Abstract
Citrus canker disease caused by Xanthomonas citri subsp. citri (Xcc) is one of the most devastating diseases affecting the citrus industry worldwide. In our previous study, the canker-resistant transgenic sweet orange (Citrus sinensis Osbeck) plants were produced via constitutively overexpressing a spermidine synthase. To unravel the molecular mechanisms underlying Xcc resistance of the transgenic plants, in the present study global transcriptional profiling was compared between untransformed line (WT) and the transgenic line (TG9) by hybridizing with Affymetrix Citrus GeneChip. In total, 666 differentially expressed genes (DEGs) were identified, 448 upregulated, and 218 downregulated. The DEGs were classified into 33 categories after Gene ontology (GO) annotation, in which 68 genes are in response to stimulus and involved in immune system process, 12 genes are related to cell wall, and 13 genes belong to transcription factors. These genes and those related to starch and sucrose metabolism, glutathione metabolism, biosynthesis of phenylpropanoids, and plant hormones were hypothesized to play major roles in the canker resistance of TG9. Semiquantitative RT-PCR analysis showed that the transcript levels of several candidate genes in TG9 were significantly higher than in WT both before and after Xcc inoculation, indicating their potential association with canker disease.
1. Introduction
Citrus canker disease caused by a biotrophic bacterium Xanthomonas citri subsp. citri (Xcc) is one of the most devastating diseases in many citrus-producing regions. This disease results in defoliation, dieback, and premature fruit drop, leading to enormous loss of yield and fruit quality [1, 2]. Once the canker-free citrus producing areas are invaded by Xcc, all the suspected and infected trees should be uprooted and burned via an eradication programme, as has been done in Florida [3]. Because of the serious destruction and recalcitrance to management, citrus canker has been regarded as a quarantine disease in many countries. Every year, millions of dollars are spent on prevention, quarantines, eradication programs, and disease control in the world [3].
Current strategies for counteracting with canker disease are primarily directed to integrated approaches such as eradication programme and use of antibiotics or bactericides [2]. However, due to the disadvantages in labor investment, safety, consistency, and stabilization these strategies are not the ultimate solutions. Moreover, their applications are often compromised by inducing adverse environmental influence and change of pathogen strains. Therefore, the most effective and economical approach for controlling canker disease relies on the production of resistant cultivars. Genetic manipulation via transforming stress-related genes is a widely employed way to create disease-resistant germplasms that are otherwise impossible for classic breeding programme, especially in citrus. At present, antibacterial peptides, R-genes, pathogenic factors, and defense-related genes have been applied to create canker-resistant germplasms in citrus [4, 5]. In our previous study, we produced transgenic sweet orange plants with less susceptibility to citrus canker via constitutively overexpressing a spermidine synthase (SPDS, EC 2.5.1.16) [6].
SPDS is a key enzyme involving in polyamine biosynthetic pathway, which converts putrescine (Put) to spermidine (Spd). Polyamines are low-molecular-weight aliphatic compounds that exist ubiquitously in all living organisms, mainly including diamine Put, triamine Spd, and tetraamine spermine (Spm). It has been well documented that polyamines are closely involved in a variety of physiological processes, including biotic stress responses. For example, the content of free and conjugated polyamines and the activities of polyamine biosynthetic and oxidative enzymes increased during the hypersensitive response (HR) of barley after the powdery mildew fungus attack [7], as well as during the formation of maize tumors induced by the the biotrophic pathogenic fungus Ustilago maydis [8]. The transcript levels of polyamine biosynthesis-related genes were also found to be accumulated in TMV-infected tobacco [9] and in U. maydis-infected maize [8]. In tobacco, polyamine oxidase (PAO) protein and the specific PAO enzymatic activities increased after infection with compatible plant-pathogenic bacterium Pseudomonas syringae pv tabaci [10]. The studies indicated that polyamines or PAO protein accumulation may be a common event for plant response to pathogens. Moreover, augmentation of endogenous polyamine level by exogenous application of polyamine enhanced host resistance to virus or to bacterial challenge [11–13]. The previous work provided numerous evidence showing that polyamines play important roles in plant pathogen responses.
The mechanisms underlying the role of polyamines in plant defense have been described in previous studies. In summary, two main mechanisms have been proposed. The first one relates to production of hydrogen peroxide (H2O2) due to PAO-mediated polyamine catabolism, triggering HR and induced tolerance to specific pathogens [6, 8–10, 12]. The second one points to the role of polyamines (especially for Spm) as signaling molecules to activate expression of pathogenesis-related proteins [14] and a subset of HR-specific genes [15, 16]. Mitsuya et al. [14] and Sagor et al. [17] found that a number of genes in Arabidopsis showed response to exogenous application of Spm and Cucumber mosaic virus (CMV) infection based on the super serial analysis of gene expression (SuperSAGE), implying that Spm-mediated signaling pathway might play a role in CMV response. Very recently, Gonzalez et al. [18] reported that a large number of differentially expressed genes were identified in Spm-overproducing transgenic Arabidopsis and Spm-decreased mutant by using microarray analysis. It should be pointed out that although many efforts have been invested, the underlying physiological and molecular mechanisms remain still elusive. The gene regulation network in polyamine involved plant pathogen response is largely unclear, particularly in perennial plants like citrus. In our previous study, ectopic expression of SPDS gene increases both Spd and Spm levels in the transgenic sweet orange and confers canker resistance. As Spm functions as a signaling molecule, it is hypothesized that the transgenic line might display an extensive transcriptional reprogramming. To address this issue and to gain new insights into the molecular mechanisms on the enhanced disease tolerance, genome-wide transcriptome analysis was conducted using the Affymetrix Citrus GeneChip microarray technology. The Affymetrix Citrus GeneChip contains 30,171 probe sets representing up to 33,879 citrus transcripts selected from citrus HarvEST EST and cDNA clustering database. The transcriptional profiling described here may contribute to explain molecular mechanism of polyamine in regulating plant pathogen response.
2. Methods and Materials
2.1. Microarray Hybridization and Data Analysis
The leaves were collected from untransformed line (WT) and the transgenic line (TG9) plant for hybridization with the Affymetrix Citrus GeneChip (Affymetrix, Santa Clara, CA, USA). In brief, 2 g leaves were sampled from uniform new flushes (about 20 days after sprout) of WT and TG9, and then immediately immersed in liquid nitrogen and stored at −80°C. All the other processes including the total RNA extraction (20 μg at least), cDNA and cRNA synthesis, cRNA fragmentation, hybridization, washing and staining, and scanning were performed by Gene Technology Company Limited of Shanghai in China. The detailed experimental procedures can be found in the GeneChip Expression Analysis Technical Manual (http://www.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf). To satisfy biological reproducibility requirements, the experiment was carried out using three independent biological replicates for both WT and TG9 (means both WT and TG9 were hybridized with microarray for three times).
The probe array was scanned with the Affymetrix GeneChip Scanner 3000, and the images were analyzed with the Affymetrix GeneChip Operating software (GCOS 1.4) to generate raw data, saved as CEL files. The CEL files were then imported into commercial Partek Genomic Suite 6.4 software (Partek Inc., St. Louis, MO) according to the way of RMA quantile normalization to obtain RMA data containing the expression values. Analysis of variance (ANOVA) was used to compare the statistical expression difference between TG9 and WT. Probe sets with a P value ≤ 0.05 and 2-fold change were considered as differentially expressed genes (DEGs) between the two groups at a statistically significant level.
