Abstract
The clinical Staphylococcus epidermidis isolate 426-3147L exhibits an unusually high resistance to linezolid that exceeds 256 μg/ml. The presence of the cfr gene, encoding the RNA methyltransferase targeting an rRNA nucleotide located in the linezolid binding site, accounts for a significant fraction of resistance. The association of cfr with a multicopy plasmid is one of the factors that contribute to its elevated expression. Mapping of the cfr transcription start sites identified the native cfr promoter. Furthermore, analysis of the cfr transcripts in Staphylococcus epidermidis 426-3147L showed that some of them originate from the upstream plasmid-derived promoters whose activity contributes to efficient cfr transcription. The genetic environment of the cfr gene and its idiosyncratic transcription pattern result in increased activity of Cfr methyltransferase, leading to a high fraction of the ribosomes being methylated at A2503 of the 23S rRNA. Curing of the Staphylococcus epidermidis 426-3147L isolate from the cfr-containing plasmid reduced the linezolid MIC to 64 μg/ml, indicating that other determinants contribute to resistance. Nucleotide sequence analysis revealed the presence of the C2534T mutation in two of the six 23S rRNA gene alleles as well as the presence of mutations in the genes of ribosomal proteins L3 and L4, which were previously implicated in linezolid resistance. Thus, the combination of resistance mechanisms operating through alteration of the drug target site appears to cause an unusually high level of linezolid resistance in the isolate.
INTRODUCTION
The oxazolidinone antibiotic linezolid was introduced into clinical practice in 2000. Linezolid is active against most Gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), enterococci, and pneumococci (1). Linezolid targets the bacterial ribosome, where it binds in the catalytic peptidyl-transferase center (PTC) and inhibits protein synthesis (2–5). Due to the synthetic nature of the drug, it was anticipated that resistance to linezolid would be slow to develop. Indeed, during the first years after the introduction of linezolid into the market, resistant isolates appeared only sporadically, and all of the reported resistance mechanisms were confined to spontaneous ribosomal mutations. Mutations in rRNA at or near the drug binding site confer various levels of resistance depending on the type of mutation and the number of mutated rrn alleles. For example, the linezolid MIC of Enterococcus faecium may vary from 8 to 64 μg/ml depending on the number of mutated alleles (6). The most frequently found linezolid resistance mutation is the G2576T transversion (Escherichia coli numbering is used here and throughout) (7, 8). Resistance mutations have also been described for several other 23S rRNA positions (e.g., T2500A, G2447T, C2534T, G2215A, and T2504A) (reviewed in references 9 and 10). Aside from rRNA mutations, mutations in ribosomal proteins L4 and L3 have been found in some of the resistant isolates. While some of these mutations (e.g., a 6-bp deletion in the L4 gene) may play a direct role in linezolid resistance (11), the alterations in the L3 protein are usually found in combination with the 23S rRNA mutations and thus may help to decrease the fitness cost associated with changes in the rRNA structure (12–15).
The synthetic drug linezolid acts upon the ribosomal PTC, which is targeted by many natural antibiotics. Not surprisingly, natural resistance mechanisms that affect the PTC could render cells insensitive not only to natural antibiotics but also to linezolid. The cfr gene, whose product, the Cfr methyltransferase, modifies a 23S rRNA residue in the PTC, represents the first of such examples (16, 17). Cfr-directed C8 methylation of A2503 of 23S rRNA renders bacteria resistant to all of the peptidyl transferase-targeting antibiotics, including linezolid (18–20). The cfr gene was originally found in the plasmid pSCFS1 in a Staphylococcus sciuri strain isolated from cattle (16). In this strain, cfr is preceded by two overlapping open reading frames (ORFs), which code for peptides of 59 and 44 amino acids, respectively (21). While the role of these ORFs in controlling cfr expression has not been elucidated, their presence argues that cfr is inducible and, similar to several other resistance genes, could be controlled by translational attenuation (22, 23). Several other staphylococcal isolates from farm animals have been reported to harbor plasmids carrying cfr, where it is often associated with mobile genetic elements, such as transposons and insertion elements (24, 25).
In 2007, the first cfr-positive clinical isolate was reported (26). In this S. aureus isolate, designated CM05, the cfr gene was located in the chromosome, and its genetic environment differed greatly from that seen in the plasmids (19, 26, 27). In the CM05 isolate, cfr is located immediately downstream of the Tn917-derived gene erm(B), which encodes a methyltransferase that modifies A2058 in 23S rRNA and confers resistance to macrolides, lincosamides, and streptogramin B. Integration of erm(B) upstream of cfr resulted in the disruption of the putative cfr promoter and the deletion of the first 61 nucleotides (nt) of ORF1; it also placed cfr under the control of the erm(B) promoter. As a result, in the CM05 strain, erm(B) and cfr form an operon, mlr, whose expression confers resistance to all clinically relevant antibiotics that target the large ribosomal subunit (19, 26). Another Tn917-derived erm(B) gene is located downstream of the cfr gene. The 1.5-kb cfr-containing region is inserted in the 23S rRNA gene of the rrn4 allele. Active transcription of the rRNA operon might contribute to cfr expression in the CM05 isolate (27).
Since 2007, cfr has been found in a number of linezolid-resistant clinical staphylococcal isolates, where it can be present on plasmids or in the chromosome (28–35). Association of cfr with mobile genetic elements and the low fitness cost associated with its expression (36) may account for its spread and maintenance in clinical pathogens.
Although cfr renders staphylococci linezolid resistant, the presence of the gene increases the MIC only severalfold, bringing it in the range of 8 to 32 μg/ml. Therefore, when the cfr-positive S. epidermidis strain 426-3147L, with a linezolid MIC exceeding 256 μg/ml, was isolated, the reasons for such a high level of resistance remained unclear (33). In this work, we examined the genetic environment of cfr in strain 426-3147L and characterized its expression. We also analyzed the presence of other genetic changes that can contribute to linezolid resistance of the 426-3147L isolate.
