Abstract
A study was conducted to determine the primary sources of fecal pollution in a subtropical watershed using host-specific assays developed in temperate regions. Water samples (n = 534) from 10 different sites along the Rio Grande de Arecibo (RGA) watershed were collected mostly on a weekly basis (54 sampling events) during 13 months. DNA extracts from water samples were used in PCR assays to determine the occurrence of fecal bacteria (Bacteroidales, Clostridium coccoides, and enterococci) and human-, cattle-, swine-, and chicken-specific fecal sources. Feces from 12 different animals (n = 340) and wastewater treatment samples (n = 16) were analyzed to determine the specificity and distribution of host-specific assays. The human-specific assay (HF183) was found to be highly specific, as it did not cross-react with nontarget samples. The cattle marker (CF128) cross-reacted to some extent with swine, chicken, and turkeys and was present in 64% of the cattle samples tested. The swine assays showed poor host specificity, while the three chicken assays showed poor host distribution. Differences in the detection of host-specific markers were noted per site. While human and cattle assays showed moderate average detection rates throughout the watershed, areas impacted by wastewater treatment plants and cattle exhibited the highest prevalence of these markers. When conditional probability for positive signals was determined for each of the markers, the results indicated higher confidence levels for the human assay and lower levels for all the other assays. Overall, the results from this study suggest that additional assays are needed, particularly to track cattle, chicken, and swine fecal pollution sources in the RGA watershed. The results also suggest that the geographic stability of genetic markers needs to be determined prior to conducting applied source tracking studies in tropical settings.
INTRODUCTION
Culturable counts of fecal indicator bacteria (FIB), such as enterococci and fecal coliforms, are used to measure fecal pollution levels in watershed systems. In order to be effective, indicators (i) should not survive for extended periods of time in the environment, (ii) should be exclusively associated with the intestinal tracts of humans and other warm-blooded animals, and (iii) should be associated with the occurrence of human enteric pathogens. However, growing evidence suggests that under some environmental conditions, FIB can survive outside the animal gut, that they are associated with a wide array of nonmammalian vertebrates and invertebrates, and that their correlation with pathogens varies significantly. Some of the evidence has been obtained from studies conducted in tropical regions (1, 2). For instance, currently used FIB can occur naturally in water accumulated in tropical epiphytic plants (3), can survive in tropical marine waters in the presence of nutrients (4), and can proliferate in tropical soils (5, 6). Studies in Mediterranean coastal areas have shown that Escherichia coli densities are often high in the sediments, serving as important sources of FIB and pathogens (7). Collectively, these studies suggest that the detection of FIB in tropical and subtropical water systems does not strictly imply fecal origin.
Identifying the primary sources of pollution is critical in implementing adequate pollution control and prevention strategies. In this regard, risk management practices in recreational waters have become critical to the public health community not only due to their importance in preventing human and ecosystem diseases but also due to the economic repercussions associated with tourism and the food industry (8). Poor microbial water quality increases the costs of services derived from water use and reuse. Hence, accurate and cost-effective fecal source identification is crucial to the implementation of management practices that can accurately and cost effectively prevent, control, and remediate fecal pollution events. Fecal source identification is challenging, as fecal pollution can occur via many different sources, including point and nonpoint sources. Point sources are relatively easy to manage because the pollutant enters the environment through an identifiable route (e.g., sewage treatment plant effluents). However, nonpoint pollution sources, such as waterfowl, water runoff from agricultural fields, and leaking septic systems, are far more difficult to manage because pollution routes are not easily identifiable, are generally diffuse, and may also be intermittent through time and space (9).
Human fecal pollution is considered to carry higher public health risks than nonhuman fecal pollution, although nonhuman sources are becoming increasingly relevant to human health due to the emergence of zoonotic pathogens. Unfortunately, traditional methods used to monitor FIB fail to discriminate between human and animal contamination. Alternative molecular techniques to identify pollution sources based on 16S rRNA genes have been developed and evaluated within the last decade (8). Library-independent assays targeting Bacteroidetes are particularly promising (10, 11, 12), as members of this phylum make up a significantly higher portion of fecal bacteria of warm-blooded animals than FIB and, since many of them are considered obligate anaerobes, they are presumed to only survive for short periods of time after released from their hosts (13, 14). Some Bacteroidales species have been shown to exhibit host specificity, and therefore, assays targeting these bacteria have been used to identify fecal sources (15, 16).
Several fecal source tracking (FST) studies have been conducted in recent years, most of which have been performed in watersheds within temperate regions (17, 18). In contrast, FST studies in the tropics are relatively few and most of them have used human-specific assays on a limited number of samples collected during short periods of time (19, 20). For example, Amador et al. (19) studied the incidence of human pollution using a Bifidobacterium adolescentis assay at several sites along two water reservoirs in Puerto Rico. While the data implicated the presence of human fecal sources in one of the reservoirs, sampling was restricted to only 1 day and an evaluation of the specificity and distribution of the host-specific markers was not conducted. In another study, the presence of Bacteroidetes was examined in human-, cattle-, and swine-specific assays in four riverine sites in Hawaii (21). Analyses were performed for water samples collected in two different years, but all of the samples were collected within four to five consecutive days, which is arguably insufficient to capture temporal trends. Additionally, a small number of fecal samples were used as part of the assay's evaluation in the latter study. Therefore, it remains unclear to what extent the particular idiosyncrasies that characterize the tropics (e.g., environmental conditions, land use, and urban spread) affect the applicability of these FST markers.
The objectives of this study were to further test the specificity of several host-specific markers by challenging them against animal feces collected in a tropical setting and to evaluate the spatial and temporal applicability of these source-specific markers across a tropical watershed impacted by different sources of fecal pollution.
MATERIALS AND METHODS
Watershed description.
