserved sequence motifs are highlighted in Figure 1A from the structure of the representative DEAD-box helicase Vasa of Drosophila. (We will return to this structure, shown in Fig. 1B, in greater detail in the next section). The SF2 proteins are ubiquitous to all kingdoms of life, and multiple paralogues are encoded by the genomes of many organisms.5,13,14 The NTPase activities of these helicases are stimulated by RNA, and while the DEAD-box proteins use ATP as substrate, DEAH proteins have been found to use other NTPs.15 The DEAD family and its close DExD/H relatives will be the main helicases discussed in this review.
Figure 1. The structure of a representative DEAD box helicase engaged with RNA and ATP, and a schematic of the principal helicase superfamily 2 (SF2) motifs. (A) A schematic to show the conserved DEAD box motif and affiliated sequence motifs. The N-terminal RecA domain is shown as a light blue bar and the C-terminal RecA domain is shown as a dark blue bar. Sequence motifs involved with binding to ATP/Mg2+ are highlighted in red, and motifs involved with the interaction with RNA are highlighted in green. (B) The crystal structure of the Drosophila Vasa helicase in complex with single strand RNA and non-hydrolysable ATP. The secondary structural elements of the helicase are represented in cartoon style, and the protein surface is semi-transparent. The conserved RNA binding motifs are shown in green and the RNA is shown in orange. Motifs involved in ATP binding and hydrolysis are colored red, non-hydrolyzable ATP is shown as magenta sticks, and the magnesium ion is shown as a yellow sphere.
It is becoming increasingly apparent that some DExD/H RNA helicases do not in fact have any unwinding activities per se; instead they perform other functions such as strand annealing,16 protein displacement17 and single-strand RNA-displacement.12 Some RNA helicases nucleate ribonucleoprotein assembly by acting as “clamps” on the RNA substrate.12 Often helicases function in multicomponent assemblies to aid RNA degradation and processing, and to act as motors for these macromolecular machines.18
Structure and Mechanism of the DExD/H-Box Motif Helicases
Helicases of superfamilies 1 and 2 contain a conserved core composed of a repeat of a domain that is a structural homolog of the bacterial enzyme for DNA recombination, RecA. The first structural view of how these domains in DEAD-box helicases engage RNA and ATP was provided by the crystal structure of the Drosophila Vasa in complex with single stranded RNA and non-hydrolyzable ATP (AMP-PNP) (Fig. 1B).19 This and subsequent crystal structures of DExD/H-box proteins, together with supporting biophysical data, indicate that the two RecA-like domains of these enzymes do not have a defined relative orientation in the apo form, and are tethered together by a flexible linker. However, with RNA and ATP binding the RecA domains come together to form an intra-domain cleft that clamps down upon the nucleotide (see the magenta stick representation of the AMP-PNP in Fig. 1B). Within this cleft, the ester linkage to the γ-phosphate of the ATP is presented for hydrolytic attack.19-24
The orientation of the domains in the ATP-bound state also generates an RNA-binding surface distal to the ATP binding site, and RNA bound here bridges the two RecA-like domains (top of Fig. 1B). The RecA domains contact mostly the phosphodiester backbone of the RNA, and interactions are mediated by a structural motif whose evolutionary origin was likely to have been an ancient anion-binding module.25 The phosphodiester backbone of the RNA has a kinked conformation that is incompatible with duplex geometry, suggesting that RNA binding by DExD/H-box family members might induce distortions that disrupt RNA secondary structure to favor unwinding. The RNA unwinding activity of the DExD/H helicases is typically restricted to less than two full turns, so it seems that the enzymes can only be weakly processive at best. This behavior might be rationalized by the crystal structure of Vasa, which suggests that the site of interaction with the RNA is not extensive. Another interesting feature of the helicases is that they don’t appear to unwind with strict polarity (i.e., 5′- > 3′, or the reverse direction),26 and this behavior might also be accounted for by the limited extent of the RNA binding surface, which might only permit the helicase to opportunistically grasp exposed single-stranded regions.
The RecA domain interactions with the RNA backbone provide no direct base recognition and therefore do not confer sequence specificity. Nonetheless the different DExD/H-box helicases have an apparent high degree of discriminating specificity for particular RNAs and play well defined roles in vivo. For instance, several yeast DEAD-box proteins have specific functions such as nuclear mRNA splicing or ribosome biogenesis, but these specificities are not recapitulated in vitro with the purified enzymes.12 This puzzling aspect of helicase specificity is most likely explained in part by the variety of domains fused to the conserved RecA core at either or both termini, which can contribute to recognition of sequence or defined structure in the RNA targets. An example of an appended domain that helps to recognize specific RNA targets is the evolutionarily conserved RRM RNA-binding motif, which confers specific recognition of 23S rRNA in bacteria through interaction with a specific base within the context of a defined RNA fold.27,28
Other appended domains, and the RecA core itself, can in some cases help recruit partner proteins that may influence specificity (Fig. 2). The auxiliary domains may form compact folds or more variable polypeptide tails that are predicted to be natively unstructured but nonetheless assist RNA-binding and strand displacement.29-31 These protein-protein interactions, in the context of multi-component assemblies such as the bacterial RNA degradosome, or eukaryotic exon-junction complex (discussed below), may control the activity of the helicases in vivo so that they do not hydrolyse NTPs in futile cycles.32,33
Figure 2. The structure of SF2 family helicases discussed in this review (Vasa, exon-junction complex (EJC), RIG-I, Mtr4, Ski2, hSuv3). For all helicases the two structurally conserved RecA core domains are shown in the same orientation as light blue (N-terminal) and dark blue (C-terminal) cartoons with semi-transparent surfaces. Where present in the crystal structures RNA is shown as an orange cartoon, and ATP/Mg2+ is shown as purple spheres. The figure highlights how auxiliary domains or protein partners can interact with the conserved RecA core of SF2 family members. Additional domains appended to or associating with the helicases are shown as cartoon representation and labeled.