MATERIALS AND METHODS
Strains.
S. epidermidis strain 426-3147L was isolated in 2007 through the LEADER Program from a blood culture of a 79-year-old female patient in Arizona (33) who had received vancomycin, cefepime, and ampicillin-sulbactam, although no linezolid use was documented. The same strain was isolated in the same facility 1 year later (30).
The cfr-positive S. sciuri strain carrying the plasmid pSCFS1 was isolated in 2000 from the nasal swab of a calf (16).
S. epidermidis ATCC 12228 is a linezolid-sensitive reference strain from the American Type Culture Collection.
DNA isolation.
S. epidermidis and S. aureus cells were grown in brain heart infusion (BHI) media (BD Diagnostics, Sparks, MD). DNA was isolated using the MasterPure Gram-positive DNA purification kit (Epicentre Biotechnologies, Madison, WI) by following the manufacturer's protocol but with the inclusion of a 5-min bead-beating step after the addition of proteinase K (included in the kit).
RNA isolation.
Total RNA was isolated from staphylococcal strains using the RNeasy Minikit (Qiagen, Germantown, MD) by following the manufacturer's protocol, with some modifications. Briefly, overnight cultures of cells grown in BHI media were diluted 1:100 in fresh media and grown to an A600 of 0.5. Five ml of cell culture was pelleted, washed with 500 μl H2O, and resuspended in 200 μl of lysis buffer (Tris-HCl, pH 7.5, 30 mM MgCl2, 30 mM NH4Cl) containing 0.5 mg/ml of lysostaphin (Sigma, St. Louis, MO). After incubation for 30 min at 37°C, 350 μl of the kit buffer RLT was added, and the remaining steps of RNA isolation were performed as instructed by the kit manual.
Plasmid isolation.
Total plasmid DNA was isolated from S. epidermidis or S. aureus using the PureYield Plasmid Midiprep system (Promega, Madison, WI), with minor modifications. Briefly, 50 ml of cells was pelleted and resuspended in 2.88 ml resuspension buffer (supplied in the kit) supplemented with 120 μg lysostaphin and 120 μg lysozyme (both from Sigma). The mixture was incubated at 37°C for 1 h, and the remaining steps were carried out by following the kit manual.
MICs.
MICs were determined by broth microdilution according to the procedure established by the Clinical and Laboratory Standards Institute (37).
Detection of cfr by PCR.
PCR for detection of the presence of cfr in staphylococci was carried out on whole cells. Colonies were picked from agar plates and incubated for 10 min in 10 μl of H2O at 100°C. After 1 min of centrifugation, 1 μl of the resulting supernatant was used as a template in the PCR that was carried out with two cfr-specific primers, GAGATAACAGATCAAGTTTTA and CGAGTATATTCATTACCTCAT, under the following conditions: 2 min at 94°C; 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 45 s; and a final extension at 72°C for 1 min.
Primer extension analysis of the A2503 modification.
Primer extension analysis of the extent of A2503 methylation was carried out as previously described (17, 26), with minor modifications. Specifically, 1 pmol of [5′-32P]-labeled DNA primer ACTGTCTCACGACGTTCT was annealed to 1.5 μg of S. aureus or S. epidermidis total RNA. The primer was extended with AMV reverse transcriptase (Seikagaku America, Falmouth, MA) in the presence of 1 mM each dGTP, dATP, and dCTP and 0.25 mM dTTP. The cDNA products were resolved in a denaturing 6% polyacrylamide sequencing gel.
Transformation of S. aureus RN4220 with the p7LC plasmid.
Total plasmid DNA isolated from 426-3147L was transformed into the S. aureus strain RN4220 by electroporation (38). Transformants were selected on BHI agar pates containing 10 μg/ml florfenicol.
Mapping the cfr transcription start site.
Twenty μg of total RNA isolated from S. sciuri, S. aureus, or S. epidermidis cells was annealed with 0.75 pmol of [5′-32P]-labeled primer ATATCATTGTTTAGTTTAAAAAAGGACAG. The primer was extended with AMV reverse transcriptase (Seikagaku America) in 0.13 M Tris-HCl, pH 8.5, 10 mM MgCl2, and 10 mM dithiothreitol (DTT) for 30 min at 40°C in the presence of 1 mM each dGTP, dATP, dCTP, and dTTP. The reactions were stopped by the addition of 1 μl of 10 N NaOH. After a 15-min incubation at 37°C, the reactions were neutralized with 1 μl of 10 N HCl. The cDNA products were ethanol precipitated and resolved in a denaturing 6% polyacrylamide sequencing gel alongside the sequencing reactions.
Curing 426-3147L of the p7LC plasmid.
A culture of 426-3147L was grown overnight at 37°C in BHI media. The culture was streaked onto BHI agar plates and incubated at 42°C for 48 h. The colonies were restreaked onto fresh BHI plates and incubated for 24 h at 42°C. Once again, the colonies were restreaked onto a fresh BHI plate and maintained at 42°C for 48 h. Colonies from the final plate were replica plated on BHI agar plates without supplement or supplemented with 20 μg/ml florfenicol.
Sequencing rrl, rplC, and rplD.
The individual 23S rRNA gene (rrl) alleles were PCR amplified using conditions described by Liakopoulos et al. (39) and sequenced. The genes rplC and rplD, encoding the ribosomal proteins L3 and L4, respectively, were PCR amplified according to Mendes et al. (34) and sequenced. Nucleotide and deduced amino acid sequences were compared to those from wild-type S. epidermidis ATCC 12228 (40).