The Río Grande de Arecibo (RGA) watershed is located along the western-central part of the north coast area of the island of Puerto Rico (Fig. 1). The RGA watershed is the third largest watershed in Puerto Rico, supplying approximately 440,488.38 m3 of water per day to the metropolitan area (the most densely populated area of Puerto Rico), and therefore, it is one of the primary sources of drinking water in Puerto Rico. Its catchment area is approximately 769 km2, with water flows running northward from the central mountain range into a coastal valley before discharging into the Atlantic Ocean. The central mountainous part of the watershed (southernmost part) is seated on impermeable basaltic rocks (22). The predominant land uses in this area are forest reserves, coffee plantations, and minor crops, such as beans, plantains, and citrus fruits. Urban development is mainly confined to the growth centers of the municipalities of Adjuntas, Utuado, and Jayuya. As the slope decreases, the basaltic bedrocks shift toward highly permeable limestone (karstic) bedrock and to a coastal alluvial plain (northernmost portion). In this area, cattle farming operations and urban development increase considerably. Most of the population of the RGA watershed is located in the coastal alluvial plains near the municipality of Arecibo (23). Three secondary sewage treatment plants discharge disinfected secondary effluents into the watershed: two drain into Río Cidra and Río Caunillas, tributaries of the RGA, while the third drains directly into the RGA.
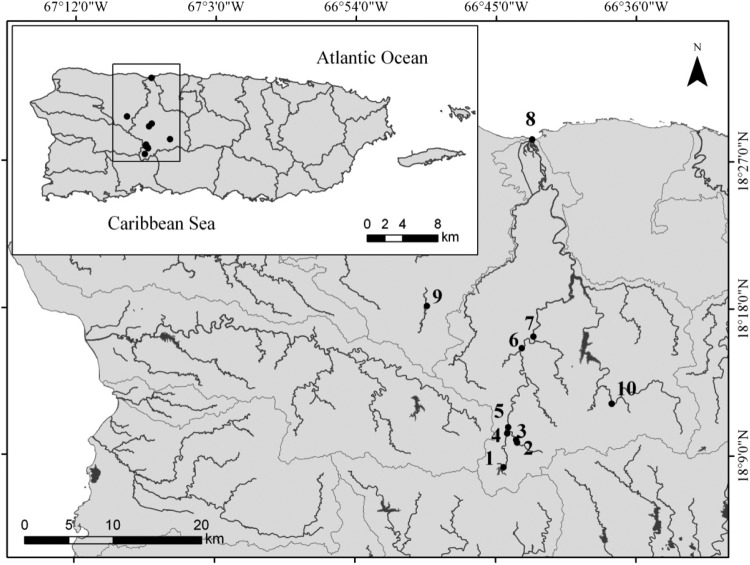
Fig 1.
Locations of sampling sites.
Sampling sites.
Ten sampling sites in the RGA watershed were selected according to previously recorded fecal pollution levels and presumed primary fecal pollution sources. Sites 1 to 4, 9, and 10 are at headwater tributaries of the RGA, whereas sites 6, 7, and 8 are located at the RGA (Fig. 1 and Table 1; also see the supplemental material). Sites 1 and 2 were located within or near the Guilarte National Forest, where human development and residences, poultry yards, and cattle farms are either scarce or absent. Sites 3 and 4 (Cidra River) and 6 and 7 (RGA) represent sites before and after wastewater treatment plants. Sites 5 to 8 are located at the RGA, downstream from sites 1 to 4. Site 5 is surrounded by woods and scattered houses that use septic tanks. Site 6 is located approximately 1.62 km upstream from site 7, and site 7 is located 120 m downstream from the sewage treatment plant. Site 8 is located right before the RGA drains into the Atlantic Ocean, close to the town center of Arecibo. Site 9 is located between a fenced farm with approximately 140 cattle on one side and human residences with septic tanks on the other side. Site 10 is located at Río Caunillas, 15 m downstream from the Jayuya sewage treatment plant effluent and surrounded by grasslands where cows and horses frequently graze.
Table 1.
Description of the Rio Grande de Arecibo watershed study sites
| Site | Location | Altitude (m) | Landscape feature |
Presumed primary fecal contamination sourcea | ||
|---|---|---|---|---|---|---|
| Forestland | Grassland | Human density | ||||
| 1 | Lago Garza, Guilarte Forest | 736 | High | Scarce | Low | Some waterfowl (heron) |
| 2 | Rio Vaca in Guilarte Forest | 542 | High | Scarce | Low | Septic tanks, some poultry |
| 3 | Upstream from Adjuntas WWTPb, Cidra River | 510 | Low | Scarce | Medium | Urban runoff, some waterfowl |
| 4 | Downstream from Adjuntas WWTP, Cidra River | 510 | Low | Scarce | Medium | Sewage, some poultry |
| 5 | Beginning of Arecibo River | 450 | Medium | Low | Medium | Urban runoff, septic tanks, some poultry |
| 6 | Upstream from Utuado WWTP | 376 | Low | Medium | High | Cattle, septic tanks |
| 7 | Downstream from Utuado WWTP | 376 | Low | Low | High | Sewage, some cattle and poultry, iguanas |
| 8 | Mouth of Arecibo River | Sea level | Low | High | High | Cattle, urban runoff |
| 9 | Criminales River | 276 | Medium | Medium | Medium | Cattle, urban runoff and septic tanks, some poultry |
| 10 | Downstream from Jayuya WWTP | 460 | High | Medium | Low | Sewage, cattle, some poultry |
There is historical knowledge that host animals are present at these sites for a significant part of the year.
WWTP, wastewater treatment plant.
Sample collection.
Water samples from sites 1 to 8 were collected from 30 October 2009 to 22 December 2010. Sites 9 and 10 were sampled from January 2010 to October 2010. Sampling was conducted weekly with the exception of October 2009, January 2010, June 2010, and December 2010 (for specific dates, see the supplemental material). Between 51 and 54 water samples were collected per site. In addition, raw sewage samples (n = 16) were collected from the Adjuntas treatment plant from August to October 2010 and used as positive controls for the human-specific assay. All samples were collected using sterile bottles and transported on ice to the laboratory at the University of Puerto Rico—Río Piedras Campus, where the samples were filtered within 6 h of collection. Water samples (100 ml) were filtered through polycarbonate membranes (0.4-μm pore size, 47-mm diameter; GE Water and Process Technologies, Trevose, PA), which were stored at −80°C until DNA extraction. To test for host specificity, fresh fecal samples from cattle, pigs, goats, horses, pigeons, monkeys, swans, fish, guineas, chicken, ducks, and turkeys were aseptically collected and stored at −80°C until further processing. All membranes and fecal samples were shipped overnight on dry ice to the EPA laboratory at the AWBERC building (Cincinnati, OH) for DNA extraction and PCR-based analyses.