2.2. Microarray Annotation and Functional Analysis
To assign putative functions of DEGs, Gene ontology (GO) term, Enzyme Commission (EC), and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation were performed using the Blast2GO [19] software. Blast2GO assigns GO annotation through three steps, blasting, mapping, and annotation. GO terms for each of the three main categories (biological process, molecular function, and cellular component) were obtained by using the combined graphs function of the software with default parameters. The KEGG analysis were performed by using the KEGG annotating function of Blast2GO software, and the annotated KEGG pathways were further manually classified according to the published KEGG pathway lists (http://www.genome.jp/kegg/pathway.html).
2.3. Semiquantitative RT-PCR Analysis
Semiquantitative RT-PCR was employed to validate the microarray results using the same set of RNA samples for the hybridization experiments. Each RNA sample was pretreated with PCR amplification-grade RNase-free DNase I (Takara, Dalian, China) at 37°C to exclude DNA contamination. cDNA synthesis was done by the ReverTra Ace-α-kit (Toyobo, Japan) following the manufacturer's instructions. Specific primers of candidate genes were designed by Primer Premier 5.0 software (PRIMER Biosoft International, Palo Alto, CA) based on the citrus consensus sequences downloaded from Affymetrix website (Table 1). Each PCR reaction was composed of 200 ng cDNA, 2.0 μL 10× reaction buffer, 1.0 mM MgCl2, 0.2 mM dNTP, 1.0 U of DNA polymerase (Taq, Fermentas) and 0.4 μM of each primer in a total volume of 20 μL. PCR amplifications were performed at 94°C for 5 min, followed by 28–32 cycles of 94°C for 40 s, 52°C for 40 s, 72°C for 40 s and 5 min extension at 72°C. An Actin gene (Table 1, [20]) was used as an internal positive control. Band intensity was quantified by Quantity One analysis software (Bio-Rad Laboratories), and the fold change was calculated by the signal intensity of TG9-specific product divided by the signal intensity of WT-specific product.
Table 1.
Primers pairs used for Genechip verification and expression analysis of candidate genes.
| Probe ID | Primers (5′–3′) | Fragment size |
|---|---|---|
| Cit.60.1.S1_at | F: TGGCGTATTGGGTGGGGCTG |
258 bp |
| R: AGATACCCCCGCCCGTGCAA | ||
| Cit.6308.1.S1_at | F: AGCCAGGGCTCGCTTTTGGG |
294 bp |
| R: TCTTGCATCCAAGCTGACACCAGT | ||
| Cit.19313.1.S1_at | F: GTGAGGTATTTCGGCGAGGGG |
348 bp |
| R: GGCTTGCGATAACAGAGTGC | ||
| Cit.5367.1.S1_at | F: GACAGCCACATTCCAAGCAG |
430 bp |
| R: TGAGGCAAGTAGCGACAACG | ||
| Cit.1406.1.S1_s_at | F: TGTTAGGTCTTTTGGTGTCTATTGTT |
369 bp |
| R: CAGCCTCAGTTTGGGCATTG | ||
| Cit.9510.1.S1_s_at | F: GCTTATGCTTCTCCCAAACGA |
450 bp |
| R: ACCAGCCAAATACTTGCTCTTC | ||
| Cit.3246.1.S1_at | F: GTGAAAGGAAACGCAACGAA |
328 bp |
| R: TCCCAGGTCCAGTTACCAATG | ||
| Cit.6097.1.S1_s_at | F: AGCCTATGTCAAAGCAATCCG |
391 bp |
| R: GCTGCTGTCTACCCCGTCTAA | ||
| Cit.5734.1.S1_at | F: GCTGCTCGCTTTGGCTTCA |
367 bp |
| R: TTTCTTCATACTCCAGGCACTCA | ||
| Cit.57.1.S1_at | F: GCTGGCGGCGGTGGAGTTAG |
408 bp |
| R: CGAAAAGCGGCCAACGCTGC | ||
| Cit.8163.1.S1_x_at | F: AGCGTGGAGAAGGCCTGGAC |
217 bp |
| R: CCGCGAACTGCCCGATCTCC | ||
| Cit.11548.1.S1_at | F: GCCTTACCTTCTCCTTCCTCAT |
590 bp |
| R: AGTCGGTGGGCAAGTCTCA | ||
| Cit.4425.1.S1_at | F: ACTCCAACACCTTTATTCCTTCAC |
337 bp |
| R: CATCTCCGCTATTGCCCACT | ||
| Cit.28117.1.S1_s_at | F: TAGACCGACTGACTGCACCAA |
239 bp |
| R: TGCGAAATACAAAATGAACCC | ||
| Cit.13055.1.S1_at | F: TGACTCCGCCGTTGTGAAGA |
253 bp |
| R: CACCCGCCGACAACATACA | ||
| Cit.20495.1.S1_at | F: TCACGGACAACGAAGACAAAG |
179 bp |
| R: TCAACCAAAGCCGAGCAA | ||
| Cit.5856.1.S1_at | F: GAAGCAACAGTTCCAGCAGC |
264 bp |
| R: CACGAAGCCATCCAGTCAATA | ||
| Cit.17124.1.S1_at | F: CAGAAGGCAGCCACGATGA |
299 bp |
| R: GATGAGGATGACGAAGAAGAAGC | ||
| Cit.2333.1.S1_at | F: GTGGAAGGGGTAACTGGGATT |
461 bp |
| R: GCAGAAGTTATTGAAAATGGGTG | ||
| Cit.6121.1.S1_at | F: AGATGAGTCACAAAGACCAGGAGG |
205 bp |
| R: CACAGGCGTCAACCAATCAAG | ||
| Cit.31932.1.S1_at | F: TGTAGTCGGTGGTGGCTGTAG |
436 bp |
| R: TGAAAAGTGGGGTGGCATT | ||
| Actin | F: CATCCCTCAGCACCTTCC |
190 bp |
| R: CCAACCTTAGCACTTCTCC |
In another experiment, semiquantitative RT-PCR was performed to evaluate the expression patterns of several genes before or after Xanthomonas axonopodis pv. citri (Xcc) inoculation. For this purpose, the leaves sampled from uniform new flushes (about 20 days after sprout) of WT and TG9 were divided into two groups, respectively. One group of leaves without Xcc inoculation (uninoculated leaves) were immediately immersed in liquid nitrogen and stored at −80°C. And another group of leaves were subjected to a pinprick inoculation with Xcc bacterial suspension as described by Fu et al. [6]. Twenty-four hours after inoculation (hpi), the whole leaves of WT and TG9 were collected and stored at −80°C. The total RNA was isolated from uninoculated (0 hpi) and inoculated (24 hpi) leaves according to Liu et al. [21]. The other processes including RNA pretreatment, cDNA synthesis, PCR amplification, and quantification of band intensity were the same as mentioned above.
3. Results
3.1. Screening of the Differentially Expressed Genes and Verifying the Microarray Data
In our previous study, we produced a SPDS-overexpressed transgenic sweet orange line with higher levels of Spd and Spm and better resistance to canker disease [6]. To reveal the molecular mechanisms underlying canker resistance in TG9, the global transcriptional profiling of TG9 and WT were compared by citrus genome Genechip analysis. After statistical analysis, 666 genes with signal ratio fold change larger than 2 or smaller than 0.5 (P value ≤ 0.05) between the TG9 and WT were identified as differentially expressed genes (DEGs). Among these genes, 448 and 218 were upregulated and downregulated, respectively (see Supplemental Tables S1 and S2 in Supplementary Material available online at doi:10.1155/2012/918136).