Nucleotide sequence accession number.
The sequence determined in the course of this work was deposited in GenBank under accession number JX910899.
RESULTS
Increased extent of A2503 modification in 426-3147L.
The PCR analysis showed the presence of the cfr gene in the genome of 426-3147L (Fig. 1A), indicating that the activity of the Cfr methyltransferase accounts for at least a fraction of the linezolid resistance. Cfr-dependent methylation of C8 of A2503 in 23S rRNA interferes with the progression of reverse transcriptase (17), which makes it possible to use the primer extension technique for the evaluation of the extent of Cfr-mediated rRNA modification (19, 26). The appearance of a strong reverse transcriptase stop at A2503 in the RNA isolated from 426-3147L, which was lacking in control RNA samples, confirmed the efficient methylation of this nucleotide by the Cfr methyltransferase in 426-3147L (Fig. 1B). The extent of A2503 methylation in 426-3147L is ca. 1.5-fold higher than that in the S. sciuri strain carrying the pSCFS1 plasmid (Fig. 1B), indicating that the cfr gene structure, its copy number, or specific expression pattern could account for the elevated level of Cfr activity in the 426-3147L strain. It should be noted, however, that primer extension analysis allows only the relative quantification of A2503 modification, and it remains unknown which fraction of 23S rRNA molecules is modified in S. sciuri/pSCFS1 or 426-3147L.
Fig 1.
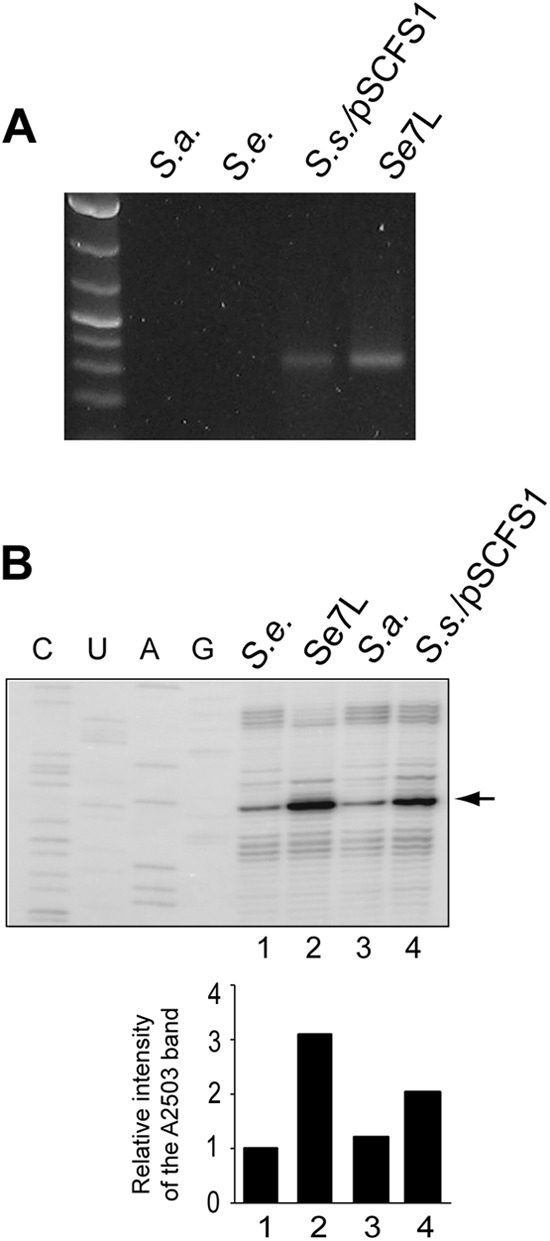
Linezolid resistance of strain 426-3147L correlates with expression of Cfr. (A) PCR analysis shows the presence of the cfr gene in 426-3147L. Linezolid-susceptible strains of S. aureus (RN4220) (S.a.) and of S. epidermidis (ATCC 12228) (S.e.) were used as negative controls. The S. sciuri strain carrying the pSCFS1 plasmid (S.s./pSCFS1) was used as a positive control. (B) Primer extension analysis of the extent of Cfr-mediated C8 methylation of A2503 in the 426-3147L strain. Controls were the following: S.e., cfr-negative S. epidermidis ATCC 12228; S.a., cfr-negative S. aureus; S.s./pSCFS1, cfr-positive S. sciuri carrying the pSCFS1 plasmid. The primer extension band corresponding to A2503 is indicated by an arrow. A weak reverse transcriptase stop in the cfr-negative control samples reflects C2 methylation of A2503 by an endogenous methyltransferase, RlmN (49). The bar graph below the gel shows the results of quantification of the band intensities.
The cfr gene is located on a plasmid in 426-3147L.
The cfr gene is often found on plasmids (16, 24, 25, 41, 42). However, in some clinical and animal staphylococcal isolates it resides in the chromosome (25–27, 43). Our preliminary analysis showed that 426-3147L carries one or several plasmids that could host the cfr gene. In order to determine whether cfr was located on a plasmid or the chromosome, total DNA or plasmid preparation from 426-3147L was digested with the HindIII restriction enzyme and, after Southern blotting, hybridized with a cfr-specific probe (Fig. 2A and B). A probe specific for the chromosomal gene se0011, encoding homoserine-o-acetyltransferase, was included in the hybridization mixture as a control. While total DNA and the plasmid-enriched fraction contained DNA bands that hybridized with both probes (indicating contamination of the plasmid preparation with chromosomal material), the relative intensity of the cfr-specific signal was significantly stronger in the plasmid-enriched fraction, indicating that the cfr gene resides on the plasmid.
Fig 2.