Assemblies with Helicases: The Bacterial RNA Degradosome
In many bacterial species, a multi-enzyme assembly known as the RNA degradosome serves as a central machinery of RNA decay and processing. The components of the degradosome cooperate to degrade most RNA transcripts and to process precursors of structured RNAs.34 In Escherichia coli and several other bacteria, the core of the complex is formed by the conserved endoribonuclease RNase E. The protein composition of the degradosome assemblies differs between evolutionarily divergent bacterial species, and some degradosomes include enzymes such as exoribonucleases and metabolic enzymes, but they often include a DEAD-box helicase that directly associates to an endoribonuclease (Fig. 3). An RNA helicase has been found to be part of the RNA degradosome in several species, including E. coli, Caulobacter crescentus and Pseudoalteromonas haloplanktis where a DEAD-box protein was shown to interact with RNase E by co-immunoprecipitation.35,36 In Pseudomonas syringae and Rhodobacter capsulatus RNA helicases were shown to interact with RNase E by co-sedimentation and co-immunopurification37,38 and in Vibrio angustum a helicase has been shown to bind to RNase E by bacterial two-hybrid analysis and co-immunoprecipitation.39
Figure 3. RNA helicases are recurrent components of diverse bacterial RNA degradosomes. The cartoon provides a schematic of the components of various bacterial RNA degradosomes where RNA helicases are proposed to form part of the complex. A degradosome has also been suggested for Bacillus subtilis, but reports conflict on whether RNase J1/J2 form part of this assembly43 and with data for different subcellular localization of RNase J1 and RNase Y.44 RNase J1/J2 are bordered with a dashed line to represent the uncertainty of whether these proteins form part of the B. subtilis degradosome. The evidence for the Staphylococcus aureus degradosome is based on yeast two-hybrid screens and awaits validation.
Numerous bacterial species lack RNase E, such as the Gram-positive model organism Bacillus subtilis, but in these organisms an RNA degradosome assembly is proposed to be organized through the endoribonuclease RNase Y, which has an entirely different fold and evolutionary ancestry from RNase E.40-42 A subject of debate is whether an RNase Y based degradosome assembly recruits the exo/endoribonucleases RNase J1/J2, with the interaction between RNase Y and RNase J1/J2 in B. subtilis being shown by Strep-protein interaction and bacterial two-hybrid experiments,40,42 but not replicated in separate yeast two-hybrid experiments43 and difficult to reconcile with observations that the subcellular localization of RNase Y and RNase J1/J2 differ in vivo (44; note that RNase Y = YmdA and RNase J1 = YkqC in this reference), although it cannot be ruled out that the fluorescent tag used has not interfered with protein-protein interactions. A putative degradosome assembly in Staphylococcus aureus based on RNase Y has been identified by two-hybrid analysis,45 but a direct interaction between RNase Y and RNase J1/J2 has not been shown.
The degradosome of Escherichia coli
The vast body of functional data on the RNA-degradosome is derived from studies on the E. coli assembly, for which the canonical components and their interaction sites are represented schematically in Figure 4. RNase E has a globular N-terminal catalytic domain46 and a natively disordered C-terminal domain that recruits the other key degradosome proteins: the DEAD-box helicase RhlB, enolase and the exoribonuclease polynucleotide phosphorylase (PNPase). The recognition sites for these partner proteins are small segments of 20 to 40 amino acids found within the C-terminal domain of RNase E, and structures are available that show how these recognize enolase47 and PNPase.48 The degradosome is directed to the cytoplasmic membrane by a small amphipathic helix in RNase E49 and possibly the catalytic domain itself.50 This localization is likely to affect the organization and activity of the degradosome. The binding site for RhlB has been mapped by limited protease digestion30 to a segment that encompasses a conserved 13-residue core motif in RNase E that is essential for the interaction with that helicase36 and which likely interacts with the C-terminal RecA domain of RhlB.30,51 The interaction between RhlB and RNase E stimulates the helicase activity of RhlB,30,52 but there are currently no structural data for the complex that help to explain how this boost occurs.
Figure 4. Domain structure of RNase E with the binding sites for RNA, helicase, enolase and PNPase. One putative protomer of the degradosome assembly is shown, and four such protomers might form the core of the canonical degradosome.34 The disordered C-terminal domain of RNase E is depicted as the thick, blue curvy line. The recognition sites for monomeric RhlB, dimeric Enolase and trimeric PNPase are small segments of 20 to 40 amino acids represented as solid blocks on the C-terminus or RNase E. The catalytic N-terminal domain of RNase E is a tetramer (blue), and one protomer is highlighted with a dotted outline. The large domain harbours the catalytic site and is within the dashed circle, with the missing wedge engaging the RNA substrate; the small domain that organizes the dimer-of-dimer interfaces is outlined with the trapezoid. PDB codes for the structrures presented: RNase E, 2BX2; Vasa, the Drosophila homolog of E. coli RhlB, 2DB3; enolase, 3H8A; PNPase, 3GME.