DNA extractions and PCR analysis.
Total DNA was extracted from filters and fecal samples (0.25 to 0.30 g) using the PowerSoil DNA isolation kit, following the manufacturer's instructions (Mo Bio Laboratories, Inc.). DNA extracts were stored at −20°C until used in PCR assays. Nine different primer sets were tested against fecal and water DNA extracts (see Table S1 in the supplemental material). Bac32F and Ccoc assays were used to test for the presence of fecal bacteria. Bac32F is specific to members of the Bacteroidetes phylum (10), while Ccoc is specific to Clostridium coccoides (24). Conventional Bac32F, Ccoc, and Eub8F (25) PCR assays (i.e., presence/absence) and an enterococcal quantitative PCR (qPCR) assay (26, 27) were used to determine the PCR inhibition potential of DNA extracts, following the approach suggested by Bustin et al. (28). Specifically, undiluted and 10-fold-diluted fecal and water DNA extracts were used as the templates to further test for PCR inhibition. For conventional PCR assays, inhibition was defined as the presence of bands in agarose gels for reactions in which the undiluted DNA extract was negative while the diluted extract was positive (see the supplemental material). For the Entero1 qPCR assay, reactions were considered inhibited if fluorescence signals were not detected or were considerably reduced in the undiluted extracts compared to their strengths in the diluted extracts (see the supplemental material for additional details). No-template controls were used to check for cross-contamination (two per 96-well PCR plate).
The HF183, CF128, and PF163 assays were used to amplify human-, ruminant-, and swine-specific Bacteroidetes species, respectively. CP2-9, CBR2-42, and CP3-49 assays were used to amplify chicken-specific fecal sources. The reaction mixtures (25 μl) contained 2.5 mM deoxynucleoside triphosphates, 0.25 μM primers, 1 μl of buffer, 1 μl of template, and 0.625 nM units of TaKaRa Ex Taq (TaKaRa Bio, Inc.). PCR amplifications were performed in a Bio-Rad Tetrad2 Peltier thermal cycler (Bio-Rad, Hercules, CA) under the following cycling conditions: one initial denaturation step at 95°C for 5 min and 35 cycles of 1 min at 95°C, 1 min at the optimum annealing temperature (see Table S1 in the supplemental material), and 1 min at 72°C. PCR products were visualized on a 1.5% agarose gel using GelStar nucleic acid gel stain (Lonza, Rockland, ME).
Statistical analysis.
Kruskal-Wallis nonparametric analyses of variance (ANOVAs) were performed to test for differences between (i) sampling site as the independent variable and PCR results from water samples per primer as the dependent variable and (ii) collection period as the independent variable and PCR results from water samples per primer as the dependent variable. Binary data (presence/absence) were used for these analyses, and differences with P values of <0.05 were considered statistically significant. One multidimensional scaling (MDS) analysis using Euclidean distances was performed per PCR assay to explore similarities among the sampling points with respect to the PCR results.
Bayes' theorem was used to estimate the confidence that a given marker was detecting that particular fecal source. Specifically, we calculated the posterior probability using the following formula: P(A|B) = [P(A) × P(B|A)]/{[P(A) × P(B|A)] + [P(A′) × P(B|A′)]} as described in Weidhaas et al. (47). This involved calculating the posterior probability [P(A|B)] by determining the ratio of true positives [P(B|A)] and false positives [P(B|A′)] in fecal samples and the ratio of water samples that tested positive [P(A)]. Additionally, we calculated the posterior probability for each marker at each site by varying the prior probability from worst-case scenario (i.e., negative signals in all samples, or 0) to best-case scenario (i.e., positive signals in all samples, or 1) as described by Lamendella et al. (29).
RESULTS
Detection of molecular markers in fecal samples.
A total of 340 fecal samples collected in Puerto Rico were processed in this study. Regardless of the animal type, PCR inhibition was considered minimal, judging by the fact that none of the fecal samples tested were negative for all of the general PCR assays (Table 2). Specifically, all fecal samples tested with the 16S rRNA gene eubacterial primers produced a PCR product. With the exception of pigeon fecal samples, most fecal samples tested were positive with the general Bacteroidales assays (i.e., 77 to 100% of detection). When DNA extracts were diluted 10-fold, 21% of the undiluted extracts were found to inhibit the PCR assays (i.e., as determined by the presence of amplification products with diluted versus undiluted extracts) (see Tables S3 and S4 in the supplemental material). PCR inhibitors were removed by diluting the extracts (i.e., amplification products were observed in the diluted extracts). The results from the enterococcal qPCR assays were similar to those obtained with the general PCR assays (see Table S4). Moreover, the number of extracts that were positive with the Entero1 qPCR assay after dilution of the extracts was greater than the number that were positive but considered to be inhibited based on the cycle threshold value. Overall, these results are in agreement with the results of Haugland et al. (30), who concluded that amplification interference could largely be addressed by diluting DNA extracts. The occurrence of the Clostridium-specific marker was also high in the samples tested (i.e., 82%). Since the Ccoc assay targets Gram-positive bacteria and the Bac32F targets Gram-negative bacteria, the high amplification rate also suggests that the DNA extraction kits used in this study are capable of recovering DNA from different phylogenetic groups.
Table 2.