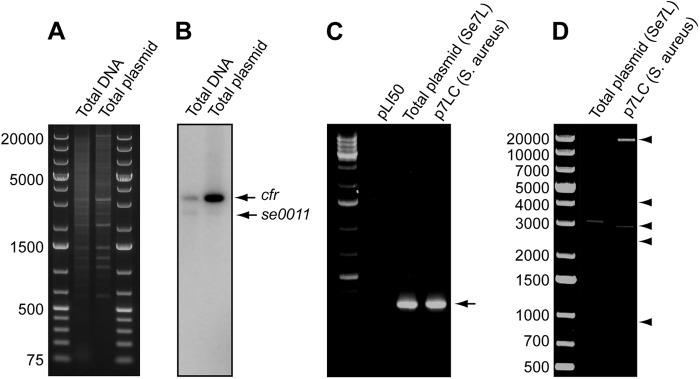
cfr gene in strain 426-3147L is located on a plasmid. (A) HindIII digest of total DNA or total plasmid preparation from 426-3147L. (B) The same gel after blotting and hybridization with the cfr-specific probe or probe for chromosomal gene se0011. (C) PCR detection of the cfr gene in the total plasmid preparation from 426-3147L or from S. aureus cells transformed with the plasmid prepared from 426-3147L. The plasmid vector pLI50 was used as a negative control. (D) HindIII digest of the total plasmid prepared from 426-3147L and from the transformed S. aureus cells. DNA bands attributed to the p7LC plasmid are indicated by arrowheads.
In order to unequivocally verify the presence of cfr on a plasmid, the plasmid preparation from 426-3147L was used to transform the S. aureus strain RN4220. Transformants were selected on plates containing 10 μg/ml florfenicol. The high florfenicol and linezolid MICs (Table 1) and PCR analysis (Fig. 2C) confirmed that S. aureus transformants acquired the cfr gene. HindIII restriction digest of the plasmid prepared from the transformed RN4220 cells (designated p7LC) generated five DNA fragments with a combined size of 30.5 kb, which provides the minimally estimated plasmid size. The p7LC HindIII fragments precisely matched a subset of HindIII fragments seen in the plasmid preparation from strain 426-3147L (Fig. 2D). The simplest explanation of this result is that the 426-3147L isolate carries two or more plasmids, one of which, p7LC, carries the cfr gene.
Table 1.
MICs of linezolid and florfenicol for different strains used in the study
| Organism and strain | MIC (μg/ml) |
|
|---|---|---|
| Linezolid | Florfenicol | |
| S. epidermidis ATCC 12228 | 2 | 4 |
| S. epidermidis 426-3147L | >256 | 1,024 |
| S. aureus RN4220 | 2 | 4 |
| S. aureus RN4220/p7LC | 8 | 512 |
| S. epidermidis 426-3147L/cured of p7LC | 64 | 16 |
Although we did not make a special effort to accurately determine the copy number of the p7LC plasmid in the 426-3147L strain, the relative intensity of the p7LC-specific bands in the HindIII digest of the total DNA preparation (Fig. 2A) argues that the plasmid copy number can be in the range of 10 to 20 copies per chromosome, accounting for a high dosage of the cfr gene and, thus, likely contributing to the elevated level of Cfr expression.
Genetic organization of cfr in the p7LC plasmid.
In order to determine the structure and genetic environment of the cfr gene in the p7LC plasmid, we sequenced a 10,125-bp segment of p7LC that included the 1,050-bp cfr gene, a 5,430-bp 5′-flanking region, and a 3,645-bp 3′-flanking region (accession no. JX910899) (Fig. 3). The nucleotide sequence of cfr is identical to that of the gene in the pSCFS1 plasmid and in the CM05 S. aureus clinical isolate (16, 26). The 367-bp upstream region of cfr, including the predicted cfr transcription start site (see below), closely matched the cfr-flanking sequence in pSCFS1, except for a 51-bp deletion and 2 single-nucleotide polymorphisms. The immediate genetic environment of cfr in p7LC resembles that discovered in the recently described 40-kb plasmid pSS-01, which is found in staphylococcal isolates from swine (accession no. JQ041372) (25). A complete Tn4001-like transposon is found upstream of cfr in the p7LC plasmid. The aminoglycoside resistance gene aacA-aphD of Tn4001 is flanked by two 1.3-kb IS256 elements that carry transposase genes. As in pSS-01, the transposase genes in the left and right IS256 elements in p7LC are not identical, although they do exhibit 87% sequence identity. Another copy of the IS256-associated transposase gene, oriented toward the cfr gene and identical in its sequence to the right (cfr-proximal) transposase gene of the Tn4001-like transposon, is found downstream of cfr (Fig. 3). Similar to pSS-01, this transposase gene is followed by a short ORF (ORF1) coding for a protein which exhibits homology to a transcription regulator of the Paenibacillus vortex mer operon (25, 44).
Fig 3.
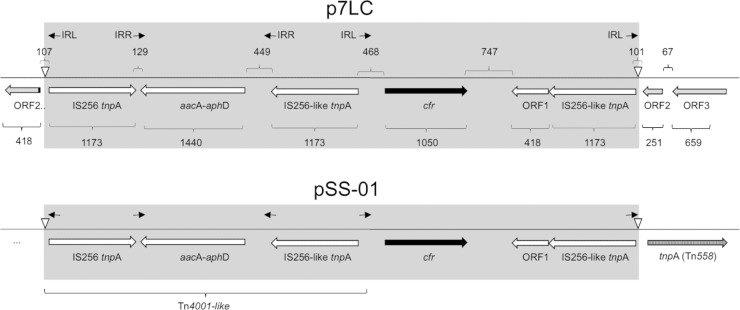
Genetic environment of cfr in p7LC. The cfr gene is shown in black. All of the other genes associated with the pSS-01-like element (gray) that are nearly identical between p7LC and pSS-01 are shown by open arrows. The genes at the insertion site of the element in p7LC are shown by diagonally hatched arrows. The Tn558 transposase gene in the 3′-flanking region of the insertion site in pSS-01 is shown vertically hatched. The extended 5′-flanking sequence in pSS-01 is unknown; the immediate 5′-flanking sequence does not show homology to the site of insertion in p7LC. The left (IRL) and right (IRR) IS256 inverted repeats are shown by arrows. The long inverted repeats at the site of the cassette insertion are indicated by open triangles. The gene sizes (in bp) are indicated below the genes; the sizes of intergenic regions are indicated above.