In order to verify the reliability of the microarray data, 9 upregulated and 1 downregulated genes were randomly selected to analyze their expression levels in TG9 and WT via semiquantitative RT-PCR using gene-specific primers (Table 1). These genes putatively encode lipid transfer protein, glutathione s-transferase, ABC transporter ATPase, phospholipase d, cell wall invertase, miraculin-like protein 2, or unknown protein. As shown in Figure 1, transcript levels of the upregulated and the downregulated genes in TG9 were significantly higher or lower than in WT. The fold changes of these genes based on the calculation from semiquantitative RT-PCR results were largely consistent with the microarray data, suggesting that the microarray data are reliable.
Figure 1.
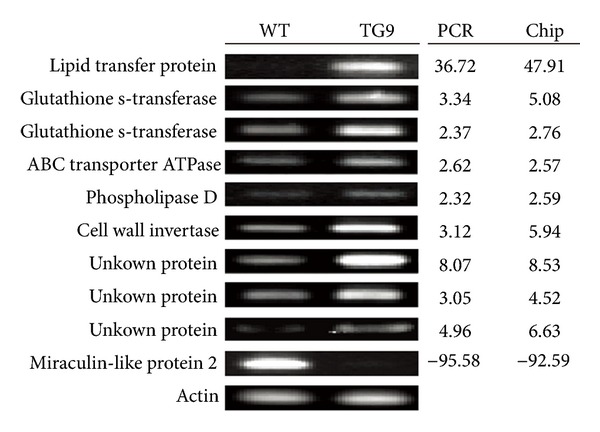
Validation of microarray results by semiquantitative RT-PCR. Ten probes putatively encoding lipid transfer protein (Cit.60.1.S1_at), glutathione s-transferase (Cit.6308.1.S1_at, Cit.9510.1.S1_s_at), ABC transporter ATPase (Cit.3246.1.S1_at), phospholipase d (Cit.6097.1.S1_s_at), cell wall invertase (Cit.5734.1.S1_at), miraculin-like protein 2 (Cit.57.1.S1_at) and unknown protein (Cit.19313.1.S1_at, Cit.5367.1.S1_at, Cit.1406.1.S1_s_at) were amplified with specific primers in WT and TG9 leaves, using Actin gene as an internal control for examining equal cDNA loading. The expression ratios between TG9 and WT were calculated by quantifying the band density using the Quantity One software.
3.2. Functional Annotation and Classification of the Differentially Expressed Genes
To further analyze the microarray data, the identified DEGs, including significantly upregulated and downregulated genes, were functionally annotated and classified using Blast2GO software. The annotated information of each gene such as sequence description, accession number of blasted gene, GO term annotation was listed in Supplemental Tables S1 and S2. The genes putatively encoding cysteine proteinase inhibitor (Cit.8163.1.S1_x_at, Cit.30421.1.S1_s_at, Cit.28011.1.S1_x_at) had the highest fold change (as high as 268.12), and the others such as lipid transfer protein (Cit.60.1.S1_at), lipid binding protein (Cit.19161.1.S1_at) and cytochrome p450 (Cit.26116.1.S1_at, Cit.4425.1.S1_at) were also upregulated to a high level (Table S1). For the downregulated genes (Table S2), the gene with the maximum fold change was miraculin-like protein. The other downregulated genes with high fold change include DNA binding protein (Cit.8142.1.S1_at, Cit.30420.1.S1_x_at), early light-inducible protein (Cit.165.1.S1_s_at), and AP2/ERF domain-containing transcription factor (Cit.11068.1.S1_at, Cit.30607.1.S1_s_at).
The functional categorization was performed according to biological process, molecular function, and cellular component using Blast2GO software. As shown in Figure 2, the biological processes of these DEGs included mainly 15 categories such as cellular process, metabolic process, response to stimulus, localization and biological regulation, and among which the genes in response to stimulus and immune system process are of interest because they may participate in canker disease resistance directly. In addition, it is intriguing to find that most of these categories contained larger number of the upregulated genes than the downregulated genes, such as cellular process, metabolic process, response to stimulus, and so forth. In the immune system process, only upregulated genes were assembled to this group. Molecular functions were primarily related to binding activity, catalytic activity, transporter activity, electron carrier activity, transcription regulator activity, and others (Figure 2). Cellular component included cell, organelle, macromolecular complex, extracellular region, membrane-enclosed lumen, and envelope. Similar to the biological process category, in molecular function and cellular component the number of upregulated genes is larger than that of downregulated genes (Figure 2).
Figure 2.
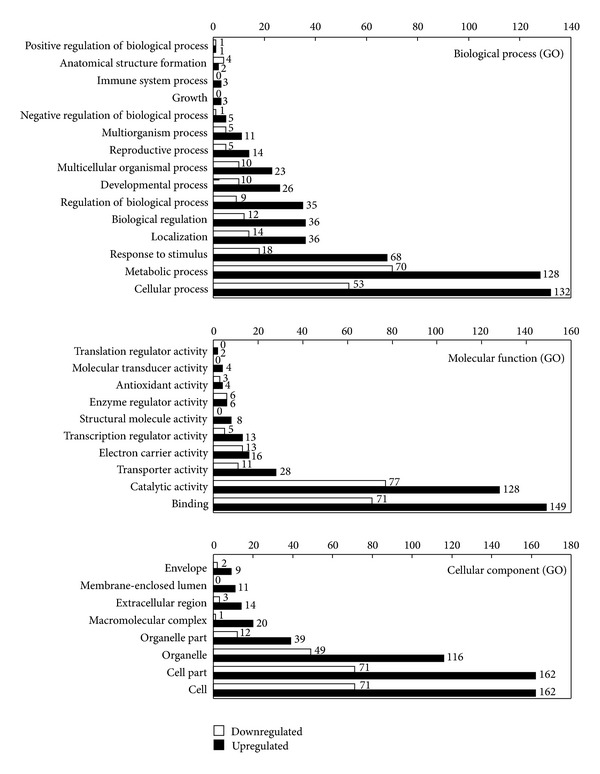
Functional categorization of upregulated and downregulated differentially expressed genes. 448 significantly upregulated and 218 significantly downregulated DEGs were categorized to biological process, molecular function, and cellular component based on GO annotation, and the represented number of each column was marked in the figure.
3.3. The Expression of Candidate Genes before and after Inoculation
According to the functional annotation and classification of the DEGs, and related studies in previous literatures, 11 genes putatively encoding cysteine proteinase inhibitor, thaumatin-like protein, cytochrome p450, aspartyl protease family protein, pyruvate kinase, pathogenesis-related protein, thioredoxin-like 5, AP2/ERF transcription factor, NADPH oxidase, TIR-NBS-LRR resistance protein, and Rubisco subunit binding protein were hypothesized to be involved in canker resistance of TG9. To answer this question and to identify canker responsive genes, the expression levels of these candidate genes were evaluated before (0 hpi) and 24 h after Xcc inoculation (24 hpi).
Before inoculation higher expression levels of the tested genes were detected in TG9 than in WT except those encode aspartyl protease family protein, NADPH oxidase, and TIS-NBS-LRR resistance protein which showed no difference between TG9 and WT (Figure 3). However, at 24 hpi the expression levels of all genes were higher in TG9 than in WT. Moreover, it is interesting to see that Xcc inoculation upregulated the genes in both TG9 and WT as compared with absence of Xcc inoculation, such as thaumatin-like protein, cytochrome p450, pathogenesis-related protein, thioredoxin-like 5, AP2/ERF transcription factor, NADPH oxidase, TIS-NBS-LRR resistance protein, and Rubisco subunit binding protein (Figure 3). Of note, the gene encoding cysteine proteinase inhibitor expressed at high levels in TG9 with or without Xcc inoculation, but it was not detected in the WT (Figure 3). Our data suggested that the expression of these candidate genes were constitutively upregulated in the transgenic line and can be further induced by the Xcc inoculation, which provides important information and evidence for its potential role in canker disease resistance.
