Abstract
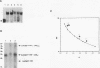
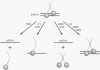
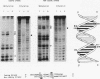
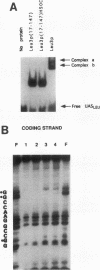
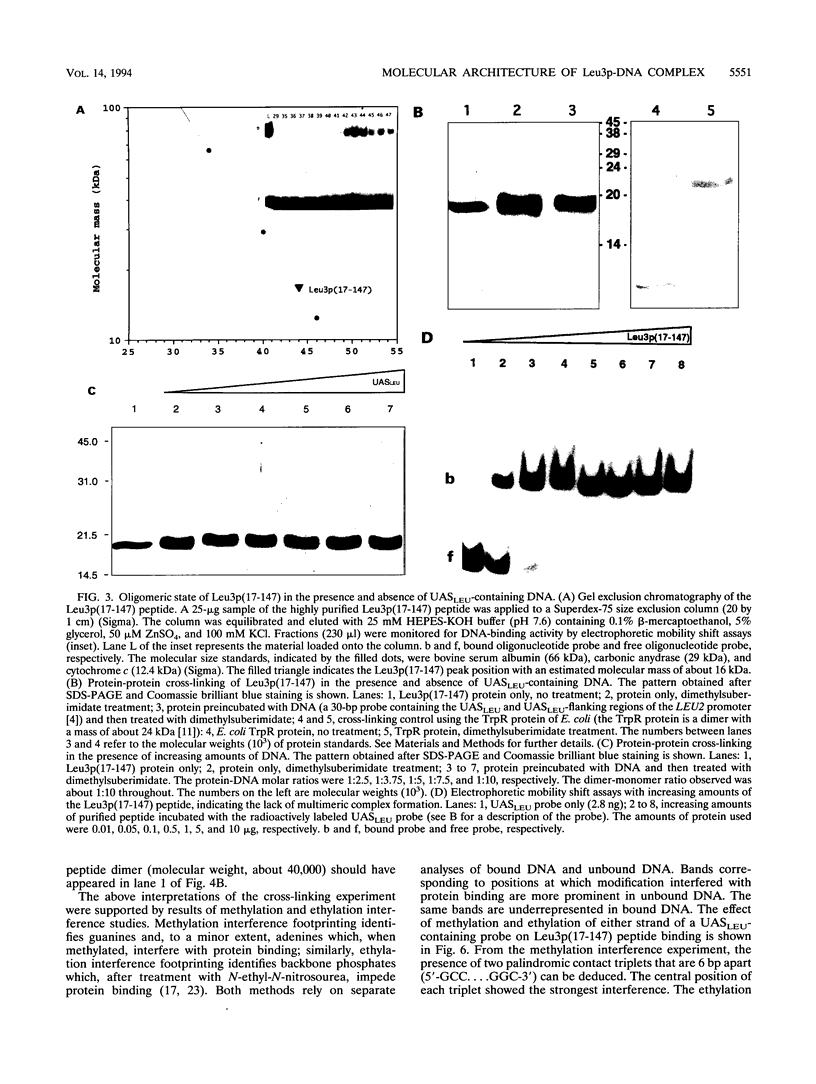
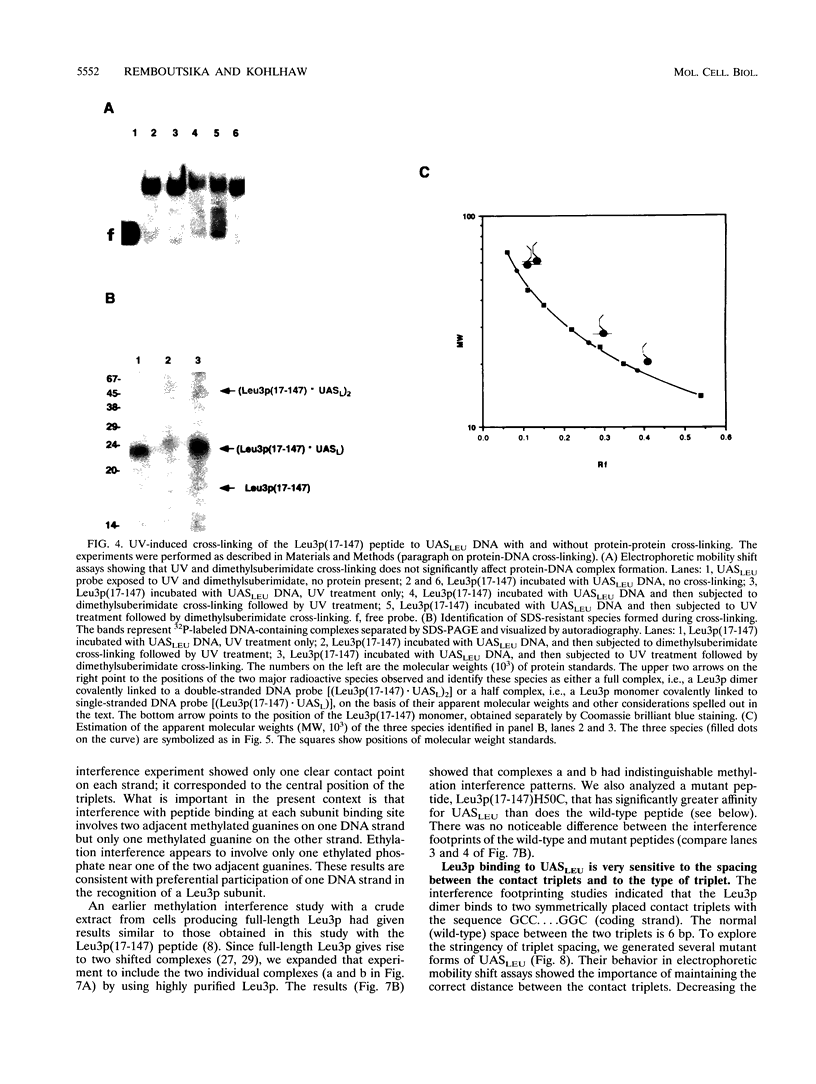
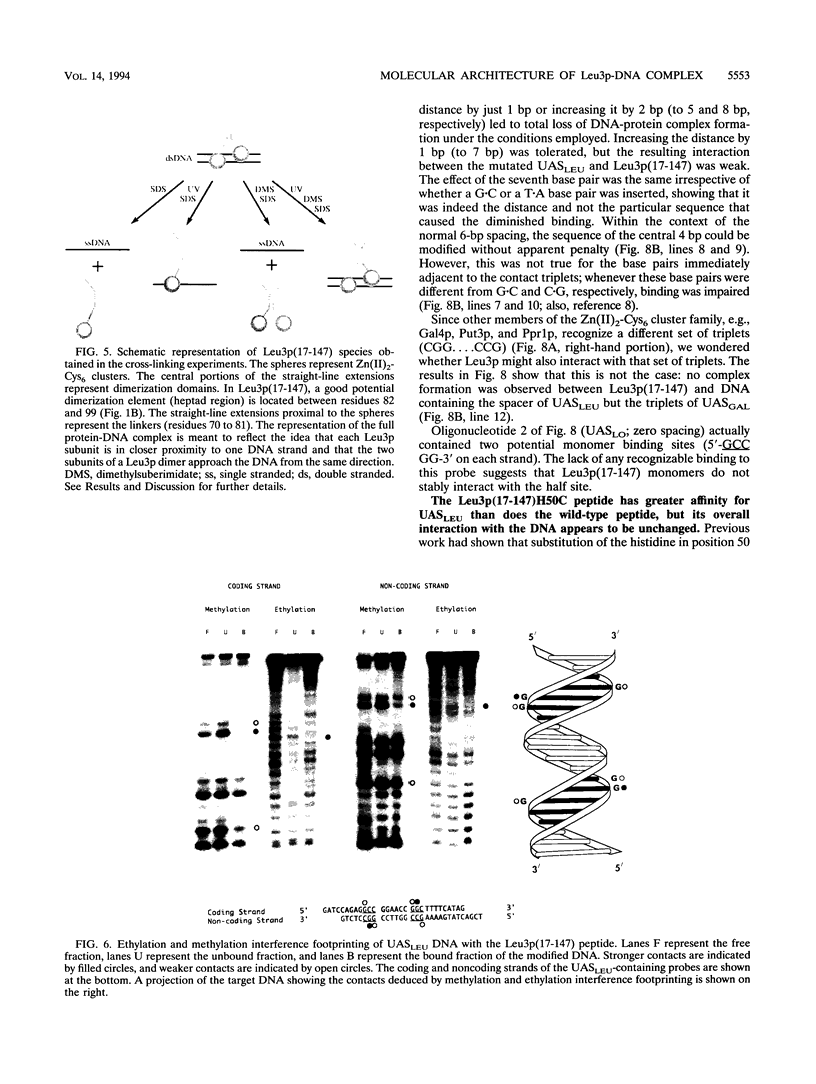
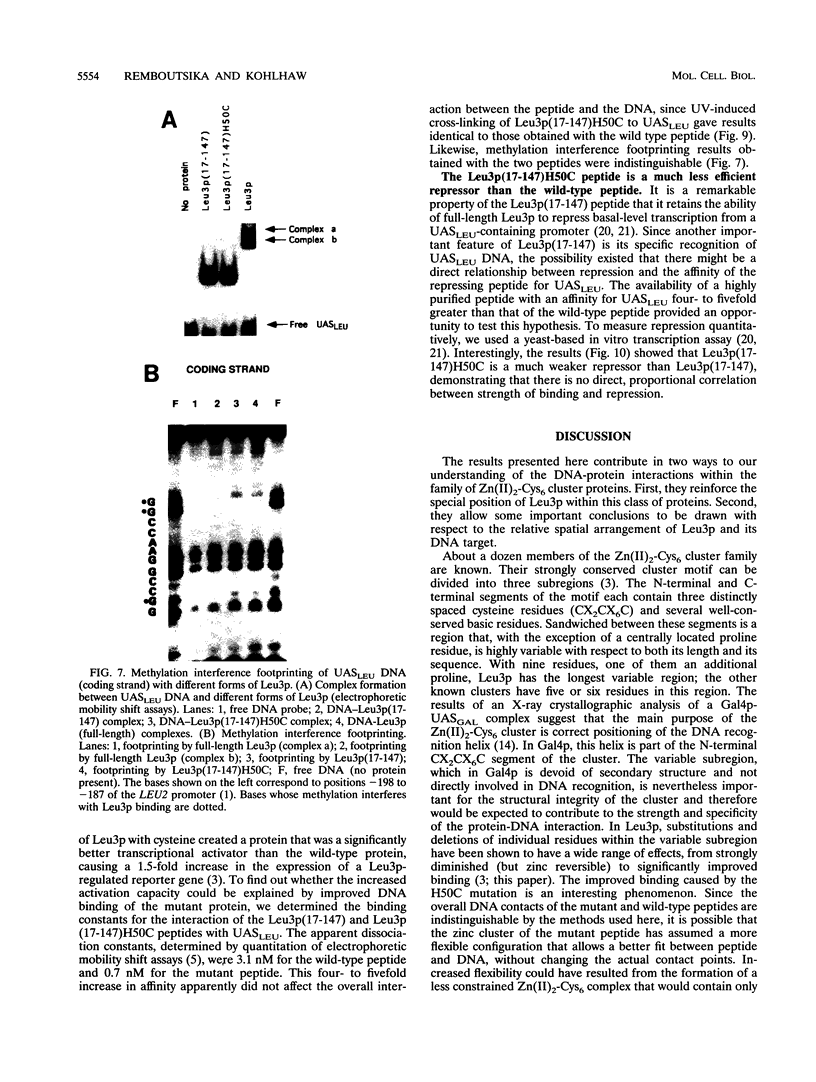
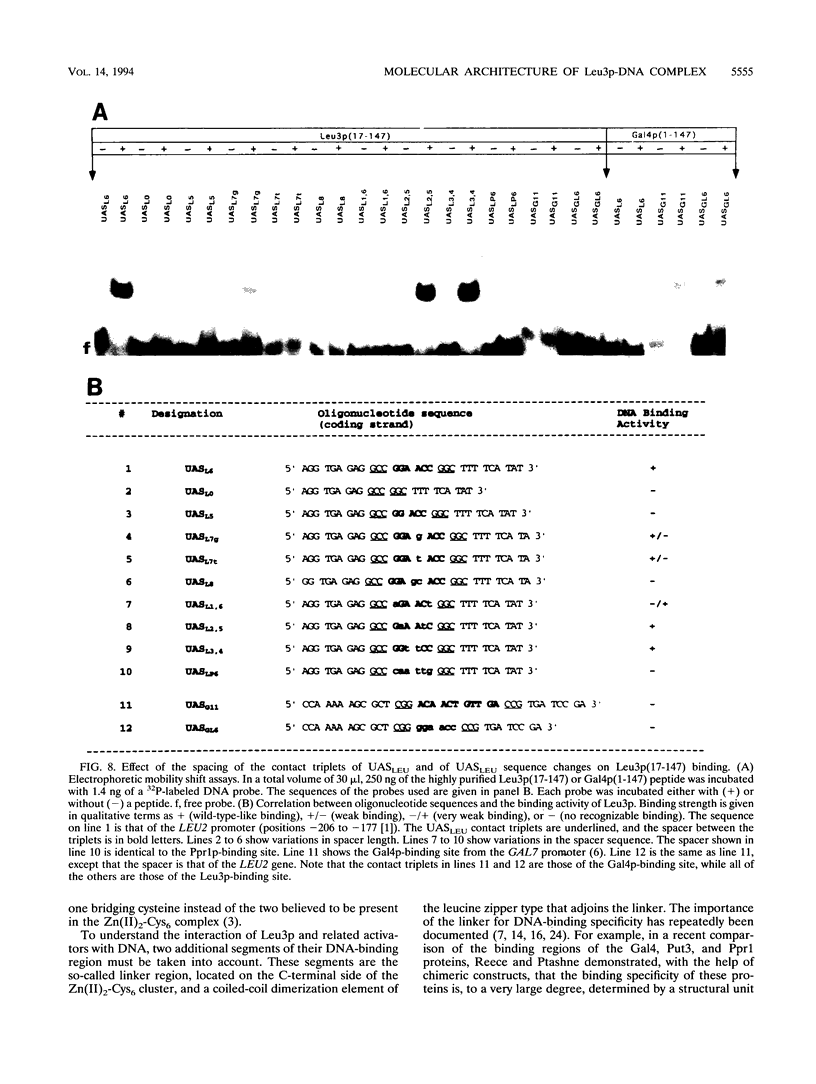
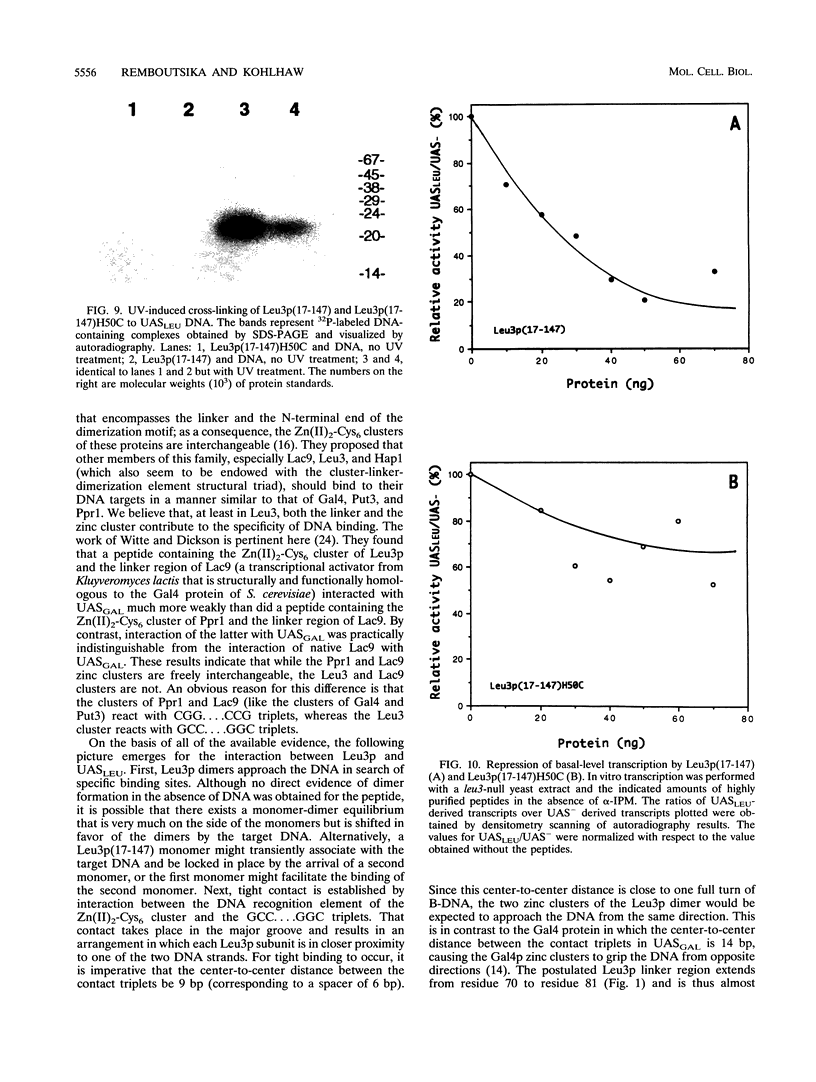
The Leu3 protein (Leu3p) of Saccharomyces cerevisiae is a pleiotropic transregulator that can function both as an activator and as a repressor of transcription. It binds to upstream promoter elements (UASLEU) with the consensus sequence 5'-GCCGGNNCCGGC-3'. The DNA-binding motif of Leu3p belongs to the family of Zn(II)2-Cys6 clusters. The motif is located between amino acid residues 37 and 67 of the 886-residue protein. In this study, we used a recombinant peptide consisting of residues 17 to 147 to explore the interaction between Leu3p and its cognate DNA. We found that the Leu3p(17-147) peptide is a monomer in the absence of UASLEU but assumes a dimeric structure