The target of insertion of the cfr-carrying genetic element is different in pSS-01 and p7LC (Fig. 3). In p7LC, the cassette is inserted in codon 84 of an ORF (ORF2) which exhibits 94% nucleotide sequence identity to the corresponding segment of a 398-codon ORF from the plasmid p18813-PO3, which is found in the S. aureus strain USA300 (45). The function of the protein encoded by ORF2 is unknown. The segment preceding ORF2 in p7LC, which includes another ORF (ORF3) coding for a hypothetical protein with unknown function, also shows high similarity (97% nucleotide identity) to the corresponding segment in p18813-PO3. The difference in the insertion loci of the cfr-carrying genetic element in pSS-01 and p7LC reveals its mobile nature and potential propensity for transposition between plasmids with various levels of species specificity.
Idiosyncratic transcription pattern of cfr in the p7LC plasmid.
In the originally described cfr-bearing plasmid pSCFS1 (S. sciuri), expression of the cfr gene was proposed to be driven by a computationally predicted Pcfr promoter located 366 bp upstream of the gene start codon (16). A similar hypothetical promoter sequence was found in the cfr-carrying plasmid pSCFS3 in Staphylococcus lentus isolates (24). Since transcription from Pcfr has not been experimentally verified, we wanted to test whether it indeed drives cfr expression and whether transcription from additional promoters could contribute to the expression of cfr in p7LC. We first tested if the hypothetical Pcfr promoter directs the transcription of cfr in the pSCFS1 plasmid. Primer extension carried out on total RNA prepared from S. sciuri cells carrying the pSCFS1 plasmid showed an indicative band corresponding to the longest cDNA product, placing the transcription start at position 5924 of the reported sequence (accession no. NC_005076) (Fig. 4), thereby confirming that the originally predicted Pcfr promoter indeed accounts for cfr expression in pSCFS1. We then proceeded with analyzing the cfr transcription pattern in the 426-3147L strain. While the band corresponding to transcripts initiated at Pcfr were clearly detected, several longer primer extension products were also observed (marked by asterisks in Fig. 4), suggesting that some of the transcripts which encompass the cfr gene in 426-3147L originated upstream of the Pcfr transcription start site.
Fig 4.
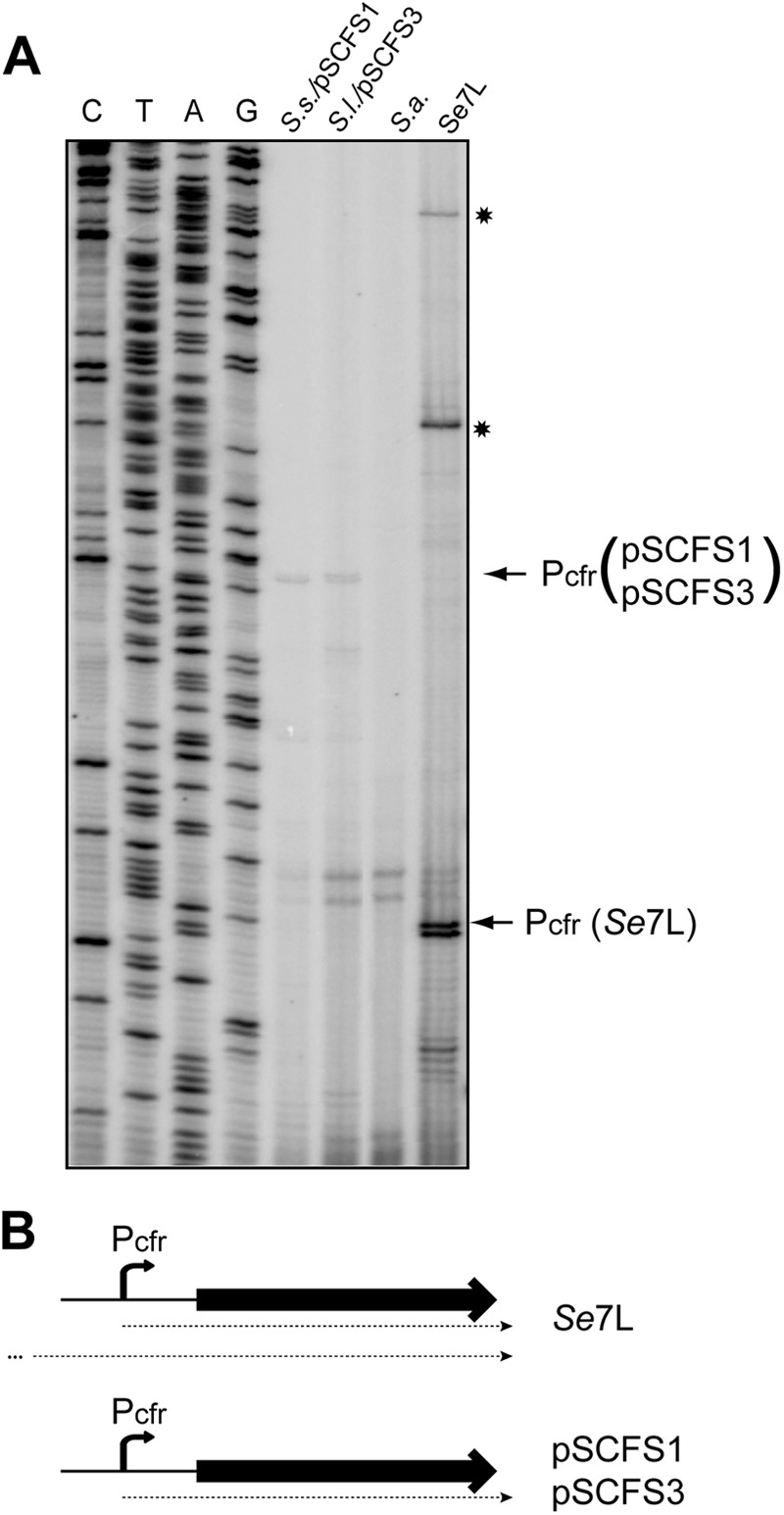
Analysis of the cfr transcription start sites. (A) Primer extension analysis was carried out on total RNA prepared from the S. sciuri strain carrying the pSCFS1 plasmid (S.s./pSCFS1), S. lentus carrying pSCFS3 (S.l./pSCFS3), S. aureus RN4220 (S.a.), or 426-3147L. The bands corresponding to cDNA products generated on transcripts initiated at Pcfr are indicated by arrows. In the 426-3147L sample, this band is shifted because of a 51-bp deletion in the cfr leader region. The cDNA bands corresponding to the longer transcript(s) present in 426-3147L are indicated by asterisks. The pSCFS1 plasmid was used to generate the sequencing ladder. (B) Schematic representation of the cfr transcripts in 426-3147L and in staphylococcal strains carrying pSCFS1 or pSCFS3.