Figure 3.
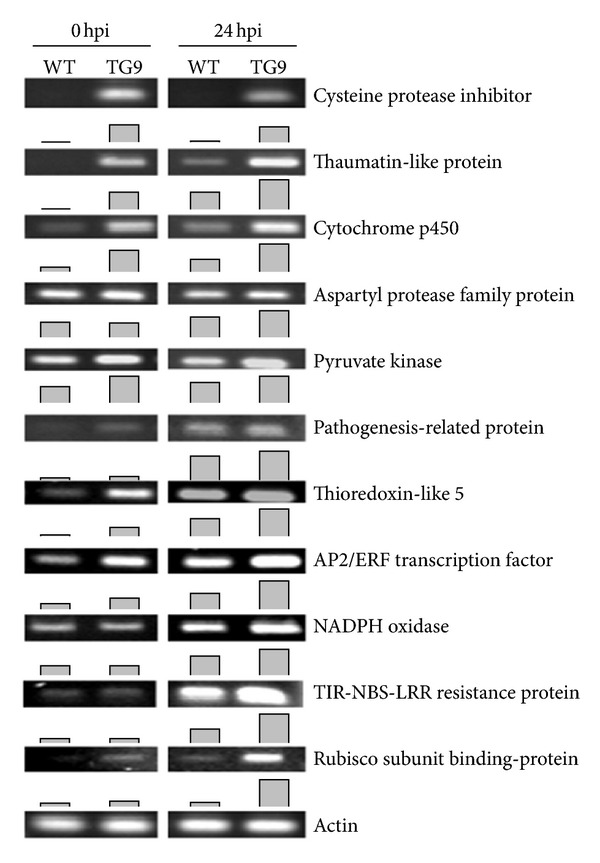
Expression of candidate genes in leaves of WT and TG9 infected or not with Xanthomonas citri subsp. Citri (Xcc). Eleven candidate genes putatively encoding cysteine proteinase inhibitor (Cit.8163.1.S1_x_at), thaumatin-like protein (Cit.11548.1.S1_at), cytochrome p450 (Cit.4425.1.S1_at), aspartyl protease family protein (Cit.28117.1.S1_s_at), pyruvate kinase (Cit.13055.1.S1_at), pathogenesis-related protein (Cit.20495.1.S1_at), thioredoxin-like 5 (Cit.5856.1.S1_at), AP2/ERF transcription factor (Cit.17124.1.S1_at), NADPH oxidase (Cit.2333.1.S1_at), TIS-NBS-LRR resistance protein (Cit.6121.1.S1_at), and Rubisco subunit binding protein (Cit.31932.1.S1_at) were assessed at 0 and 24 h post inoculation (hpi) in WT and TG9 leaves via semiquantitative RT-PCR. Column diagram was the quantification data of corresponding bands using Quantity One software.
4. Discussion
In our previous study, ectopic expression of a polyamine biosynthetic gene (MdSPDS1) in sweet orange confers citrus canker resistance, and transcript levels of several defense-related genes were induced in the transgenic line [6]. Therefore, we speculate that global transcriptional levels of transgenic plants are regulated due to the overexpression of MdSPDS1, which may explain at the transcriptional level the enhanced resistance in the transgenic plants. To confirm this hypothesis, global transcriptional profiling of WT and TG9 was compared through hybridizing with Affymetrix Citrus GeneChip in the present study. Genechip, a high-throughput and effective technology for studying global transcriptional profiling, has been widely used for deciphering molecular responses to abiotic and biotic stresses and comparing transcriptome under different treatments [22–24].
In the current study, 666 genes were identified as DEGs, accounting for 1.97% of all transcripts in the citrus genechip. Among these DEGs, the number of upregulated genes was about twice as that of the downregulated ones, in line with the microarray data overexpressing a spermidine synthase gene in Arabidopsis [25]. This result indicated that overexpression of a polyamine biosynthetic gene may lead to more prominent induction of the global transcript level. To further understand these DEGs, the functional annotation and classification were conducted using Blast2GO software. Out of the DEGs 60.66% were annotated, 39.34% upregulated, and 21.32% downregulated. Those genes without an annotation, including NoBLAST, NoMapping, and NoAnnotation, may be attributed to scarcity of enough amount of information of selected database or parameter setting. The phenomenon is not distinct as it has been also reported in previous studies [26, 27]. After GO annotation, all these DEGs were classified into 33 categories involved in biological process, molecular function, and cellular component. Moreover, the KEGG pathways of upregulated DEGs were annotated for further understanding participant metabolic and cellular processes. Three categories and annotated KEGG pathways were subjected to more detailed discussion as follow.
4.1. Genes Involved in Stimulus Response and Immune System Process
According to the GO annotation, 68 upregulated genes were shown to be involved in stimulus response and immune system process (Table 2). Based on this functional classification, we speculate that these genes have important relevance to canker disease. For instance, the gene (Cit.8878.1.S1_at) encoding major allergen pru had the highest fold change (8.63). This gene was also annotated as stress-related protein and pathogenesis-related (PR) protein 10, indicating that it may be associated with the pathogen defense. In this category, other genes include thaumatin-like protein, β-1,3-glucanase, AP2/ERF domain-containing transcription factor, ATP-binding cassette (ABC) transporter, copper/zinc superoxide dismutase, disease resistance protein, glutathione-S-transferase, and aspartyl protease family protein.
Table 2.
Significantly upregulated genes in response to stimulus and involved in immune system process.