The identification of the RNA helicase RhlB as part of the E. coli RNA degradosome, nearly two decades ago, led to the proposal that its role is to aid the processing of RNA substrates within the assembly (ref.53; see also ref. 54). As RNase E specifically cleaves single stranded substrates, and the channel for RNA in PNPase is only sufficiently wide to accommodate single-stranded RNA (as we will describe further below), it seems reasonable that the unwinding or remodelling activities of RhlB are required to improve the efficiency of mRNA degradation. Indeed, the ATPase activity of RhlB has been shown to be required for the degradation by PNPase of structured RNA transcripts with a repetitive element that forms stable stem-loop structures.30,55 The binding site for RhlB in RNase E is flanked by RNA-binding domains (Fig. 4; termed RBD for “RNA binding domain” and AR2 for “arginine rich region 2”). The cooperation of RhlB with those two domains is crucial to stimulate ATP-dependent RNA degradation. Additionally the arginine rich tail on the C-terminus of RhlB, which is predicted to be structurally disordered, contributes to the interaction between RNA substrates and this helicase.30
The potential role of RhlB in aiding the degradation of RNA by the RNases of the degradosome assembly is summarized schematically in Figure 5. The model proposes that RNA elements with regions of secondary structure are bound to the RNA degradosome cooperatively through the combined contributions of the RBD, AR2 and RhlB. Based on studies of other DEAD box helicases, ATP-binding by RhlB may be necessary and sufficient for RNA binding or unwinding of the RNA, but hydrolysis of the ATP is the license for product release for the next cycle of RNA unwinding.12 Dissociation of phosphate or ADP favors release of the RNA from the helicase, which is required for continuing the cycle. It is unlikely that the process of RNA unwinding will be a futile cycle in which the RNA abruptly snaps back into its native conformation; instead it is likely that the newly unwound RNA substrate will be efficiently cleaved by both RNase E and PNPase.
Figure 5. A model for the interplay of RNA helicases and ribonucleases in the RNA degradosome. (Top) The degradosome (enolase is not depicted for clarity) primed for RNA substrate capture. (Middle) Structured RNA is captured by the RNA binding elements of RNase E (RBD and AR2) and the C-terminal arginine rich extension of RhlB. ATP binding and hydrolysis promotes cycles of RNA unwinding and remodelling the RNA interactions with RBD and AR2 by RhlB. (Bottom) As RNA unwinding by RhlB proceeds the RNA substrate can be efficiently cleaved by RNase E and/or PNPase. The model is based on inferences from experimental data.12,32
Under stress conditions, some of the four other DEAD box helicases present in E. coli may substitute for RhlB in the degradosome. For instance, CsdA has been shown to associate with RNase E under cold shock conditions.56 Additionally, RhlE has been shown to be able to interact with RNase E in vitro and this helicase can also aid the degradation of mRNA by PNPase.57 Swapping helicases perhaps modifies the degradosome activities or substrate preferences.
Helicases as Threading Machines: Cooperation with the Exoribonuclease Polynucleotide Phosphorylase and the RNA Exosome
The degradosome’s exoribonuclease PNPase cannot by itself degrade structured RNA, and as mentioned above the DEAD-box helicase RhlB aids the ribonuclease in the degradation of such species.53 RhlB facilitates formation of single stranded RNA, which PNPase processively cleaves in the 3′ to 5′ direction by phosphorolysis. PNPase plays many roles in RNA metabolism, including contribution to the decay of bulk RNA, quality control of rRNA, cold shock response, and the stability of small regulatory RNA,2,58-63 and some of these activities are likely to require partnership with the helicase.
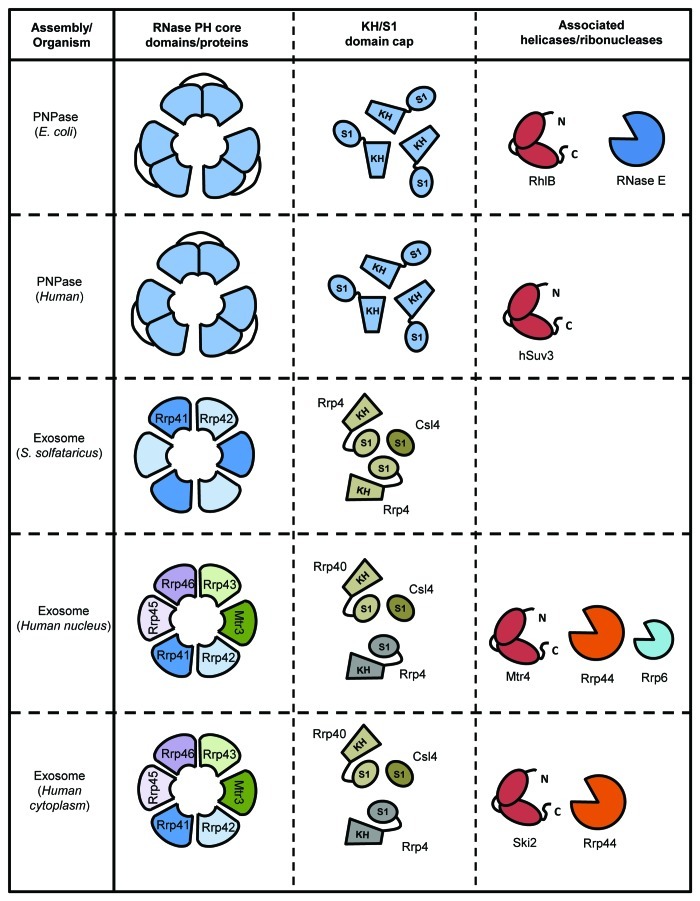
PNPase assembles into a stable homotrimer with a ring-like quaternary architecture that forms a narrow central processing channel through which the substrate threads to reach the three active sites (ref. 64; see Fig. 4). Another, more flexible ring is formed over the entrance to the channel by the arrangement of RNA binding S1 and KH domains; these help to capture and guide the RNA into the channel.65 PNPase is found in all bacterial species examined thus far, and additionally in some eukaryotic organelles including chloroplasts of some plants, where the enzyme is implicated in maintaining phosphate homeostasis66 and mammalian mitochondria, where PNPase is thought to have a role in transporting RNA into the organelle.67 Structurally homologous to PNPase are the archaeal and eukaryotic exosomes, which also share the chamber-like quaternary architecture. The structurally conserved organization of RNase PH domains capped with KH and S1 domains in PNPase and exosome complexes is summarized schematically in Figure 6.