Host specificity and distribution of MST markers used in this study
| Source of fecal samples (total no. of samples) | % of tested fecal samples positive for marker (no. of samples tested against marker) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Uni8F | Ccoc | Bac32F | HF183 | CF128 | PF163 | CBR2-42 | CP2-9 | CP3-49 | |
| Cow (66) | 100 | 86 | 77 | 0 | 64 | 0 | 0 | NDa | ND |
| Goat (32) | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 0 | 0 |
| Horse (28) | 100 | 100 | 100 | 0 | 0 | 100 | 0 | 0 | 0 |
| Swine (30) | 100 | 100 | 100 | 0 | 42 | 100 | 0 | 0 | 0 |
| Monkey (9) | 100 | 100 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fish (13) | 100 | 69 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pigeon (11) | 82 | 91 | 50 | 0 | 0 | 0 | ND | ND | 0 |
| Chicken (97) | 100 | 90 | 80 | 0 | 7 | 0 | 37 | 16 (63) | 14 (64) |
| Guinea fowl (11) | 100 | 100 | 100 | 0 | 0 | ND | 18 | 0 | 0 |
| Duck (16) | 100 | 69 | 100 | 0 | 0 | ND | 38 | 0 | 6 |
| Turkey (5) | 100 | 80 | 100 | 0 | 100 | ND | 100 | 100 | 80 |
| Swan (22) | 100 | 54 | 100 | 0 | 0 | ND | 27 | 4.5 | 0 |
| WWTP (16) | 100 | 100 | 100 | 75 | 25 | 80 (5) | 13 | 9 (11) | 9 (11) |
ND, not determined.
The level of host specificity was different for each marker, and host distribution also varied between the assays (Table 2). For example, the human marker was highly host specific (100%) and was present in 75% of the sewage samples. The cow-specific assay marker (CF128) was positive for 64% of the cow fecal samples, and it cross-amplified with all turkey samples tested, to a relatively high extent with swine feces (42%), and to lesser extents with wastewater treatment plant samples (25%) and chicken feces (7%). The swine-specific marker was positive for all swine fecal samples tested, although it cross-amplified with all horse and goat fecal samples and most wastewater treatment plant samples (80%). Each of the chicken markers (such as CBR2-42, CP2-9, and CP3-49) showed a low level of sensitivity (i.e., 14 to 37%), and they showed different levels of cross-amplification with wastewater treatment plant samples and guinea fowl, swan, duck, and turkey fecal samples.
Detection of molecular markers in water samples.
A total of 534 water samples were processed in this study (Table 3). Amplification products were obtained with undiluted DNA extracts challenged against the general eubacterial assay Eub8F (100%), the Bacteroidetes assay Bac32 (96%), and the C. coccoides assay (81%). These assays cover a wide diversity of bacteria, including numerically dominant fecal bacteria groups, and therefore, the results suggest that the DNA was of good quality and that PCR inhibitors were removed in most samples. Further evidence of PCR inhibition was determined using diluted water DNA extracts. Specifically, only 2 to 6% of all water DNA extracts was inhibited in samples from 8 of the 10 sites, while no PCR inhibition was detected in samples from the other two sites (see Table S2 in the supplemental material). In all cases, diluting the samples 10-fold removed the observed PCR inhibition.
Table 3.
Distribution of molecular markers used in this study in different sites within the RGA watershed
| Site | % of tested water samples positive for marker (no. of samples tested) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ccoc | Uni8F | Bac32F | HF183 | CF128 | PF163 | CBR2-42 | CP2-9 | CP3-49 | |
| 1 | 78 (54) | 100 (54) | 94 (54) | 6 (54) | 6 (54) | 0 (33) | 6 (54) | 3 (39) | 0 (39) |
| 2 | 82 (54) | 100 (54) | 94 (54) | 6 (54) | 4 (54) | 0 (33) | 2 (54) | 0 (39) | 3 (39) |
| 3 | 81 (53) | 100 (53) | 93 (53) | 17 (53) | 8 (53) | 3 (32) | 2 (53) | 0 (38) | 0 (38) |
| 4 | 82 (54) | 100 (54) | 96 (54) | 54 (54) | 2 (54) | 24 (33) | 6 (54) | 0 (39) | 0 (39) |
| 5 | 65 (54) | 100 (54) | 94 (54) | 13 (54) | 6 (54) | 6 (33) | 2 (54) | 0 (39) | 0 (39) |
| 6 | 78 (54) | 100 (54) | 96 (54) | 7 (54) | 6 (54) | 6 (33) | 0 (54) | 0 (39) | 0 (39) |
| 7 | 87 (52) | 100 (52) | 96 (52) | 46 (52) | 25 (52) | 23 (31) | 2 (52) | 3 (37) | 0 (37) |
| 8 | 78 (54) | 100 (54) | 96 (54) | 19 (54) | 17 (54) | 12 (33) | 2 (54) | 0 (39) | 3 (39) |
| 9 | 88 (51) | 100 (51) | 92 (51) | 12 (51) | 14 (51) | 3 (31) | 0 (51) | 0 (37) | 0 (37) |
| 10 | 74 (54) | 100 (54) | 94 (54) | 46 (54) | 2 (54) | 12 (34) | 0 (54) | 3 (40) | 0 (40) |
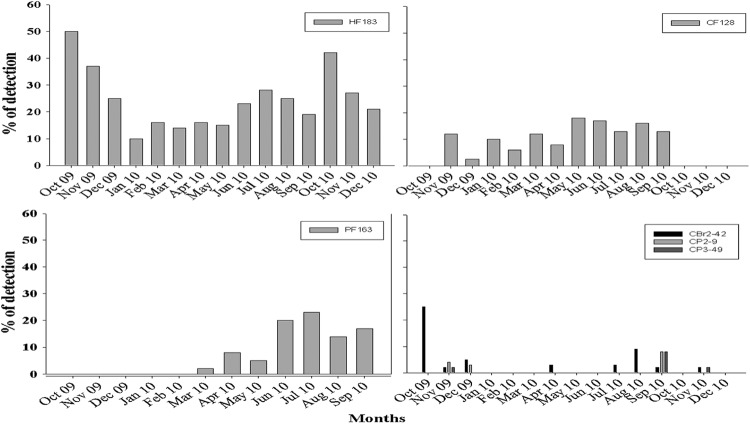
The water samples yielded significantly different amplification results across study sites with the HF183 assay (Kruskal-Wallis test, H9,436 = 96.71; P ≤ 0.001). For example, detection across study sites ranged from 4 to 54%. Sites 1 and 2 exhibited the lowest frequencies of amplification (6% for both sides), while sites 4, 7, and 10 exhibited the highest detection frequencies (54%, 46%, and 46%, respectively) (see Fig. S3 in the supplemental material). Similarly, the detection frequencies of HF183 were also significantly different among collection periods (Kruskal-Wallis test, H14,571 = 17.56644; P = 0.0016). Samples from October 2009 and October 2010 had the highest detection frequencies (50% and 42%, respectively), while samples collected in March 2010 had the lowest detection frequency (28%) (Fig. 2). Detection of the HF183 marker varied across sites over time (Kruskal-Wallis test, H139,546 = 215.7655; P < 0.001), with sites 1, 2, and 6 showing the lowest numbers of signals throughout the study (see Fig. S3). Human-specific signals occurred only once during October and November 2010 at site 1, once in February and October 2010 at site 2, twice in November 2009 at site 6, and once in June 2010 and October 2010 at site 6. In contrast, the detection frequencies at sites 4, 7, and 10 were the highest throughout the study period. Specifically, at site 4, HF183 occurred throughout the sampling period (13 months), with detection frequencies ranging from 75% (3 of 4 samples) during November 2009 to 25% (1 of 4) in April 2010 (see Fig. S3). Positive signals for HF183 were observed at sites 7 and 10 during most of the months (12 and 10 months, respectively), ranging from 25 to 75% of the samples during these periods. At sites 3, 5, 8, and 9, detection of HF183 varied considerably throughout the study, although signals were detected at least once during November 2009.