| Gene ID | Seq. description | Hit ACC | Fold change |
|---|---|---|---|
| Cit.8878.1.S1_at | Major allergen Pru | ABK06393 | 8.63139 |
| Cit.11548.1.S1_at | Thaumatin-like protein | Q9SMH2 | 5.84254 |
| Cit.4426.1.S1_at | Homogentisate geranyl geranyl transferase | XP_002282953 | 5.63043 |
| Cit.9703.1.S1_at | Beta-1,3-glucanase | CAA03908 | 5.45386 |
| Cit.29880.1.S1_at | ATP binding | XP_002517441 | 5.3541 |
| Cit.4504.1.S1_at | Thiamin biosynthesis protein | XP_002525602 | 4.83609 |
| Cit.14918.1.S1_at | Basic 7s globulin 2 precursor small | XP_002517165 | 4.6577 |
| Cit.21616.1.S1_at | abc transporter | CBI40242 | 4.6402 |
| Cit.9706.1.S1_s_at | Beta-1,3-glucanase | ABQ45848 | 4.54013 |
| Cit.38637.1.S1_at | Protein | XP_002517054 | 4.25433 |
| Cit.28117.1.S1_s_at | Aspartyl protease family protein | ABK28718 | 3.93758 |
| Cit.3949.1.S1_s_at | Copper zinc superoxide dismutase | ACC93637 | 3.85253 |
| Cit.17438.1.S1_at | Protein | XP_002284819 | 3.69927 |
| Cit.12005.1.S1_s_at | ATP-binding cassette | CBI40242 | 3.65824 |
| Cit.6364.1.S1_s_at | Peptidase m | XP_002518664 | 3.64541 |
| Cit.4504.1.S1_s_at | Thiamin biosynthesis protein | XP_002525602 | 3.60992 |
| Cit.11209.1.S1_s_at | Nematode-resistance protein | XP_002268520 | 3.47979 |
| Cit.17124.1.S1_at | AP2/ERF domain-containing transcription factor | NP_182011 | 3.3661 |
| Cit.35636.1.S1_s_at | Hypothetical protein | XP_002262662 | 3.30867 |
| Cit.9584.1.S1_x_at | Glutathione s-transferase | XP_002273830 | 3.00065 |
| Cit.9587.1.S1_at | Glutathione s-transferase | XP_002273830 | 2.91727 |
| Cit.916.1.S1_at | Protein | XP_002525204 | 2.89418 |
| Cit.31147.1.S1_at | Disease resistance protein | CAN77656 | 2.82347 |
| Cit.9704.1.S1_at | Beta-1,3-glucanase | ABQ45848 | 2.79617 |
| Cit.20853.1.S1_at | Citrate synthase | ACU42176 | 2.79073 |
| Cit.9510.1.S1_s_at | Glutathione s-transferase | XP_002530205 | 2.75554 |
| Cit.17124.1.S1_s_at | AP2/ERF domain-containing transcription factor | NP_182011 | 2.72797 |
| Cit.21717.1.S1_at | Wound-induced protein win2 | XP_002319077 | 2.68663 |
| Cit.28472.1.S1_at | Protein | XP_002320004 | 2.68478 |
| Cit.17374.1.S1_at | Calcium binding protein | ABK06394 | 2.6752 |
| Cit.23704.1.S1_at | Aspartyl protease family protein | ABK28718 | 2.64168 |
| Cit.9587.1.S1_x_at | Glutathione s-transferase | XP_002273830 | 2.62727 |
| Cit.2809.1.S1_s_at | AP2 domain-containing transcription factor | XP_002281709 | 2.62632 |
| Cit.6280.1.S1_at | bt4 protein binding transcription regulator | XP_002304319 | 2.60135 |
| Cit.12004.1.S1_at | ATP-binding cassette | CBI30263 | 2.57256 |
| Cit.12589.1.S1_at | Syntaxin | XP_002326741 | 2.52495 |
| Cit.32844.1.S1_s_at | Glutathione s-transferase | XP_002520166 | 2.52047 |
| Cit.31254.1.S1_at | ATP-binding cassette | CAN77838 | 2.47584 |
| Cit.12560.1.S1_s_at | erd15 protein | XP_002268033 | 2.45013 |
| Cit.38633.1.S1_at | Transparent testa 12 | XP_002314825 | 2.43996 |
| Cit.24178.1.S1_at | sec12-like protein 1 | CBI40184 | 2.43778 |
| Cit.3550.1.S1_at | Ankyrin repeat-containing | XP_002526791 | 2.38623 |
| Cit.28117.1.S1_at | Aspartyl protease family protein | ABK28718 | 2.32863 |
| Cit.9584.1.S1_s_at | Glutathione s-transferase | XP_002273830 | 2.32824 |
| Cit.13439.1.S1_at | DNA binding | XP_002512121 | 2.32431 |
| Cit.31932.1.S1_at | Rubisco subunit binding-protein beta | XP_002514548 | 2.26026 |
| Cit.4131.1.S1_at | Pyridoxal kinase | CBI33550 | 2.25534 |
| Cit.7994.1.S1_at | Protein | XP_002511077 | 2.22751 |
| Cit.5117.1.S1_at | Universal stress protein | XP_002515296 | 2.2129 |
| Cit.4146.1.S1_at | Serine palmitoyltransferase | CBI15735 | 2.21272 |
| Cit.10854.1.S1_s_at | Protein | XP_002514963 | 2.20543 |
| Cit.21798.1.S1_at | Glucosyl transferase | ACS87992 | 2.17486 |
| Cit.465.1.S1_s_at | Thiamin biosynthetic enzyme | XP_002305603 | 2.17393 |
| Cit.1610.1.S1_at | Peptidyl-prolyl cis-trans isomerase-like protein | XP_002271056 | 2.17052 |
| Cit.11040.1.S1_at | Hydroxyacylglutathione hydrolase | XP_002329233 | 2.1672 |
| Cit.2333.1.S1_at | NADPH oxidase | XP_002511059 | 2.15579 |
| Cit.13055.1.S1_at | Pyruvate kinase | NP_001065749 | 2.14823 |
| Cit.26113.1.S1_at | Phototropic-responsive nph3 family protein | CAN63893 | 2.11134 |
| Cit.30576.1.S1_at | Guanylyl cyclase | XP_002277052 | 2.11123 |
| Cit.836.1.S1_s_at | Protein | XP_002520818 | 2.09287 |
| Cit.14371.1.S1_at | Homogentisic acid geranylgeranyl transferase | BAH10642 | 2.08956 |
| Cit.23824.1.S1_at | Protein | XP_002263043 | 2.08815 |
| Cit.32832.1.S1_at | Protein | XP_002269885 | 2.07239 |
| Cit.5555.1.S1_at | cop9 complex subunit | XP_002302493 | 2.05901 |
| Cit.9144.1.S1_at | ATP-dependent clp | XP_002511102 | 2.05338 |
| Cit.1554.1.S1_at | Membrane protein | CBI27668 | 2.03856 |
| Cit.35569.1.S1_s_at | Syntaxin | XP_002326741 | 2.02263 |
| Cit.2495.1.S1_at | RNA binding protein rp120 | XP_002511064 | 2.01664 |
Thaumatin-like protein (TLPs), categorized under the PR5 family, can be induced by various stresses, such as salinity, wound, and pathogen infection [28]. In addition, in vitro bioassays have shown that TLPs possess antifungal activity [29]. In the present study, expression analysis of TLPs before or after Xcc inoculation showed that TG9 had significant higher transcript levels than WT, in particularly after Xcc inoculation (Figure 3), indicating the function of TLPs on canker disease resistance. β-1,3-glucanase, hydrolyzing the 1,3-β-D-glucosidic linkages of β-1,3-glucan, belongs to PR2 family, has been shown to play a crucial role in plant pathogen defense [30–32]. AP2/ERF domain-containing transcription factor is an important plant-specific transcription factor, which has been suggested to play a critical role in stress response. Overexpression of AP2/ERF induced several PR genes expression and enhanced disease resistance in tobacco [33, 34]. In this study, AP2/ERF was induced by Xcc inoculation, and TG9 had a higher transcript level than WT (Figure 3), suggesting that AP2/ERF domain-containing transcription factor may be presumably implicated in citrus canker disease. ABC transporter, a membrane protein, exists in bacteria, fungi, animals, and plants and acts on absorption and secretion of many substrates. Martinoia et al. [35] reported that ABC transporter possibly involved in secretion of antimicrobial compounds and detoxification of some toxic metabolites in plant defence.