Figure 6. A schematic representation of the domain architecture of PNPase and exosome ring-like assemblies. Fused domains or separate proteins forming the ring-like core are shown in the left column, the arrangement of S1 and KH domain containing proteins forming the “cap” is shown the second column. Additional RNases and/or helicases known to associate with the assembly are shown in the final column.
The eukaryotic exosome was discovered during a search for yeast factors that play roles in RNA processing and degradation.68,69 Its chamber is formed from nine distinct protein subunits that share homology with the corresponding domains of PNPase. In the eukaryotic nucleus, the exosome is required for the processing of rRNAs, small nuclear RNAs, small nucleolar RNAs, and long non-coding RNAs (lncRNAs).70 In the cytoplasm, the exosome participates in bulk mRNA turnover and the degradation of mRNAs with a premature stop codon.
While bacterial PNPase and the archaeal exosome harbour phosphorolytic exo-ribonuclease activity, the eukaryotic exosome has lost this catalytic capacity. Instead, ribonuclease activity is conferred by additional protein components that interact with the nine-subunit exosome core. In both the nuclear and the cytoplasmic forms, the exosome’s endo- and exo-ribonuclease activities are largely attributable to the enzyme Rrp44 that binds to one face of the exo-9 channel (namely, the opposite face to which the S1 and KH like domains associate; see Fig. 7B). Additionally, in the nucleus Rrp6, a homolog of bacterial distributive exo-ribonuclease RNase D, associates with the exosome. Recently, it has been shown that despite lacking ribonuclease activity, the exo-9 channel is still the entry point for RNA substrate, and that the exo-9 channel itself modulates the RNase activities of both Rrp44 and Rrp6.71
Figure 7. The potential cooperation of helicases with processing chambers. The helicase in the RNA degradosome (RhlB) and the TRAMP (Trf4/Air2/Mtr4p Polyadenylation) complex,70 may help to generate single stranded RNA that can be fed into the narrow central channels of PNPase and the exosome, respectively. The crystal structures are shown for the trimeric polynucleotide phosphorylase of C. crescentus [PDB 4AIM (A)] and the heterohexameric eukaryotic exosome [PDB 2NN6 (B)]. The core of the two enzymes, shown in blue, is formed by RNase PH-like subunit that forms the central channel. The KH domains are shown in green and the S1 domains are in salmon. The path of the single stranded RNA is inferred from structural data for the bacterial PNPase65 and the exosome.73,75
Predicted to occur in numerous species,72 the archaeal exosome, like its eukaryotic counterpart, has a nine subunit chamber-like architecture. The archaeal assembly is constructed of three hetero-dimers of Rrp41-Rrp42 and a trimeric cap of Rrp4 or Csl4.73 Both Rrp4 and Csl4 have an N-terminal all β-domain and a central S1-RNA binding domain. Additionally, Rrp4 has a C-terminal KH domain, while Csl4 has a C-terminal zinc-ribbon domain.74 Although the majority of studies on the archaeal exosome have focused on assemblies with a homotrimeric Rrp4 or Csl4 cap, it has recently been shown that hetero-trimeric caps are found in vivo, and it has been suggested that such caps endow the exosomes with capacity to bind different RNA substrates.75
In both PNPase and the exosome, the entrance to the central channel narrows to form an aperture just wide enough for single-stranded RNA substrate to feed through (Fig. 7).64,76,77 The narrow shape suggests a necessity for structured RNA substrates to be un-wound prior to processing by the degradative machinery. As discussed in the previous section, PNPase as part of the RNA degradosome is in proximity to the DEAD-box helicase RhlB. Like the protein complexes formed by PNPase, both the cytoplasmic and nuclear exosomes are known to associate with peripheral proteins assemblies containing RNA helicases. These exosome-associated complexes act as adaptors to bring RNAs to the exosome for processing or degradation.
In the cytoplasm the exosome interacts with the Ski complex,78 comprising the proteins Ski2 (helicase), Ski3 (tetratrico-peptide repeat protein -TPR) and Ski8 (WD40 protein). The Ski complex physically interacts with the exosome via the protein Ski7.79 In the nucleus, the exosome interacts with the TRAMP complex (Trf4/Air2/Mtr4p Polyadenylation complex) comprising poly(A) polymerase (Trf4/Trf5), a zinc finger protein (Air1/Air2), and the DExH helicase Mtr4. This assembly contributes to RNA quality control by recognizing aberrant transcripts, tagging them with polyA tails, and directing them to the exosome. The helicase Mtr4 is required for nearly all of the functions of the exosome in the yeast nucleus,80 including ribosome biogenesis.81 The human homolog of Mtr4 is a component of two distinct complexes that interact with the exosome.82 The potential role of helicases to unwind structured RNA substrates prior to channelling into the narrow processing channels of PNPase and the exosome is summarized schematically in Figure 7.