when the DNA is present. Results of protein-DNA cross-linking and methylation and ethylation interference footprinting experiments show that the Leu3p(17-147) dimer interacts symmetrically with two contact triplets separated by 6 bp and suggest that the peptide approaches its target DNA in such a way that each subunit is positioned closer to one DNA strand than to the other. The binding of Leu3p is strongly affected by the spacing between the contact triplets of the UASLEU and by the type of triplet. Binding occurs when the triplets are 6 bp apart (normal spacing) but fails to occur when the triplets are 0, 5, or 8 bp apart. Weak binding occurs when the triplets are 7 bp apart. Binding does not occur when the UASLEU triplets (GCC....GGC) are replaced with triplets found in the UAS elements for Gal4p, Put3p, and Ppr1p (CGG....CCG). The apparent Kd for the normal Leu3p(17-147)-UASLEU complex is about 3 nM. A mutant form of Leu3p(17-147) in which the histidine at position 50 has been replaced with cysteine binds UASLEU with significantly greater affinity (apparent Kd of about 0.7 nM), even though the interaction between the mutant peptide and target DNA appears to be unchanged. Interestingly, repression of basal-level transcription, which is a hallmark property of the wild-type Leu3p(17-147) peptide, is largely lost with the mutant peptide, indicating that there is no direct correlation between strength of binding and repression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Bai Y. L., Kohlhaw G. B. Manipulation of the 'zinc cluster' region of transcriptional activator LEU3 by site-directed mutagenesis. Nucleic Acids Res. 1991 Nov 11;19(21):5991–5997. doi: 10.1093/nar/19.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisco P. R., Kohlhaw G. B. Regulation of yeast LEU2. Total deletion of regulatory gene LEU3 unmasks GCN4-dependent basal level expression of LEU2. J Biol Chem. 1990 Jul 15;265(20):11667–11675. [PubMed] [Google Scholar]
- Cao Z., Umek R. M., McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991 Sep;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Chasman D. I., Kornberg R. D. GAL4 protein: purification, association with GAL80 protein, and conserved domain structure. Mol Cell Biol. 1990 Jun;10(6):2916–2923. doi: 10.1128/mcb.10.6.