4.2. Cell Wall-Related Genes
Plant cell wall is a battlefield of host-pathogen interaction because the invasion of pathogen must first break the physical barrier of plant cell wall, which constitutes the first line of pathogen defence [36–38]. In this study, 12 cell wall-related genes were significantly upregulated in TG9, including cysteine proteinase inhibitor, basic 7s globulin 2 precursor small subunit, glutathione-S-transferase, and some hypothetical cell wall proteins (Table 3). Cysteine proteinase inhibitor, a proteinase inhibitor, is categorized under the PR6 family, which plays important roles in plant defence [28, 39]. As shown in Figure 3, the gene encoding cysteine proteinase inhibitor (Cit.8163.1.S1_x) was upregulated in TG9 before and after Xcc inoculation, but it was not detected in WT. Solomon et al. [40] reported that cysteine proteinase inhibitor can regulate the process of programmed cell death (PCD), an important step in HR. Therefore, cysteine proteinase inhibitor can possibly enhance disease resistance via regulating HR in plants, in agreement with an apparent HR of TG9 leaves in our previous work [6]. Among the 12 cell wall-related genes, Cit.14918.1.S1_at was annotated as basic 7s globulin 2 precursor small subunit (the similarity was 100%) by blast2go or annotated as xyloglucan-specific endoglucanase inhibitor protein (XEGIP, the similarity was 99%) in NCBI database. XEGIP has been shown to be the newest class of plant-derived proteins that inhibit pathogen-secreted cell wall degrading enzymes [41, 42]. Cernadas et al. [22] also reported that canker disease inoculation induced upregulation of endoglucanase inhibitor protein in sweet orange. Therefore, we can speculate that the gene XEGIP is possibly involved in canker disease resistance via suppressing the plant cell wall degradation in the transgenic line. Glutathione-S-transferase (GST), a protein with multiple functions, is closely associated with detoxification of some hydrophobic and electrophilic compounds, transport of auxin and phenylpropanoids, and activation of phenylpropanoid metabolism as signalling molecules [43, 44]. GST has been proposed as a marker gene for pathogen reactions, and an increase in transcript level of GST gene has been shown to be relevant to pathogen challenge [28]. In this study, the transgenic line had higher expression levels of GST, suggesting that the former may exhibit a better detoxification and regulation capacity under biotic stress when compared with the wild type, leading to less serious damage.
Table 3.
Significantly upregulated cell wall-related genes.
| Gene ID | Seq. description | Hit ACC | Fold change |
|---|---|---|---|
| Cit.8163.1.S1_x_at | Cysteine proteinase inhibitor | P37842 | 268.1240 |
| Cit.30421.1.S1_s_at | Cysteine proteinase inhibitor | AAG38521 | 94.58230 |
| Cit.28011.1.S1_x_at | Cysteine proteinase inhibitor | AAG38521 | 85.76650 |
| Cit.14918.1.S1_at | Basic 7s globulin 2 precursor small subunit | XP_002517165 | 4.65770 |
| Cit.35636.1.S1_s_at | Hypothetical protein | XP_002262662 | 3.30867 |
| Cit.9510.1.S1_s_at | Glutathione s-transferase | XP_002530205 | 2.75554 |
| Cit.14913.1.S1_at | Class III chitinase | XP_002276365 | 2.68317 |
| Cit.30421.1.S1_x_at | Cysteine proteinase inhibitor | AAG38521 | 2.47166 |
| Cit.9064.1.S1_x_at | 40s ribosomal protein s9 | XP_002511557 | 2.06885 |
| Cit.9144.1.S1_at | ATP-dependent clp | XP_002511102 | 2.05338 |
| Cit.2495.1.S1_at | RNA binding protein rp120 | XP_002511064 | 2.01664 |
| Cit.9064.1.S1_s_at | 40s ribosomal protein s9 | XP_002511557 | 2.00929 |
4.3. Transcription Factors Pertinent to Pathogen Attack
It is well known that transcription factors (TFs) play a crucial role in abiotic and biotic stresses via regulating a series of downstream target genes. In this study, 13 TFs were identified in the upregulated genes, including AP2/ERF, MADS, BT4 (BTB and TAZ domain protein 4) protein, NAC, MYB, and several other unnamed TFs (Table 4). In an earlier work, Kasukabe et al. [25] reported that TFs like AP2/ERF, NAC, and MYB were upregulated in the transgenic Arabidopsis thaliana plants overexpressing FSPDS gene relative to the wild type. Moreover, 47 TFs, such as NAC, MYB, WRKY, and bZIP, were found to be upregulated in SPMS-overexpressing transgenic Arabidopsis [18]. Our data and earlier results demonstrate that modification of the polyamine synthesis may cause the transcription reprogramming in the transgenic plants, which may be ascribed to the regulatory role of polyamines. TG9 contained higher level of spermine, which has been proposed as a signal molecule in previous studies [11, 14]. The upregulation of an array of the TFs suggests that the transgenic plants possess a robust system of transcriptional modulation towards the disease tolerance by regulating a large spectrum of relevant target genes of different TFs. Implication of the corresponding TFs in biotic stress has been experimentally corroborated in earlier studies [45–50]. For example, Nakashima et al. [48] reported transgenic rice transformed with an NAC gene displayed enhanced resistance to blight disease. In another work, Vailleau et al. [50] showed that overexpression of an MYB gene in Arabidopsis thaliana and tobacco conferred resistance to both bacteria and fungus.
Table 4.
Significantly upregulated transcription factor related genes.
| Gene ID | Seq. description | Hit ACC | Fold change |
|---|---|---|---|
| Cit.17124.1.S1_at | AP2/ERF domain-containing transcription factor | NP_182011 | 3.36610 |
| Cit.15253.1.S1_s_at | MADS-domain transcription factor | XP_002273223 | 3.21612 |
| Cit.17124.1.S1_s_at | AP2/ERF domain-containing transcription factor | NP_182011 | 2.72797 |
| Cit.2809.1.S1_s_at | AP2 domain-containing transcription factor | XP_002281709 | 2.62632 |
| Cit.6280.1.S1_at | BT4 (BTB and TAZ domain protein 4) protein binding transcription regulator | XP_002304319 | 2.60135 |
| Cit.39092.1.S1_at | NAC domain protein; IPR003441 | XP_002300866 | 2.30719 |
| Cit.29898.1.S1_at | Unnamed protein product | CBI21863 | 2.20763 |
| Cit.3005.1.S1_at | FLC-like 1 splice variant 4 | ACB72865 | 2.14257 |
| Cit.8950.1.S1_at | AP2/ERF domain-containing transcription factor | ABB89755 | 2.13449 |
| Cit.30576.1.S1_at | Guanylyl cyclase | XP_002277052 | 2.11123 |
| Cit.14044.1.S1_at | Transcription factor, putative | XP_002514876 | 2.06065 |
| Cit.15941.1.S1_at | Chromatin remodeling complex subunit | ABA18099 | 2.05183 |
| Cit.35206.1.S1_at | MYB family transcription factor | FR828559 | 2.00112 |
Apart from the abovementioned TFs, it is interesting to find that the TFs involved in flowering regulation, such as MADS and FLC (flowering locus C) [51], were also upregulated in the TG9. Upregulation of MADS and FLC in TG9 suggests that overexpression of MdSPDS1 has led to alteration of gene network associated with flowering in the transgenic plant. Although it will need time to compare the flowering dynamics between the transgenic line and WT, the polyamines have been shown to participate in the physiological process of flowering in plants [52, 53].
4.4. Potential KEGG Pathways Involved in Defence
To further understand the metabolic and cellular processes involved in defence, KEGG pathways of the upregulated DEGs were annotated by Blast2GO software. As shown in Supplemental Table S3, the annotated KEGG pathways included carbohydrate metabolism, energy metabolism, lipid metabolism, nucleotide metabolism, amino acid metabolism, cofactors and vitamins metabolism, biosynthesis of polyketides, terpenoids, alkaloids, hormones and other secondary metabolites, and phosphatidylinositol signaling transduction process.