The crystal structures of the helicase components of the cytoplasmic Ski complex (Ski2) and the nuclear TRAMP complex (Mtr4) have been recently solved.83-85 Both proteins have a classical DEAD-box helicase core, but additionally both proteins have a structurally similar insert within a helical bundle domain appended to the helicase core (Fig. 2). The insertion forms an elongated structure protruding from the helicase core, with a β-barrel domain sitting atop two pairs of anti-parallel α-helices, and it has a role in RNA binding, but not formation of the Ski/TRAMP complexes. In addition, the barrel like KOW domain appended to the elongated helical structure in Mtr4 has been shown to be important for activation of exosome function in vitro.31
There are other links between RNA helicases and components of exosome/PNPase like machineries. In human mitochondria the partner of PNPase is likely to be DExH/D helicase hSuv3.86,87 The crystal structure of the human helicase hSuv3p, thought to interact with human PNPase has also recently been solved and is shown in Figure 2.88 The hSuv3 protein has a similar duplicated RecA-like core with additional strands and auxiliary α-helical domains at both termini, and the C-terminal domain helps to cradle the single stranded RNA on the surface of the RecA-like domains. Much like the Ski2 and Mtr4 helicases, a prominent feature of this structure is an extended helical protrusion emanating from the N-terminal RecA-like domain. Unlike the exosome associating helicases this protrusion is not capped with a barrel like domain, but the conservation of the helical protrusion may suggest a role for this domain in the association with PNPase.
In the yeast Saccharomyces cerevisiae the nuclear exosome component Rrp6 has recently been shown to be genetically linked to the DEAD-box helicase Dbp2, and a double deletion of these two genes results in a synthetic lethal phenotype.89 Dbp2 is the yeast ortholog of the mammalian helicase p68, which has well defined roles in ribosome biogenesis, and transcriptional and co-transcriptional processes with RNA polymerase II.90 Also, the S. cerevisiae Suv3 homolog forms a degradosome complex with the 3′- > 5′ exoribonuclease Dss1 (related to bacterial RNase II).91,92 The mitochondria of Trypanosoma brucei also has a degradosome like assembly, with an RNase II-related exoribonuclease associated with an RNA helicase.93
Helicases in Eukaryotic mRNA Degradation Pathways
In eukaryotes, one mRNA decay pathway involves deadenylation of the 3′ end of the transcript, followed by de-protection of the 5′ end of the RNA by removing the 7-methylguanosine cap, which licenses 5′- > 3′ exoribonuclease degradation. In this pathway, the RNA helicase Dhh1/Rck/DDX6 is a cofactor that promotes the decapping process, and recent evidence suggests that this results from the action of the helicase to slow ribosome transit rates on the transcript, making them more vulnerable to decapping.94 The Dhh1 homolog in trypanosomes affects post-transcriptional regulation of many developmentally important transcripts in an ATPase dependent manner,95 and the mechanism, hypothetically, might involve ribosome transit. In an interesting parallel, the RhlB helicase of the degradosome has been found to help recruit the degradosome to polysomes in E. coli, and it is proposed that this interaction may help sRNA mediated silencing of target transcripts.96
The human DEAH helicase RHAU (DHX36, G4R1) is involved in AU-rich element mediated mRNA decay.97 The AU-rich element is a degradation signal in the 3′ untranslated region and is found in many unstable mRNAs. RHAU interacts with deadenylases and the human exosome, and it enhances mRNA decay in an ATP-dependent fashion. Curiously, this helicase can resolve RNA G-quadruplexes,98 which are unusually stable nucleic acid structures; it therefore seems likely that the helices must employ a special unwinding mechanism to cope with such a substrate.
Helicase Activity in Surveillance: The Case of Non-Sense Mediated Decay
The intricate machinery of non-sense mediated decay (NMD) illustrates the contributions of DExD/H helicases to RNA surveillance and to their actions in nucleating and remodelling ribonucleoprotein complexes.
The NMD decay pathway was originally discovered as an evolutionary conserved eukaryotic mechanism to detect and degrade mRNAs with a premature stop codon. Subsequently, NMD has been shown to be responsible for modulating the steady-state level of mRNAs generally, and it triggers mRNA decay when translation is inefficient. NMD is promoted by long 3′ untranslated regions or when a stop codon is present at least 50–54 nucleotides upstream of an exon-to-exon splice junction in metazoa. The components of the NMD machinery include kinases, an endonuclease (Smg6) and the superfamily 1 helicase Upf1. The helicase Upf1 is the central and required component of the NMD machinery. An mRNA tagged by the NMD machinery is cleaved by the endonuclease Smg6, followed by disassembly of the NMD complex by Upf1 in an ATP-dependent process.99
NMD can be triggered by the inappropriate location of the exon-junction complex (EJC), which forms at exon-exon splice junctions and includes a DEAD-box helicase. The EJC is not strictly required for NMD, but enhances NMD when positioned downstream of the terminating ribosome, and it represents an evolutionary tinkering of the NMD pathway. The EJC comprises four proteins [MAGO, Y14, Barentz and the DEAD-box Helicase eIF4AIII (eukaryotic initiation factor 4AIII)] (Fig. 2). The eIF4AIII engages mRNA, and hydrolysis of ATP by the helicase is controlled by interactions with partner proteins.33 In effect, the helicase acts as an ATP-dependent clamp that nucleates assembly of a ribonucleoprotein complex.12 The EJC forms a complex with the proteins Upf2 and Upf3 on the mRNA. A model for surveillance complex formation during non-sense mediated decay proposes the formation of a complex of the EJC with the UPF assembly.100 Premature translation termination recruits to the ribosome Upf1, a SF1 RNA helicase. If recruitment occurs at a spacing of less than 30 nucleotides from a downstream Upf2-Upf3-EJC complex, then a kinase is recruited that phosphorylates the helicase, triggering binding of Upf2-Upf3-EJC complex. The mRNP is remodelled by the activity of Upf1, which unwinds RNA in a cofactor dependent fashion and is followed by endonucleolytic cleavage of the mRNA by the endonuclease Smg6. Allosteric RNA clasping domains of Upf1 can open and shut to engage and disengage the RNA substrate and thereby control the helicase activity. Upf2 acts as the activating switch for the process.101
Helicases in Riboregulation
RNA interference (RNAi) is the mechanism by which short double stranded RNAs trigger silencing of cognate genes.102 The process involves cleavage of the short duplex by the RNase III enzyme Dicer to generate small RNAs that are 21–23 nucleotides in length.103 These RNA species are incorporated into the multi-enzyme RNA induced silencing complexes (RISC) where they guide the gene silencing machinery to target transcripts. The human Dicer protein contains an N-terminal helicase domain that belongs to the RIG-I family of DEAD-box helicases (Fig. 2), that includes pattern recognition receptor of the innate immune system (104; see also http://www.rnahelicase.org/rig.htm). Several functions have been proposed for this helicase domain, including substrate binding in humans,105 recognition of precursors of microRNA in Drosophila melanogaster,106 and catalysis of translocation on long double stranded RNA substrates in C. elegans and D. melanogaster.107,108 Despite the different roles suggested for Dicer helicase in diverse species, the overall domain architecture of Dicer is conserved, and it has been proposed that binding of RNA substrate by Dicer results in a marked conformational change where the helicase clamps down onto the double stranded RNA in a manner similar to RIG-I109 (as shown in the top right panel, Fig. 2).