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton J. C., Johnston S. A. Altering DNA-binding specificity of GAL4 requires sequences adjacent to the zinc finger. Nature. 1989 Aug 31;340(6236):724–727. doi: 10.1038/340724a0. [DOI] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988 Jul;8(7):2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadhavi P. L., Raine A. R., Alefounder P. R., Laue E. D. Complete assignment of the 1H NMR spectrum and secondary structure of the DNA binding domain of GAL4. FEBS Lett. 1990 Dec 10;276(1-2):49–53. doi: 10.1016/0014-5793(90)80504-c. [DOI] [PubMed] [Google Scholar]
- Gardner K. H., Pan T., Narula S., Rivera E., Coleman J. E. Structure of the binuclear metal-binding site in the GAL4 transcription factor. Biochemistry. 1991 Nov 26;30(47):11292–11302. doi: 10.1021/bi00111a015. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Yost H. J., Casadaban M. J. Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature. 1984 Feb 23;307(5953):740–742. doi: 10.1038/307740b0. [DOI] [PubMed] [Google Scholar]
- Reece R. J., Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993 Aug 13;261(5123):909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Kohlhaw G. B. Purification and structural characterization of transcriptional regulator Leu3 of yeast. J Biol Chem. 1993 Feb 5;268(4):2505–2512. [PubMed] [Google Scholar]
- Sze J. Y., Remboutsika E., Kohlhaw G. B. Transcriptional regulator Leu3 of Saccharomyces cerevisiae: separation of activator and repressor functions. Mol Cell Biol. 1993 Sep;13(9):5702–5709. doi: 10.1128/mcb.13.9.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science. 1992 Nov 13;258(5085):1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- Tu H., Casadaban M. J. The upstream activating sequence for L-leucine gene regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Jul 11;18(13):3923–3931. doi: 10.1093/nar/18.13.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Hillen W. DNA contacts probed by modification protection and interference studies. Methods Enzymol. 1991;208:365–379. doi: 10.1016/0076-6879(91)08020-i. [DOI] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. The C6 zinc finger and adjacent amino acids determine DNA-binding specificity and affinity in the yeast activator proteins LAC9 and PPR1. Mol Cell Biol. 1990 Oct;10(10):5128–5137. doi: 10.1128/mcb.10.10.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wilson S., Walker B., Dawid I., Paisley T., Zimarino V., Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987 Nov 27;238(4831):1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Zhou K. M., Bai Y. L., Kohlhaw G. B. Yeast regulatory protein LEU3: a structure-function analysis. Nucleic Acids Res. 1990 Jan 25;18(2):291–298. doi: 10.1093/nar/18.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. M., Kohlhaw G. B. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J Biol Chem. 1990 Oct 15;265(29):17409–17412. [PubMed] [Google Scholar]
- Zhou K., Brisco P. R., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 1987 Jul 10;15(13):5261–5273. doi: 10.1093/nar/15.13.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]