Starch and sucrose not only serve as typical carbon and energy sources, but also play important roles in plant defense. Sucrose has been recognized as an endogenous signal to induce defense responses against pathogens [54]. Recently, Singh and Shah [55] reported that starch and sucrose contents significantly accumulated in the green peach aphid infested Arabidopsis and tomato leaves. Starch accumulation has been suggested to facilitate the host plant to generate a secondary sink that suppresses the insect to manipulate host metabolism [55]. Improved metabolism of starch and sucrose in TG9 possibly provides energy and signaling to the plant against Xcc attack. Over the past three decades, glutathione was gradually known to be involved in plant defense reactions and as a signaling molecule to induce various defense genes, and it has also been reported to crosstalk with a variety of hormone-related defense signaling, such as salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and abscisic acid (ABA) [56]. Höller et al. [57] reported that enhanced glutathione metabolism was correlated with sulfur-induced resistance in tobacco mosaic virus-infected tobacco plants. Therefore, we could speculate that enhanced glutathione metabolism in TG9 is a potential mechanism for the enhanced Xcc resistance. On the other hand, biosynthesis of other compounds, such as cytochrome P450, phenylpropanoids, and plant hormones has been well documented to be involved in pathogen defense. For example, cytochrome P450 plays critical roles in the biosynthesis of defense-related compounds, hormones, and signaling molecules [58]. And it is interesting to find that a number of cytochrome P450-related genes were induced in Xcc-inoculated Meiwa kumquat (Fortunella crassifolia) in our previous study [59]. Phenylpropanoids, such as lignin and lignans, coumarins, and flavonoids, can function as preformed and inducible antimicrobial compounds or as signal molecules in plant-microbe interactions [60]. The phytohormones, including SA, JA, ET, and ABA, have been well known as important signaling molecular to induce plant defense reactions after pathogens attack [61]. Based on these illustrations, it is surmised that the pathways related to starch and sucrose metabolism, glutathione metabolism, metabolism of xenobiotics by cytochrome P450, biosynthesis of phenylpropanoids, and plant hormones may be significantly modulated in TG9, constituting an important defence against the pathogen attack.
5. Conclusion
Global transcriptional profiling was compared between WT and TG9 by hybridizing with Affymetrix Citrus GeneChip in this study. In total, 666 DEGs were identified, including 448 upregulated genes and 218 downregulated genes. After functional annotation and classification, the DEGs implicated in stimulus response and immune system process, cell wall and transcriptional regulation, and cellular and metabolism processes, such as starch and sucrose metabolism, glutathione metabolism, biosynthesis of phenylpropanoids, and plant hormones were hypothesized to play major roles in the canker resistance of TG9. Our data suggest that genetic engineering of a polyamine biosynthetic gene has a profound impact on the transcriptome of the transgenic plants. In the future, extra work is required to verify the function of the DEGs in the canker tolerance of the transgenic line. The present work lays groundwork for deciphering the molecular events of the transgenic line and for tapping desirable genes that hold great potential for genetic engineering aiming at improving biotic stress tolerance.
Supplementary Material
Supplemental Table S1: List of significantly upregulated genes in TG9 as comparing with WT. Fold-change = TG9/WT.
Supplemental Table S2: List of significantly downregulated genes in TG9 as compared with WT. Fold-change = TG9/WT.
Supplemental Table S3: KEGG pathways of the upregulated DEGs in TG9.
Acknowledgments
This work is financially supported by National Science Foundation of China (30921002), Special Fund for Agro-scientific Research in the Public Interest (201003067), the National High Technology Research and Development Program (863 Program) of China (2011AA100205), and Hubei Provincial Natural Science Foundation (2009CDA080).
References
- 1.Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Molecular Plant Pathology. 2003;4(3):141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 2.Graham JH, Gottwald TR, Cubero J, Achor DS. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Molecular Plant Pathology. 2004;5(1):1–15. doi: 10.1046/j.1364-3703.2004.00197.x. [DOI] [PubMed] [Google Scholar]
- 3.Das AK. Citrus canker—a review. Journal of Applied Horticulture. 2003;5(1):52–60. [Google Scholar]
- 4.Barbosa-Mendes JM, Filho FDAAM, Filho AB, Harakava R, Beer SV, Mendes BMJ. Genetic transformation of Citrus sinensis cv. Hamlin with hrpN gene from Erwinia amylovora and evaluation of the transgenic lines for resistance to citrus canker. Scientia Horticulturae. 2009;122(1):109–115. [Google Scholar]
- 5.Mendes BMJ, Cardoso SC, Boscariol-Camargo RL, Cruz RB, Mourão Filho FAA, Filho AB. Reduction in susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis expressing the rice Xa21 gene. Plant Pathology. 2010;59(1):68–75. [Google Scholar]
- 6.Fu XZ, Chen CW, Wang Y, Liu JH, Moriguchi T. Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H2O2 production and transcriptional alteration. BMC Plant Biology. 2011;11(article 55) doi: 10.1186/1471-2229-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley T, Walters DR. Polyamine metabolism in barley reacting hypersensitively to the powdery mildew fungus Blumeria graminis f. sp. hordei . Plant Cell and Environment. 2002;25(3):461–468. [Google Scholar]
- 8.Rodríguez-Kessler M, Ruiz OA, Maiale S, Ruiz-Herrera J, Jiménez-Bremont JF. Polyamine metabolism in maize tumors induced by Ustilago maydis . Plant Physiology and Biochemistry. 2008;46(8-9):805–814. doi: 10.1016/j.plaphy.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Yoda H, Yamaguchi Y, Sano H. Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiology. 2003;132(4):1973–1981. doi: 10.1104/pp.103.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moschou PN, Sarris PF, Skandalis N, et al. Engineered polyamine catabolism preinduces tolerance of tobacco to bacteria and oomycetes. Plant Physiology. 2009;149(4):1970–1981. doi: 10.1104/pp.108.134932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamakawa H, Kamada H, Satoh M, Ohashi Y. Spermine is a salicylate-independent endogenous inducer for both tobacco acidic pathogenesis-related proteins and resistance against tobacco mosaic virus infection. Plant Physiology. 1998;118(4):1213–1222. doi: 10.1104/pp.118.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marina M, Maiale SJ, Rossi FR, et al. Apoplastic polyamine oxidation plays different roles in local responses of tobacco to infection by the necrotrophic fungus Sclerotinia sclerotiorum and the biotrophic bacterium Pseudomonas viridiflava . Plant Physiology. 2008;147(4):2164–2178. doi: 10.1104/pp.108.122614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzarato L, Trebbi G, Pagnucco C, Franchin C, Torrigiani P, Betti L. Exogenous spermidine, arsenic and β-aminobutyric acid modulate tobacco resistance to tobacco mosaic virus, and affect local and systemic glucosylsalicylic acid levels and arginine decarboxylase gene expression in tobacco leaves. Journal of Plant Physiology. 2009;166(1):90–100. doi: 10.1016/j.jplph.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuya Y, Takahashi Y, Berberich T, et al. Spermine signaling plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus . Journal of Plant Physiology. 2009;166(6):626–643. doi: 10.1016/j.jplph.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y, Berberich T, Miyazaki A, Seo S, Ohashi Y, Kusano T. Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. The Plant Journal. 2003;36(6):820–829. doi: 10.1046/j.1365-313x.2003.01923.x. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi Y, Uehara Y, Berberich T, et al. A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. The Plant Journal. 2004;40(4):586–595. doi: 10.1111/j.1365-313X.2004.02234.x. [DOI] [PubMed] [Google Scholar]