Fig 2.
Monthly detection frequencies in water samples of markers targeting human (HF183)-, cow (CF128)-, swine (PF163)-, and chicken (CBR2-42, CP2-9, and CP3-49)-specific assays.
CF128 detection in water samples also varied significantly across study sites (Kruskal-Wallis test, H9,534 = 33.74; P = 0.0001), but overall, it was detected with much less frequency than the human marker. Sites 7, 8, and 9 had the highest occurrence of CF128 among all the study sites, with 14 to 25% (13 of 52 samples) (see Fig. S4 in the supplemental material), while it was rarely detected at sites 4 and 10 (1 of 54 samples) (see Fig. S4). Detection at the other sites ranged from 6% to 10% (see Fig. S4). Likewise, the detection frequencies of CF128 varied significantly over time (Kruskal-Wallis test, H14,571 = 27.52017; P = 0.0165). Monthly detections ranged between 18% (7 of 39) in May 2010 to 0% from October to December 2010 (see Fig. S4). Detection of CF128 across sites also varied significantly over time (Kruskal-Wallis test, H139,546 = 186.5752; P = 0.0044). Site 7 had the highest variation frequencies over time, with positive signals in 8 of the 13 months sampled. In contrast, only once throughout the study (September 2010) was CF128 detected at site 4.
The detection frequencies of PF163 in water samples varied significantly across study sites (Kruskal-Wallis test, H9,324 = 27.09; P = 0.0013), ranging from 0 at sites 1 and 2 to 23 to 24% at sites 7 and 4, respectively (see Fig. S5 in the supplemental material). However, no significant variation occurred over time (see Fig. S5) or between sites and time (Kruskal-Wallis test, H14,336 = 0.000000; P = 1.000). The detection frequencies of the three chicken markers tested in water samples did not show significant differences across sites or months or between the interaction of site/month (Fig. 2), probably due to the low detection rates, which never exceeded more than 6% of the water samples tested per site.
DISCUSSION
We evaluated the performance of some of the most frequently used source tracking assays in one of the most important watersheds in Puerto Rico. To our knowledge, this represents the largest fecal source tracking study in the tropics in terms of the number of fecal samples challenged (340), animals tested for host specificity and host distribution (12 different hosts), number of water samples tested (534), and overall duration of the study (13 months). The presence of Bacteroidetes in most of the fecal samples (>95%) suggested that methods targeting this bacterial phylum may be used to track sources of fecal pollution in tropical waters (Table 2). Host-specific Bacteroidetes signals were detected at all sites, although their relative occurrence varied by site and by month (Table 3). Similarly, signals for C. coccoides were noted in most fecal sample types (82%), also suggesting that clostridium-based assays may be an alternate option for FST studies. Interestingly, from an ecological standpoint, the results indicate that C. coccoides is a member of different gut environments and not exclusively associated with humans as previously implied (31, 32).
The human marker (HF183) was found to be highly specific, which is in agreement with several studies (33, 34). In contrast, the specificity and host distribution of the cattle-specific marker (CF128) were relatively modest. CF128 detected 64% (42 of 66) of cattle fecal DNA extracts, a result that is lower than the results of previous studies, which have reported values greater than 90% (11, 12, 35, 36). PCR inhibition could not explain the relatively lower sensitivity results with the cattle assay, as only 17% of the samples that yielded negative signals with the CF128 assay were also negative with the general Bacteroidetes or the C. coccoides markers (see the supplemental material). Additionally, all of the cow fecal samples were positive for the general eubacterial assay (Eub8F). The CF128 assay failed to discriminate cattle fecal DNA extracts from swine (42%), chicken (17%), and turkey (100%). These results are not surprising, as the CF128 assay has been reported to cross-react with fecal DNA extracts from swine (35, 37) and other ruminants, including deer, elk, goat, llama, sheep, caribou, and bison (38). Therefore, the CF128 assay is not strictly host specific and should be complemented with other cattle fecal markers when trying to identify cattle pollution sources in environmental waters.
All the pig feces samples tested were positive with the PF163 assay, although the assay cross-reacted with DNA extracts from goat, horse, and wastewater. High sensitivity and relatively low specificity have also been reported previously for this marker. For instance, the sensitivities of PF163 have ranged from 70 to 100%, while it cross-amplified with cattle, chicken, raccoon, and horse fecal DNA (29). These results suggest that the bacterial populations targeted by PF163 are also widely distributed in animals (including humans). Recent phylogenetic analyses have further confirmed that PF163 markers are shared by multiple hosts (39). Thus, the detection of the PF163 marker in environmental waters in Puerto Rico does not indicate fecal contamination exclusively from swine. Additional studies are needed to fully determine the performance of this assay in other tropical settings.