Riboregulation is also an important aspect of gene regulation in bacteria, especially in their multifaceted responses to stress. This mode of regulation is mediated by small non-coding RNAs of 50 to 300 nucleotides that share imperfect sequence complementarity with the target RNAs. Although there is no definitive evidence that helicases are involved in this process, indirect evidence suggests that they could play a role. In this regard, it is interesting to note that truncations of RNase E that lose the degradosome scaffolding domain, including the helicase binding site, are deficient for some small RNA mediated responses. Deletion of this portion of Salmonella RNase E (residues 702 to 1061) weakens the repression of the mRNA encoding the outer membrane protein ompD by the sRNA MicC.110 Other studies show that similar deletions decrease the degradation rate of sRNAs,62 and diminish the effectiveness for target gene silencing.111,112 Some portion of the C-terminal domain of RNase E may be important for mediating the repression effects of sRNAs, and one model proposes that these domains may help to recruit the complex of sRNA with the RNA chaperone Hfq.113,114 Binding data suggest that RNA can bridge between Hfq and the RNA-binding domains that are located in the C-terminal half of RNase E.115 The interaction of these RNA-binding domains may help to present the seed region of sRNA, and also assist the delivery of the target to the catalytic domain of RNase E for cleavage. We envisage a mechanism for this action in which sRNA activates the catalytic domain of RNase E, while other components of the degradosome assembly including RhlB might interact with the target site to aid presentation to the active site.116 As mentioned above, RhlB has been found to help recruit the degradosome to polysomes in E. coli, where it may help sRNA mediated silencing of target transcripts.96
Summary and Perspective
RNA helicases have been shown to contribute to RNA decay and processing in diverse biological systems. Representative examples of the interplay between RNA helicases and RNA processing enzymes have been discussed here, and are summarized in Table 1. We have described how the RhlB helicase of the E. coli degradosome contributes to the degradation of structured RNA intermediates by unwinding and/or remodelling, so that such species become suitable single-stranded substrates for RNase E and PNPase. Similarly, a helicase may play a similar role to favor substrate feeding to the eukaryotic exosome (Fig. 7).
Table 1. A selection of representative DExD/H RNA helicases associated with RNA degradation.
| Helicase | Roles | Reference |
|---|---|---|
| Upf1 |
Non-sense mediated decay in yeast and metazoans. |
99–101 |
| eIF4AIII |
Component of the EJC, involved in non-sense mediated decay in vertebrates; see Figure 2. |
33 |
| Mtr4 |
Component of the adaptor for feeding RNA to the nuclear exosome; see Figures 2, 6 and 7. |
31, 80–84 |
| Ski2 |
Component of the adaptor for feeding RNA to the cytoplasmic exosome; see Figures 2 and 6. |
78, 79, 85 |
| hSuv3 |
Potential adaptor for human PNPase; see Figures 2 and 6. |
86–88 |
| Dhh1/Rck/DDX6 |
Activation of transcript decapping in metazoans, acting as a general translation repressor. |
94 |
| RHAU |
AU-rich element mediated mRNA decay in metazoans; may unwind guanine tetraplex RNA structures. |
97, 98 |
| RhlB |
Component of the degradosome of gamma-proteobacteria; see Figures 3, 4, 5 and 7. |
34–39 |
| CshA | Component of the degradosome of gram-positive bacteria; see Figure 3 | 40–42 |
RNase E of the E. coli degradosome is also involved in the processing of precursors of structured RNAs, such as tRNAs, rRNA and 6S RNA. The question naturally arises as to why the latter structured substrates are processed, while the former are degraded. In these cases, and more generally in cases where a helicase participates in quality control checks of RNA, the mechanism might involve a test of the extent of exposed single-stranded region. For instance, if the substrate has a footprint of fewer than 8 nucleotides on the helicase, then it may lead to displacement, while those with a greater footprint cannot be displaced.16,117,118 Perhaps instead they might be unfolded elsewhere - the unfolded structure for instance would have such footprints on the RBD and AR2 sites in the context of the E. coli RNA degradosome (Fig. 4).