The sizes of the additional transcription products suggest that they were initiated within the Tn4001-like sequence whose right boundary is located 127 bp upstream from the cfr promoter start site. The right-most tnp gene of Tn4001 (proximal to cfr) and the aminoglycoside resistance gene aacA-aphD of Tn4001 are oriented in the direction opposite that of cfr (Fig. 3), so that promoters responsible for their transcription cannot contribute to cfr expression. Therefore, either a cryptic right-wise-oriented promoter within the cfr-proximal part of the Tn4001-like transposon or transcription from the promoter that drives expression of the left-most tnp gene of Tn4001 accounts for additional transcripts that carry the cfr sequence. Irrespective of the exact location of the additional transcriptional start site(s), the presence of an additional promoter(s) that contributes to the transcription of cfr in the p7LC plasmid in the 426-3147L strain (and possibly in pSS-01) could lead to an elevated level of cfr mRNA and more efficient expression of the methylase than that of the strains where cfr is expressed exclusively from the Pcfr promoter.
Determinants other than cfr contribute to the high level of linezolid resistance in the 426-3147L strain.
Our analysis of the cfr genetic organization in the 426-3147L strain revealed that association of the gene with a multicopy p7LC plasmid and transcription from additional promoters could enhance its expression. However, none of these factors seem to be significant enough to account for the very high linezolid MIC of the 426-3147L strain (>256 μg/ml). Indeed, the linezolid MIC of S. aureus RN4220 transformed with p7LC reached only 8 μg/ml. Although this difference could be attributed to the genetic background of the host or to a different copy number of p7LC in S. epidermidis and S. aureus, it was likely that other resistance determinants were operating in the 426-3147L isolate. To verify this possibility, the 426-3147L cells were cured of the cfr-carrying p7LC plasmid. The loss of p7LC was confirmed by cfr-specific PCR. The cured strain exhibited a linezolid MIC of 64 μg/ml, which was notably reduced compared to the original, cfr-positive cells but substantially higher than the MIC of the linezolid-susceptible S. epidermidis control strain ATCC 12228 (Table 1). This result clearly showed that additional resistance mechanisms played a role in linezolid resistance of 426-3147L.
Besides the presence of the cfr gene, linezolid resistance can be conferred by mutations in 23S rRNA as well as in ribosomal proteins L3 and L4 (reviewed in references 9 and 10). To test for the presence of the resistance mutations, we sequenced domain V of each of the six individual rrl alleles of the 23S rRNA gene in 426-3147L as well as the genes rplC and rplD, encoding ribosomal proteins L3 and L4. The C2534T mutation (E. coli numbering) was identified in two of the six rrl alleles. This mutation was previously found in linezolid-resistant isolates of S. epidermidis (15, 46). Three missense mutations, altering amino acid residues His146Gln, Val154Leu, and Ala157Arg, were identified in the rplC gene, and the gene rplD had an insertion of a glycine codon (GGT) between the Gly71 codon and Arg72 codon. Although the direct contribution of the 23S rRNA mutation C2534T or the identified ribosomal protein mutations in linezolid resistance remains to be elucidated, their occurrence in the genes previously implicated in resistance to the drug and the general proximity to the site of linezolid binding in the ribosome is highly suggestive of their role in increasing the linezolid MIC of the 426-3147L isolate.
DISCUSSION
In this paper, we have described a combination of mechanisms that account for the exceptionally high resistance of the clinical 426-3147L strain to linezolid. Of note, the finding of identical isolates of 426-3147L in the same health care facility in subsequent years stresses the ability of this strain to persist in a clinical setting, suggesting low combined fitness cost of the resistance mechanisms in 426-3147L and highlighting the importance of understanding the nature of linezolid resistance (30, 47).