The chicken-specific markers (CP2-9, CP3-49, and CBR2-42) showed the lowest levels of sensitivity and specificity among the host-specific markers tested in this study. For instance, the three chicken-specific markers tested in this study did not amplify more than a third of the chicken fecal samples tested. Similar results were reported when these markers were challenged against fecal DNA extracts from chickens in the United States and China (40). In addition, one of the markers (CBR2-42) failed to discriminate between fecal DNA extracts from chicken and other avian species. These markers were developed using cryptic metagenomic sequences that are likely to be associated with single-copy targets. Metagenomic markers used in previous source tracking studies have also been detected at lower frequencies than were obtained in 16S rRNA gene-based assays (36), suggesting that in cases where the contamination is relatively low, multiple-copy-gene assays might be better in environmental applications. Our data suggest that better assays are needed to detect the presence of poultry pollution in tropical waters. On the other hand, the low detection of signals is compatible with the absence of sizeable poultry operations in the watershed and low numbers of ranging chickens in areas surrounding the watershed.
The frequent detection of Bacteroidetes and C. coccoides and the levels of enterococci and thermotolerant coliforms in the water samples strongly suggest that there is a history of fecal pollution in the study sites (Table 3; also see Fig. S1 and S2 in the supplemental material). With respect to human fecal sources, of the RGA sites, water samples from sites 1 and 2 were associated with the lowest numbers of positive HF183 signals. This low level of detection suggests a relatively low human fecal impact, which is in agreement with the low human density associated with these sites compared to the human densities of other sites within the watershed. In contrast, the highest levels of HF183 detection occurred in sites downstream from sewage treatment effluents. Moreover, HF183 was detected every month in this watershed, further implicating human pollution as RGA's primary fecal pollution source. Cattle and swine markers were also detected at most sites but at a much lower frequency. The incidence of the cattle marker is compatible with the presence of free-range cattle grazing in areas near sites 6 to 9. The detection of swine signals (PF163) is puzzling, as there is not a sizeable population of pigs and there are no confined swine operations in this watershed. It should be noted that PF163 cross-reacted with wastewater samples and that the highest detection levels were in sites downstream from wastewater treatment plants, suggesting that most of these signals are associated with false-positive signals.
We applied Bayesian statistics to determine the confidence of using these markers in this watershed. In this study, the confidence of a human signal being the product of human sources was very high (i.e., 1.0) (see Table S5 in the supplemental material). Similar results have been noted with this assay on different continents, suggesting its value across different geographic locations (34, 41). With the exception of sites 1 and 2, the detection frequencies of human signals were lower than expected, considering the residential density associated with most of these sites. These results suggest that the populations targeted by HF183 have a poor fate (survival) and/or transport in this watershed, and therefore, other bacterial populations might be better indicators of human pollution. Detection of the cattle marker was also expected to be higher in this watershed, considering the presence of cattle operations in the watershed. On the other hand, the CF128 assay showed a low confidence value (i.e., 0.33), similar to those calculated for the results obtained by Fremaux et al. (35). The confidence value in the latter study increased when we pooled all ruminant data (i.e., from 0.30 to 0.69). These results further confirm that CF128 is not a good universal cattle assay marker. In our study, factors that contributed to the relatively low confidence level of detection of CF128 signals were the presence of false positives (i.e., swine, chicken, and turkey) and false negatives (i.e., 36%). While the CF128 marker has been shown to cross-amplify with feces from goat, moose, deer, and elk (35) as well as cattle, no other major ruminant inhabits the island of Puerto Rico, further bringing into question the value of the CF128 assay in tropical settings. However, it should be noted that the confidence value in our study increased for sites presumed to be impacted by cattle (i.e., 0.64). Hotspots may be detected in simple case scenarios by this assay, despite the assay's relatively poor performance at discriminating between different ruminant sources.
Differences in conditional probability for the same assay in different studies suggest that some markers are not universally useful and that there is a need to thoroughly validate assays prior to using them in applied studies. In this regard, many culture-independent microbial source tracking (MST) assays are not different than culture-dependent approaches from the standpoint that it is difficult to achieve high confidence levels in light of the low level of host preferential distribution. Thus, there is a need for developing better host-specific assays, as many of them can exhibit relatively high levels of cross-reactivity, low levels of host sensitivity, and low detection limits. Indeed, Liu et al. (42) recently showed that the cattle assay used in this study had a high rate of false positives and proposed a new reverse primer to use in conjunction with the original CF128F primer to alleviate the problem of low specificity. Future studies should further examine the value of this assay in other subtropical and tropical locations.
Interest in applying source tracking assays has grown steadily in recent years, in great part due to the high level of host specificity of the markers reported in the literature and their detection in environmental waters. The results from this study revealed some of the potential weaknesses associated with the application of FST assays in the tropics using PCR assays/molecular markers developed in temperate regions. Geographic differences in assay performance may be due to differences in host specificity. Thus, prior to using assays in environmental applications, host specificity tests need to be performed with feces collected from the same geographic location as the study sites, as the host stability of most MST markers on a global basis is poorly understood. In our study, assay markers that have been reported to be highly specific (i.e., for cattle and chicken) were shown to cross-amplify with other fecal sources. In most studies, MST assays have been evaluated with a small number of fecal samples. In fact, the number of studies with a comprehensive temporal component is relatively small, and more importantly, most have been conducted with samples collected in temperate regions. To date, most source-tracking studies conducted in tropical waters have not comprehensively accounted for seasonal variations, have been conducted using a small number of sites, and have not evaluated the host specificity and host distribution of the assays using a significant number of fecal samples from targeted and nontargeted hosts (19, 43).