It seems likely that the fold of the RNA is a likely determinant of the pathway it will follow in the presence of helicase. One striking feature of RNA folds is the complexity of its three dimensional structure. In terms of intricacy and compact nature, the three-dimensional structures of folded RNAs rival those of proteins. RNA folds are stabilized by numerous types of tertiary interactions, and curiously even random sequence RNA seems to have greater propensity for forming compact objects compared with proteins. The fold of an RNA can be strongly influenced by its interactions with proteins and from base-pairing interactions with trans-acting RNA species, and this presents some knotty puzzles for understanding how they might be recognized with specificity. For instance, how are folded RNAs recognized and distinguished from improperly folded species? Moreover, targets of non-coding RNAs are recognized through cognate base-pairing, but these can be influenced by tertiary interactions or association with protein partners. The subtlety of the recognition has been highlighted with the recent finding that a small RNA can lose its capacity to regulate translation by the substitution of a single canonical A:U base pair with a G:U wobble base pair within a duplex region of 12 complementary base pairs.119 This corresponds to small discrimination energy that cannot account for the differences in activity of the cognate and near-cognate sRNAs. We suggest that a similar situation is likely to occur in the discrimination of RNAs for quality control mechanisms. One solution to this puzzle of specificity is for recognition to occur out of equilibrium, with energy dependent proofreading steps.80 This would bear some analogy to the processes of kinetic proofreading that is at play for translational fidelity.120 RNA helicases are well suited for this role, and may drive systems out of equilibrium, so that the RNA substrates of the helicases may be directed toward degradative pathways if they fail fidelity tests. This hypothesis awaits experimental validation.
The conservation of DExD/H-box helicases in assemblies from phylogenetically diverse lineages implicates a biological importance of their contributions to these machineries. It is becoming clear that regulation of genetic information in organisms from all phyla involves complex, densely interwoven networks of transcription factors as well as riboregulators. In the course of evolution of such networks, there must have arisen a demand for precision and accuracy in recognition and action of nucleic acid mediated processes, including the specific processing and degradation of RNAs. The DExD/H-box helicases may have met this need in numerous ways. The fusion of auxiliary protein domains to the N or C terminus of the RecA core, and their recruitment in complex RNA processing and degradative machineries may have conferred these ancient proteins with increased specificity during the evolution of regulatory complexity. As we move to understand the role of helicases in complex machinery, there will likely be a rich harvest of insight into how biological systems achieve exquisite and rapid responses in the face of diverse and varying stimuli.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are supported by the Wellcome Trust. We thank Martyn Symmons, Gadi Schuster, A.J. Carpousis, Mark Carrington and Patrick Linder for invaluable comments and stimulating discussions. We thank the anonymous reviewers for helpful comments. Resources: the RNA helicase database (www.rnahelicase.org/index.html).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/22270
References
- 1.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deutscher MP. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol Transl Sci. 2009;85:369–91. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–36. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 6.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 8.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol. 1993;3:419–29. doi: 10.1016/S0959-440X(05)80116-2. [DOI] [Google Scholar]
- 10.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–51. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabhi M, Tuma R, Boudvillain M. RNA remodeling by hexameric RNA helicases. RNA Biol. 2010;7:655–66. doi: 10.4161/rna.7.6.13570. [DOI] [PubMed] [Google Scholar]
- 12.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–16. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 13.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–64. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner NK, Cordin O, Banroques J, Doère M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–38. doi: 10.1016/S1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 16.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 17.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–8. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–62. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 19.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 20.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 22.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–54. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 24.Fan JS, Cheng Z, Zhang J, Noble C, Zhou Z, Song H, et al. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol. 2009;388:1–10. doi: 10.1016/j.jmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Milner-White EJ, Pietras Z, Luisi BF. An ancient anion-binding structural module in RNA and DNA helicases. Proteins. 2010;78:1900–8. doi: 10.1002/prot.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA. 2011;2:135–52. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;324:625–36. doi: 10.1016/S0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 28.Hardin JW, Hu YX, McKay DB. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J Mol Biol. 2010;402:412–27. doi: 10.1016/j.jmb.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banroques J, Cordin O, Doère M, Linder P, Tanner NK. Analyses of the functional regions of DEAD-box RNA “helicases” with deletion and chimera constructs tested in vivo and in vitro. J Mol Biol. 2011;413:451–72. doi: 10.1016/j.jmb.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 30.Chandran V, Poljak L, Vanzo NF, Leroy A, Miguel RN, Fernandez-Recio J, et al. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J Mol Biol. 2007;367:113–32. doi: 10.1016/j.jmb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holub P, Lalakova J, Cerna H, Pasulka J, Sarazova M, Hrazdilova K, et al. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res. 2012;40:5679–93. doi: 10.1093/nar/gks223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Górna MW, Pietras Z, Tsai YC, Callaghan AJ, Hernández H, Robinson CV, et al. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA. 2010;16:553–62. doi: 10.1261/rna.1858010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, et al. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: the structure and function of the multi-enzyme RNA degradosome. Q Rev Biophys. 2012;45:105–45. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 35.Hardwick SW, Chan VS, Broadhurst RW, Luisi BF. An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 2011;39:1449–59. doi: 10.1093/nar/gkq928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aït-Bara S, Carpousis AJ. Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria. J Bacteriol. 2010;192:5413–23. doi: 10.1128/JB.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purusharth RI, Klein F, Sulthana S, Jäger S, Jagannadham MV, Evguenieva-Hackenberg E, et al. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005;280:14572–8. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- 38.Jäger S, Fuhrmann O, Heck C, Hebermehl M, Schiltz E, Rauhut R, et al. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001;29:4581–8. doi: 10.1093/nar/29.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erce MA, Low JK, Wilkins MR. Analysis of the RNA degradosome complex in Vibrio angustum S14. FEBS J. 2010;277:5161–73. doi: 10.1111/j.1742-4658.2010.07934.x. [DOI] [PubMed] [Google Scholar]
- 40.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, et al. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–60. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehnik-Habrink M, Pförtner H, Rempeters L, Pietack N, Herzberg C, Stülke J. The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol Microbiol. 2010;77:958–71. doi: 10.1111/j.1365-2958.2010.07264.x. [DOI] [PubMed] [Google Scholar]