One of the key factors of linezolid resistance of the 426-3147L strain is the presence of the cfr gene, which encodes the methyltransferase targeting an rRNA residue located in the linezolid binding site. The presence of the cfr gene on a multicopy plasmid, p7LC, increases the gene dosage to ca. 20 copies per cell. The close physical association of the cfr gene in p7LC with the Tn4001-like transposon apparently increases the rate of its transcription, because the cfr-specific transcripts originate not only at the authentic Pcfr promoter but also within the transposon sequence (Fig. 4). The combination of these factors likely accounts for the enhanced expression of Cfr methyltransferase in the 426-3147L strain, a high degree of Cfr-mediated modification of A2503, and the resulting high level of linezolid resistance (Fig. 1B and Table 1).
In the first cfr-positive clinical MRSA isolate, cfr was genetically and transcriptionally linked to the erm(B) gene. Coexpression of the Cfr and Erm methyltransferases within the mlr operon rendered cells resistant to all of the clinically relevant antibiotics targeting the large ribosomal subunit, including drugs belonging to the macrolide, lincosamide, pleuromutilin, phenicol, oxazolidinone, and streptogramin families of compounds (19, 26). In 426-3147L, the presence of the Tn4001-like transposon upstream of cfr, which carries the aminoglycoside resistance gene aacA-aphD, links together resistance genes acting upon both ribosomal subunits. Given that the same Tn4001-cfr element is found on different plasmids within different genetic environments (Fig. 3), it is reasonable to expect dissemination of this element to other hosts and genetic locales.
The cfr gene is not the sole determinant of linezolid resistance in 426-3147L. Curing the strain of the cfr-containing p7LC plasmid reduced resistance from >256 to 64 μg/ml, indicating that other mechanisms contribute to the very high linezolid MIC of the original isolate. We found the C2534T mutation in two alleles of rRNA genes in the 426-3147L strain, as well as several mutations in proteins L3 and L4. The C2534T mutation has been found previously in S. epidermidis where, when it is present in two alleles, as is the case for the 426-3147L isolate, it increased the linezolid MIC to 8 μg/ml (48). The mutations found in 426-3147L in genes of proteins L3 and L4 are located close to the position where mutations were identified in other linezolid-resistant staphylococcal isolates (11–14, 28). These mutations can either increase the level of cfr-conferred linezolid resistance, as was previously noted for the ribosomal protein L3 mutations (28), or reduce the fitness cost associated with rRNA mutations or cfr-mediated rRNA modification.
The high level of linezolid resistance of the 426-3147L isolate seems to be accounted for by a combination of resistance mechanisms operating through the alteration of the drug target. The combined action of the cfr-mediated modification of an rRNA residue and mutations in rRNA and ribosomal proteins can be sufficient to provide the level of resistance observed in the isolate. Nevertheless, we cannot rule out the involvement of other resistance mechanisms, such as drug efflux or drug modification. Altogether, our findings reinforce a growing understanding that in spite of the synthetic nature of linezolid, the pathogens targeted by this antibiotic have sufficient genetic flexibility to acquire very significant levels of resistance by combining the effects of several different resistance mechanisms.
ACKNOWLEDGMENTS
We thank Ehsan Tavassoli for help in the early stages of this project.
This work was supported in part by a grant from the National Institutes of Health (R01AI072445; to A.S.M.).
Footnotes
Published ahead of print 17 December 2012
REFERENCES
- 1. Brickner SJ, Barbachyn MR, Hutchinson DK, Manninen PR. 2008. Linezolid (ZYVOX), the first member of a completely new class of antibacterial agents for treatment of serious gram-positive infections. J. Med. Chem. 51:1981–1990 [DOI] [PubMed] [Google Scholar]
- 2. Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42:3251–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leach KL, Swaney SM, Colca JR, McDonald WG, Blinn JR, Thomasco LM, Gadwood RC, Shinabarger D, Xiong L, Mankin AS. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol. Cell 26:393–402 [DOI] [PubMed] [Google Scholar]
- 4. Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U. S. A. 105:13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ippolito JA, Kanyo ZF, Wang D, Franceschi FJ, Moore PB, Steitz TA, Duffy EM. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51:3353–3356 [DOI] [PubMed] [Google Scholar]
- 6. Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, Venkataraman L, Moellering RC, Ferraro MJ. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358:207–208 [DOI] [PubMed] [Google Scholar]
- 8. Meka VG, Gold HS. 2004. Antimicrobial resistance to linezolid. Clin. Infect. Dis. 39:1010–1015 [DOI] [PubMed] [Google Scholar]
- 9. Shaw KJ, Barbachyn MR. 2011. The oxazolidinones: past, present, and future. Ann. N. Y. Acad. Sci. 1241:48–70 [DOI] [PubMed] [Google Scholar]
- 10. Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Bottger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob. Agents Chemother. 54:4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Locke JB, Hilgers M, Shaw KJ. 2009. Mutations in ribosomal protein L3 are associated with oxazolidinone resistance in staphylococci of clinical origin. Antimicrob. Agents Chemother. 53:5275–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob. Agents Chemother. 53:5265–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosowska-Shick K, Julian KG, McGhee PL, Appelbaum PC, Whitener CJ. 2010. Molecular and epidemiologic characteristics of linezolid-resistant coagulase-negative staphylococci at a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 68:34–39 [DOI] [PubMed] [Google Scholar]
- 15. Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. 2010. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob. Agents Chemother. 54:742–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarz S, Werckenthin C, Kehrenberg C. 2000. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44:2530–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064–1073 [DOI] [PubMed] [Google Scholar]