- 17.Sagor GHM, Cong RZ, Berberich T, Takahashi H, Takahashi Y, Kusano T. Spermine signaling in defense reaction against avirulent viral pathogen in Arabidopsis thaliana . Plant Signaling and Behavior. 2009;4(4):316–318. doi: 10.4161/psb.4.4.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez ME, Marco F, Minguet EG, et al. Perturbation of Spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis defense against Pseudomonas viridiflava . Plant Physiology. 2011;156:2266–2277. doi: 10.1104/pp.110.171413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 20.Pillitteri LJ, Lovatt CJ, Walling LL. Isolation and characterization of LEAFY and APETALA1 homologues from Citrus sinensis L. Osbeck ‘Washington’. Journal of the American Society for Horticultural Science. 2004;129(6):846–856. [Google Scholar]
- 21.Liu YZ, Liu Q, Tao NG, Deng XX. Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck) Journal of Huazhong Agricultural University. 2006;25:300–304. [Google Scholar]
- 22.Cernadas RA, Camillo LR, Benedetti CE. Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii . Molecular Plant Pathology. 2008;9(5):609–631. doi: 10.1111/j.1364-3703.2008.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao W, Wang X, Song C, Wang Y, Ren Y, Wang J. Transcriptome analysis of Hpa1(Xoo) transformed cotton revealed constitutive expression of genes in multiple signalling pathways related to disease resistance. Journal of Experimental Botany. 2010;61(15):4263–4275. doi: 10.1093/jxb/erq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun W, Xu X, Zhu H, et al. Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant and Cell Physiology. 2010;51(6):997–1006. doi: 10.1093/pcp/pcq056. [DOI] [PubMed] [Google Scholar]
- 25.Kasukabe Y, He L, Nada K, Misawa S, Ihara I, Tachibana S. Overexpression of Spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana . Plant and Cell Physiology. 2004;45(6):712–722. doi: 10.1093/pcp/pch083. [DOI] [PubMed] [Google Scholar]
- 26.Peatman E, Terhune J, Baoprasertkul P, et al. Microarray analysis of gene expression in the blue catfish liver reveals early activation of the MHC class I pathway after infection with Edwardsiella ictaluri . Molecular Immunology. 2008;45(2):553–566. doi: 10.1016/j.molimm.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Nishiuchi S, Fujihara K, Liu S, Takano T. Analysis of expressed sequence tags from a NaHCO3-treated alkali-tolerant plant, Chloris virgata . Plant Physiology and Biochemistry. 2010;48(4):247–255. doi: 10.1016/j.plaphy.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 28.Jwa NS, Agrawal GK, Tamogami S, et al. Role ofdefense/stress-related marker genes, proteins andsecondary metabolites indefining rice self-defense mechanisms. Plant Physiology and Biochemistry. 2006;44(5-6):261–273. doi: 10.1016/j.plaphy.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Hejgaard J, Jacobsen S, Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Letters. 1991;291(1):127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 30.Jongedijk E, Tigelaar H, van Roekel JSC, et al. Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica. 1995;85(1–3):173–180. [Google Scholar]
- 31.Lusso M, Kuć J. The effect of sense and antisense expression of the PR-N gene for β-1,3-glucanase on disease resistance of tobacco to fungi and viruses. Physiological and Molecular Plant Pathology. 1996;49(4):267–283. [Google Scholar]
- 32.Enrique R, Siciliano F, Favaro MA, et al. Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri . Plant Biotechnology Journal. 2011;9(3):394–407. doi: 10.1111/j.1467-7652.2010.00555.x. [DOI] [PubMed] [Google Scholar]
- 33.Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13(5):1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, Chen M, Li L, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. Journal of Experimental Botany. 2009;60(13):3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinoia E, Klein M, Geisler M, et al. Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta. 2002;214(3):345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- 36.Vorwerk S, Somerville S, Somerville C. The role of plant cell wall polysaccharide composition in disease resistance. Trends in Plant Science. 2004;9(4):203–209. doi: 10.1016/j.tplants.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Cantu D, Vicente AR, Labavitch JM, Bennett AB, Powell ALT. Strangers in the matrix: plant cell walls and pathogen susceptibility. Trends in Plant Science. 2008;13(11):610–617. doi: 10.1016/j.tplants.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Lagaert S, Beliën T, Volckaert G. Plant cell walls: protecting the barrier from degradation by microbial enzymes. Seminars in Cell and Developmental Biology. 2009;20(9):1064–1073. doi: 10.1016/j.semcdb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Ryan A. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annual Review of Phytopathology. 1990;28:425–449. [Google Scholar]
- 40.Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. The Plant Cell. 1999;11(3):431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin Q, Bergmann CW, Rose JKC, et al. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. The Plant Journal. 2003;34(3):327–338. doi: 10.1046/j.1365-313x.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- 42.Juge N. Plant protein inhibitors of cell wall degrading enzymes. Trends in Plant Science. 2006;11(7):359–367. doi: 10.1016/j.tplants.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47(1):127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- 44.Dean JD, Goodwin PH, Hsiang T. Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. Journal of Experimental Botany. 2005;56(416):1525–1533. doi: 10.1093/jxb/eri145. [DOI] [PubMed] [Google Scholar]
- 45.Collinge M, Boller T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Molecular Biology. 2001;46(5):521–529. doi: 10.1023/a:1010639225091. [DOI] [PubMed] [Google Scholar]
- 46.Hegedus D, Yu M, Baldwin D, et al. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Molecular Biology. 2003;53(3):383–397. doi: 10.1023/b:plan.0000006944.61384.11. [DOI] [PubMed] [Google Scholar]
- 47.Ren T, Qu F, Morris TJ. HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. The Plant Cell. 2000;12(10):1917–1925. doi: 10.1105/tpc.12.10.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakashima K, Tran LSP, van Nguyen D, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal. 2007;51(4):617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y, Klessig DF. Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14972–14977. doi: 10.1073/pnas.93.25.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(15):10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans PT, Malmberg RL. Do polyamines have roles in plant development. Annual Review of Plant Physiology and Plant Molecular Biology. 1989;40:235–269. [Google Scholar]
- 53.Liu JH, Moriguchi T. Changes in free polyamines and gene expression during peach flower development. Biologia Plantarum. 2007;51(3):530–532. [Google Scholar]
- 54.Moghaddam MRB, van den Ende W. Sugars and plant innate immunity. Journal of Experimental Botany. 2012;63(11):3989–3998. doi: 10.1093/jxb/ers129. [DOI] [PubMed] [Google Scholar]
- 55.Singh V, Shah J. Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signaling and Behavior. 2012;7(6):605–607. doi: 10.4161/psb.20066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghanta S, Chattopadhyay S. Glutathione as a signaling molecule another challenge to pathogens. Plant Signaling and Behavior. 2011;6(6):783–788. doi: 10.4161/psb.6.6.15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Höller K, Király L, Künstler A, et al. Enhanced glutathione metabolism is correlated with sulfur-induced resistance in tobacco mosaic virus-infected genetically susceptible Nicotiana tabacum plants. Molecular Plant-Microbe Interactions. 2010;23(11):1448–1459. doi: 10.1094/MPMI-05-10-0117. [DOI] [PubMed] [Google Scholar]
- 58.Schuler MA, Werck-Reichhart D. Functional genomics of P450s. Annual Review of Plant Biology. 2003;54:629–667. doi: 10.1146/annurev.arplant.54.031902.134840. [DOI] [PubMed] [Google Scholar]
- 59.Fu XZ, Gong XQ, Zhang YX, Wang Y, Liu JH. Different transcriptional response to Xanthomonas citri subsp. citri between Kumquat and Sweet Orange with contrasting canker tolerance. PLos one. 2012;7(7) doi: 10.1371/journal.pone.0041790.0041790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naoumkina MA, Zhao Q, Gallego-Giraldo L, Dai X, Zhao PX, Dixon RA. Genome-wide analysis of phenylpropanoid defence pathways. Molecular Plant Pathology. 2010;11(6):829–846. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bari R, Jones JDG. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1: List of significantly upregulated genes in TG9 as comparing with WT. Fold-change = TG9/WT.
Supplemental Table S2: List of significantly downregulated genes in TG9 as compared with WT. Fold-change = TG9/WT.
Supplemental Table S3: KEGG pathways of the upregulated DEGs in TG9.