The lower amplification of assay markers in some of the sites tested may be due to the survival of the host-specific bacterial populations in tropical waters. Differences in survival have been noted for E. coli and enterococci in the tropics and in temperate regions (44, 45). Interestingly, several studies have suggested that conventional indicators can survive longer in tropical waters than in temperate waters. The availability of carbon sources has been implicated in prolonged survival and seasonal variations (4). No information is available on the survival of Bacteroidetes in the tropics, information that is needed to further assess their value as indicators of fecal pollution. It should be noted that when the targeted population is low, the presence of inhibitors could result in the samples being considered negative for a given marker after DNA extract dilution. Thus, how PCR inhibitors affect assay detection limits with tropical water samples should be carefully addressed in future studies. Nonetheless, it is possible that new bacterial targets and markers are needed for tropical and subtropical zones (46). In this regard, further phylogenetic analyses will aid in identifying new targets for method development (16). The latter analyses must be applied to other emerging source tracking targets, such as Brevibacterium spp. (47), Catellicoccus marimammalium (25), Lachnospiraceae (48), and Methanobrevibacter spp. (17).
In spite of the limitations associated with the host-specific markers tested, the results from this study suggest that this watershed is regularly impacted by fecal pollution, primarily by human and cattle fecal sources. Specifically, the human-specific marker showed results consistent with the overall predictable sources, where sites near low-level human development and density yielded the lowest detection frequencies and sites impacted by sewage treatment plants yielded the highest detection frequencies among the study sites. While presence/absence assays are useful at detecting primary fecal sources, quantitative assays can theoretically provide a better idea of the overall loads of a particular fecal source. Several qPCR-based MST assays are available in the literature but have yet to be tested in tropical waters. Therefore, future studies should test the applicability of these quantitative MST assays in tropical settings and, more importantly, determine if there is a correlation between MST signals and fecal loadings of currently used indicators. This is necessary to establish regulatory thresholds and to determine how a water system should be managed to significantly reduce indicator bacterial loads.
Supplementary Material
ACKNOWLEDGMENTS
H.R. was funded via a National Research Council fellowship. The U.S. Environmental Protection Agency, through its Office of Research and Development and the RARE program, funded, managed, and collaborated in the research described herein.
This work has been subjected to the U.S. Environmental Protection Agency's administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 4 January 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03070-12.
REFERENCES
- 1. Shibata T, Solo-Gabriele HM, Sinigalliano CD, Gidley ML, Plano LR, Fleisher JM, Wang JD, Elmir SM, He G, Wright ME, Abdelzaher AM, Ortega C, Wanless D, Garza AC, Kish J, Scott T, Hollenbeck J, Backer LC, Fleming LE. 2010. Evaluation of conventional and alternative monitoring methods for a recreational marine beach with nonpoint source of fecal contamination. Environ. Sci. Technol. 44:8175–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solo-Gabriele HM, Wolfert MA, Desmarais TR, Palmer CJ. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rivera SC, Hazen TC, Toranzos GA. 1988. Isolation of fecal coliforms from pristine sites in a tropical rain forest. Appl. Environ. Microbiol. 54:513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santo Domingo JW, Fuentes FA, Hazen TC. 1989. Survival and activity of Streptococcus faecalis and Escherichia coli in petroleum-contaminated tropical marine waters. Environ. Pollut. 56:263–281 [DOI] [PubMed] [Google Scholar]
- 5. Byappanahalli M, Fujioka R. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50:27–32 [PubMed] [Google Scholar]
- 6. Betancourt WQ, Fujioka RS. 2006. Bacteroides spp. as reliable marker of sewage contamination in Hawaii's environmental waters using molecular techniques. Water Sci. Technol. 54:101–107 [DOI] [PubMed] [Google Scholar]
- 7. Luna GM, Vignaroli C, Rinaldi C, Pusceddu A, Nicoletti L, Gabellini M, Danovaro R, Biavasco F. 2010. Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Appl. Environ. Microbiol. 76:5659–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santo Domingo JW, Bambic DG, Edge TA, Wuertz S. 2007. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 41:3539–3552 [DOI] [PubMed] [Google Scholar]
- 9. Stewart JR, Gast RJ, Fujioka RS, Solo-Gabriele HM, Meschke JS, Amaral-Zettler LA, del Castillo E, Polz MF, Collier TK, Strom MS, Sinigalliano CD, Moeller PDR, Holland AF. 2008. The coastal environment and human health: microbial indicators, pathogens, sentinels and reservoirs. Environ. Health 7(Suppl 2):S3 doi:10.1186/1476-069X-7-S2-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lamendella R, Santo Domingo JW, Oerther DB, Vogel JR, Stoeckel DM. 2007. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analyses of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651–660 [DOI] [PubMed] [Google Scholar]
- 13. Bae S, Wuertz S. 2009. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 43:4850–4859 [DOI] [PubMed] [Google Scholar]
- 14. Kreader CA. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl. Environ. Microbiol. 64:4103–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dick LK, Bernhard AE, Brodeur TJ, Santo Domingo JW, Simpson JM, Walters SP, Field KG. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamendella R, Santo Domingo JW, Kelty C, Oerther DB. 2008. Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74:575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ufnar J, Wang SY, Christiansen J, Yampara-Iquise H, Carson C, Ellender RD. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J. Appl. Microbiol. 101:44–52 [DOI] [PubMed] [Google Scholar]
- 18. Walters SP, Field KG. 2007. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter. Environ. Sci. Technol. 41:1856–1862 [DOI] [PubMed] [Google Scholar]