- 42.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, et al. RNase Y in Bacillus subtilis: a Natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–41. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathy N, Hébert A, Mervelet P, Bénard L, Dorléans A, Li de la Sierra-Gallay I, et al. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010;75:489–98. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- 44.Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology. 2006;152:2895–907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- 45.Roux CM, DeMuth JP, Dunman PM. Characterization of components of the Staphylococcus aureus mRNA degradosome holoenzyme-like complex. J Bacteriol. 2011;193:5520–6. doi: 10.1128/JB.05485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–91. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 47.Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradosome. J Mol Biol. 2006;358:8–15. doi: 10.1016/j.jmb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009;389:17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murashko ON, Kaberdin VR, Lin-Chao S. Membrane binding of Escherichia coli RNase E catalytic domain stabilizes protein structure and increases RNA substrate affinity. Proc Natl Acad Sci U S A. 2012;109:7019–24. doi: 10.1073/pnas.1120181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liou G-G, Chang H-Y, Lin C-S, Lin-Chao S. DEAD box RhlB RNA helicase physically associates with exoribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNase E. J Biol Chem. 2002;277:41157–62. doi: 10.1074/jbc.M206618200. [DOI] [PubMed] [Google Scholar]
- 52.Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–81. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 54.Khemici V, Poljak L, Toesca I, Carpousis AJ. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc Natl Acad Sci U S A. 2005;102:6913–8. doi: 10.1073/pnas.0501129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–21. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 57.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol Microbiol. 2004;54:1422–30. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 58.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–51. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Awano N, Inouye M, Phadtare S. RNase activity of polynucleotide phosphorylase is critical at low temperature in Escherichia coli and is complemented by RNase II. J Bacteriol. 2008;190:5924–33. doi: 10.1128/JB.00500-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci U S A. 2003;100:6388–93. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohanty BK, Kushner SR. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol Microbiol. 2000;36:982–94. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- 62.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–64. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–89. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–26. doi: 10.1016/S0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 65.Hardwick SW, Gubbey T, Hug I, Jenal U, Luisi BF. Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol. 2012;2:120028. doi: 10.1098/rsob.120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, et al. Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol. 2009;151:905–24. doi: 10.1104/pp.109.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–67. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–66. doi: 10.1016/S0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 69.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmid M, Jensen TH. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip Rev RNA. 2010;1:474–85. doi: 10.1002/wrna.24. [DOI] [PubMed] [Google Scholar]
- 71.Wasmuth EV, Lima CD. Exo- and Endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the non-catalytic core and central channel. Mol Cell. 2012:In press. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–52. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–6. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Büttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–71. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 75.Witharana C, Roppelt V, Lochnit G, Klug G, Evguenieva-Hackenberg E. Heterogeneous complexes of the RNA exosome in Sulfolobus solfataricus. Biochimie. 2012;94:1578–87. doi: 10.1016/j.biochi.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 76.Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, et al. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–42. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–59. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 78.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–93. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–76. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 81.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–40. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–37. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 83.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107:12139–44. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–16. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–34. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284:20812–21. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, et al. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38:279–98. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jedrzejczak R, Wang J, Dauter M, Szczesny RJ, Stepien PP, Dauter Z. Human Suv3 protein reveals unique features among SF2 helicases. Acta Crystallogr D Biol Crystallogr. 2011;67:988–96. doi: 10.1107/S0907444911040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cloutier SC, Ma WK, Nguyen LT, Tran EJ. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem. 2012;287:26155–66. doi: 10.1074/jbc.M112.383075. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) Am J Transl Res. 2010;2:223–34. [PMC free article] [PubMed] [Google Scholar]
- 91.Malecki M, Jedrzejczak R, Stepien PP, Golik P. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J Mol Biol. 2007;372:23–36. doi: 10.1016/j.jmb.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 92.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012;40:1728–36. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mattiacio JL, Read LK. Evidence for a degradosome-like complex in the mitochondria of Trypanosoma brucei. FEBS Lett. 2009;583:2333–8. doi: 10.1016/j.febslet.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci. 2010;123:699–711. doi: 10.1242/jcs.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tsai Y-C, Du D, Domínguez-Malfavón L, Dimastrogiovanni D, Cross J, Callaghan AJ, et al. Recognition of the 70S ribosome and polysome by the RNA degradosome in Escherichia coli. Nucleic Acids Res. 2012:In press. doi: 10.1093/nar/gks739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–11. doi: 10.1016/S1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 98.Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Melero R, Buchwald G, Castaño R, Raabe M, Gil D, Lázaro M, et al. The cryo-EM structure of the UPF-EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol. 2012;19:498–505, S1-2. doi: 10.1038/nsmb.2287. [DOI] [PubMed] [Google Scholar]
- 101.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, et al. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 102.Sijen T, Kooter JM. Post-transcriptional gene-silencing: RNAs on the attack or on the defense? Bioessays. 2000;22:520–31. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 103.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 104.Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol. 2009;9:85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–43. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011;18:1153–8. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 107.Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, et al. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42:172–84. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, et al. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–99. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–40. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–6. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 111.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prévost K, Desnoyers G, Jacques JF, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–96. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol. 2004;54:1063–75. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- 114.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol. 2011;79:419–32. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 115.Worrall JA, Górna M, Crump NT, Phillips LG, Tuck AC, Price AJ, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382:870–83. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–51. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 118.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, et al. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–4. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 119.Papenfort K, Podkaminski D, Hinton JC, Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci U S A. 2012;109:E757–64. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2979–86. doi: 10.1098/rstb.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]