- 18. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kehrenberg C, Ojo KK, Schwarz S. 2004. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 54:936–939 [DOI] [PubMed] [Google Scholar]
- 22. Weisblum B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramu H, Mankin A, Vazquez-Laslop N. 2009. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 71:811–824 [DOI] [PubMed] [Google Scholar]
- 24. Kehrenberg C, Schwarz S. 2006. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 50:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y, Zhang W, Wang J, Wu C, Shen Z, Fu X, Yan Y, Zhang Q, Schwarz S, Shen J. 2012. Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob. Agents Chemother. 56:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, Quinn J, Mankin AS. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Locke JB, Rahawi S, Lamarre J, Mankin AS, Shaw KJ. 2012. Genetic environment and stability of cfr in methicillin-resistant Staphylococcus aureus CM05. Antimicrob. Agents Chemother. 56:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Locke JB, Morales G, Hilgers M, G CK, Rahawi S, Jose Picazo J, Shaw KJ, Stein JL. 2010. Elevated linezolid resistance in clinical cfr-positive Staphylococcus aureus isolates is associated with co-occurring mutations in ribosomal protein L3. Antimicrob. Agents Chemother. 54:5352–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bongiorno D, Campanile F, Mongelli G, Baldi MT, Provenzani R, Reali S, Lo Russo C, Santagati M, Stefani S. 2010. DNA methylase modifications and other linezolid resistance mutations in coagulase-negative staphylococci in Italy. J. Antimicrob. Chemother. 65:2336–2340 [DOI] [PubMed] [Google Scholar]
- 30. Farrell DJ, Mendes RE, Ross JE, Jones RN. 2009. Linezolid surveillance program results for 2008 (LEADER Program for 2008). Diagn. Microbiol. Infect. Dis. 65:392–403 [DOI] [PubMed] [Google Scholar]
- 31. Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Pelaez B, Andrade R, de la Torre MA, Fereres J, Sanchez-Garcia M. 2010. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin. Infect. Dis. 50:821–825 [DOI] [PubMed] [Google Scholar]
- 32. Mendes RE, Deshpande L, Rodriguez-Noriega E, Ross JE, Jones RN, Morfin-Otero R. 2010. First report of Staphylococcal clinical isolates in Mexico with linezolid resistance caused by cfr: evidence of in vivo cfr mobilization. J. Clin. Microbiol. 48:3041–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mendes RE, Deshpande LM, Castanheira M, Dipersio J, Saubolle M, Jones RN. 2008. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 52:2244–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G, Jones RN. 2010. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J. Antimicrob. Chemother. 65:2329–2335 [DOI] [PubMed] [Google Scholar]
- 35. Jones RN, Ross JE, Bell JM, Utsuki U, Fumiaki I, Kobayashi I, Turnidge JD. 2009. Zyvox Annual Appraisal of Potency and Spectrum program: linezolid surveillance program results for 2008. Diagn. Microbiol. Infect. Dis. 65:404–413 [DOI] [PubMed] [Google Scholar]
- 36. LaMarre JM, Locke JB, Shaw KJ, Mankin AS. 2011. Low fitness cost of the multidrug resistance gene cfr. Antimicrob. Agents Chemother. 55:3714–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. CLSI 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. CLSI document M7-A7, vol 26 CLSI, Wayne, PA [Google Scholar]
- 38. Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133–138 [DOI] [PubMed] [Google Scholar]
- 39. Liakopoulos A, Neocleous C, Klapsa D, Kanellopoulou M, Spiliopoulou I, Mathiopoulos KD, Papafrangas E, Petinaki E. 2009. A T2504A mutation in the 23S rRNA gene responsible for high-level resistance to linezolid of Staphylococcus epidermidis. J. Antimicrob. Chemother. 64:206–207 [DOI] [PubMed] [Google Scholar]
- 40. Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593 [DOI] [PubMed] [Google Scholar]
- 41. Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, Arias CA. 2012. Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis. Antimicrob. Agents Chemother. 56:3917–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shore AC, Brennan OM, Ehricht R, Monecke S, Schwarz S, Slickers P, Coleman DC. 2010. Identification and characterization of the multidrug resistance gene cfr in a Panton-Valentine leukocidin-positive sequence type 8 methicillin-resistant Staphylococcus aureus IVa (USA300) isolate. Antimicrob. Agents Chemother. 54:4978–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kehrenberg C, Aarestrup FM, Schwarz S. 2007. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 51:483–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, Nikolaev V, Mugasimangalam RC, Bransburg-Zabary S, Gutnick DL, Lancet D, Ben-Jacob E. 2010. Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics 11:710 doi:10.1186/1471-2164-11-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kennedy AD, Porcella SF, Martens C, Whitney AR, Braughton KR, Chen L, Craig CT, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2010. Complete nucleotide sequence analysis of plasmids in strains of Staphylococcus aureus clone USA300 reveals a high level of identity among isolates with closely related core genome sequences. J. Clin. Microbiol. 48:4504–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhu W, Tenover FC, Limor J, Lonsway D, Prince D, Dunne WM, Jr, Patel JB. 2007. Use of pyrosequencing to identify point mutations in domain V of 23S rRNA genes of linezolid-resistant Staphylococcus aureus and Staphylococcus epidermidis. Eur. J. Clin. Microbiol. Infect. Dis. 26:161–165 [DOI] [PubMed] [Google Scholar]
- 47. Farrell DJ, Mendes RE, Ross JE, Sader HS, Jones RN. 2011. LEADER Program results for 2009: an activity and spectrum analysis of linezolid using 6,414 clinical isolates from 56 medical centers in the United States. Antimicrob. Agents Chemother. 55:3684–3690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liakopoulos A, Spiliopoulou I, Damani A, Kanellopoulou M, Schoina S, Papafragas E, Marangos M, Fligou F, Zakynthinos E, Makris D, Protonotariou E, Tsiapara F, Filos K, Diza E, Anastassiou ED, Petinaki E. 2010. Dissemination of two international linezolid-resistant Staphylococcus epidermidis clones in Greek hospitals. J. Antimicrob. Chemother. 65:1070–1071 [DOI] [PubMed] [Google Scholar]
- 49. Toh SM, Xiong L, Bae T, Mankin AS. 2008. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14:98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]