- 19. Amador JA, Sotomayor-Ramírez D, Martínez G, Chen L, Bachoon D. 2008. Tracking human faecal contamination in tropical reservoirs in Puerto Rico. Lakes Reserv. Res. Manag. 13:301–317 [Google Scholar]
- 20. Bonkosky M, Hernández-Delgado EA, Sandoz B, Robledo IE, Norat-Ramírez J, Mattei H. 2009. Detection of spatial fluctuations of non-point source fecal pollution in coral reef surrounding waters in southwestern Puerto Rico using PCR-based assays. Mar. Poll. Bull. 58:45–54 [DOI] [PubMed] [Google Scholar]
- 21. Boehm AB, Yamahara KM, Walters SP, Layton BA, Keymer DP, Thompson RS, Knee KL, Rosener M. 2011. Dissolved inorganic nitrogen, soluble reactive phosphorous, and microbial pollutant loading from tropical rural watersheds in Hawaii to the coastal ocean during non-storm conditions. Estuaries Coasts 34:925–936 [Google Scholar]
- 22. Renken RA, Ward WC, Gill IP, Gómez-Gómez F, Rodríguez-Martínez J. 2002. Geology and hydrogeology of the Caribbean islands aquifer system of the Commonwealth of Puerto Rico and the U.S. Virgin Islands. Regional Aquifer System Analysis. USGS Professional Paper 1419. U.S. Geological Survey, U.S. Department of the Interior. http://pubs.usgs.gov/pp/pp1419/pdf/BOOK.PDF
- 23. Martinuzzi S, Gould WA, Ramos-González OM. 2007. Land development, land use and urban sprawl in Puerto Rico integrating remote sensing and population census data. Landsc. Urban Plan. 79:288–297 [Google Scholar]
- 24. Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, Oyaizu H, Tanaka R. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryu H, Griffith JF, Khan IU, Hill S, Edge TA, Toledo-Hernandez C, Gonzalez-Nieves J, Santo Domingo J. 2012. Comparison of gull feces-specific assays targeting the 16S rRNA genes of Catellicoccus marimammalium and Streptococcus spp. Appl. Environ. Microbiol. 78:1909–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 27. Ryu H, Henson M, Elk M, Toledo-Hernandez C, Griffith J, Blackwood D, Noble R, Gourmelon M, Glassmeyer S, Santo Domingo J. 2013. Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl. Environ. Microbiol. 79:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. Minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 29. Lamendella R, Santo Domingo JW, Yannarell AC, Ghosh S, Di Giovanni G, Mackie RI, Oerther DB. 2009. Evaluation of swine-specific PCR assays used for fecal source tracking and analysis of molecular diversity of swine-specific “Bacteroidales” populations. Appl. Environ. Microbiol. 75:5787–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haugland RA, Siefring S, Lavender J, Varma M. 2012. Influences of sample interference and interference controls on quantification of enterococci fecal indicator bacteria in surface water samples by the qPCR method. Water Res. 46:5989–6001 [DOI] [PubMed] [Google Scholar]
- 31. Hayashi H, Sakamoto M, Kitahara M, Benno Y. 2006. Diversity of the Clostridium coccoides group in human fecal microbiota as determined by 16S rRNA gene library. FEMS Microbiol. Lett. 257:202–207 [DOI] [PubMed] [Google Scholar]
- 32. Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahmed W, Goonetilleke A, Powell D, Chauhan K, Gardner T. 2009. Comparison of molecular markers to detect fresh sewage in environmental waters. Water Res. 43:4908–4917 [DOI] [PubMed] [Google Scholar]
- 34. Ahmed W, Stewart J, Powell D, Gardner T. 2008. Evaluation of Bacteroides markers for the detection of human faecal pollution. Lett. Appl. Microbiol. 46:237–242 [DOI] [PubMed] [Google Scholar]
- 35. Fremaux B, Gritzfeld J, Boa T, Yost CK. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 43:4838–4849 [DOI] [PubMed] [Google Scholar]
- 36. Lee YJ, Molina M, Santo Domingo JW, Willis JD, Cyterski M, Endale DM, Shanks OC. 2008. Temporal assessment of the impact of exposure to cow feces in two watersheds by multiple host-specific PCR assays. Appl. Environ. Microbiol. 74:6839–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gourmelon M, Caprais MP, Ségura R, Le Mennec C, Lozach S, Piriou JY, Rincé A. 2007. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl. Environ. Microbiol. 73:4857–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gawler AH, Beecher JE, Brandão J, Carroll NM, Falcão L, Gourmelon M, Masterson B, Nunes B, Porter J, Rincé A, Rodrigues R, Thorp M, Walters JM, Meijer WG. 2007. Validation of host-specific Bacteriodales 16S rRNA genes as markers to determine the origin of faecal pollution in Atlantic Rim countries of the European Union. Water Res. 41:3780–3784 [DOI] [PubMed] [Google Scholar]
- 39. Lamendella R, Li KC, Oerther D, Santo Domingo JW. 2013. Molecular diversity of Bacteroidales in fecal and environmental samples and swine-associated subpopulations. Appl. Environ. Microbiol. 79:816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu J, Santo Domingo J, Shanks OC. 2007. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 41:3561–3574 [DOI] [PubMed] [Google Scholar]
- 41. Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890–901 [DOI] [PubMed] [Google Scholar]
- 42. Liu R, Chan CF, Lun CH, Lau SC. 2012. Improving the performance of an end-point PCR assay commonly used for the detection of Bacteroidales pertaining to cow feces. Appl. Microbiol. Biotechnol. 93:1703–1713 [DOI] [PubMed] [Google Scholar]
- 43. Bachoon DS, Miller CM, Green CP, Otero E. 2010. Comparison of four polymerase chain reaction methods for the rapid detection of human fecal pollution in marine and inland waters. Int. J. Microbiol. 2010:pii595692 doi:10.1155/2010/595692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carrillo M, Estrada E, Hazen TC. 1985. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl. Environ. Microbiol. 50:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pérez-Rosas N, Hazen TC. 1988. In situ survival of Vibrio cholerae and Escherichia coli in tropical coral reefs. Appl. Environ. Microbiol. 54:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson KL, Whilock JE, Harwood VJ. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weidhaas J, Macbeth T, Olsen R, Harwood V. 2011. Correlation of quantitative PCR for a poultry-specific Brevibacterium marker gene with bacterial and chemical indicators of water pollution in a watershed impacted by land application of poultry litter. Appl. Environ. Microbiol. 77:2094–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Newton RJ, VandeWalle J, Borchardt MA, Gorelick MH, McLellan SL. 2011. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Appl. Environ. Microbiol. 77:6972–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.