Abstract
Microbes associated with marine sponges are considered important producers of bioactive, structurally unique polyketides. The synthesis of such secondary metabolites involves type I polyketide synthases (PKSs), which are enzymes that reach a maximum complexity degree in bacteria. The Haplosclerida sponge Arenosclera brasiliensis hosts a complex microbiota and is the source of arenosclerins, alkaloids with cytotoxic and antibacterial activity. In the present investigation, we performed high-throughput sequencing of the ketosynthase (KS) amplicon to investigate the diversity of PKS genes present in the metagenome of A. brasiliensis. Almost 4,000 ketosynthase reads were recovered, with about 90% annotated automatically as bacterial. A total of 235 bacterial KS contigs was rigorously assembled from this sequence pool and submitted to phylogenetic analysis. A great diversity of six type I PKS groups has been consistently detected in our phylogenetic reconstructions, including a novel and A. brasiliensis-exclusive group. Our study is the first to reveal the diversity of type I PKS genes in A. brasiliensis as well as the potential of its microbiome to serve as a source of new polyketides.
INTRODUCTION
Many marine sponges are known as bacteriosponges because they host dense microbial communities (1, 2). The advent of massively parallel 16S rRNA gene tag pyrosequencing (3–5) led to the discovery that the bacterial communities in marine sponges are highly diverse, almost exclusively found in sponges, and mostly (approximately 72%) species specific (3, 6).
The synthesis and/or accumulation of secondary metabolites by marine sponges or their symbionts may provide chemical defenses against pathogens, predators, and competitors (7). These secondary metabolites may also act as chemical mediators and can therefore be considered key factors in the coevolution of sponges and their symbiotic microbiome (8, 9). Sponges from the order Haplosclerida, for example, are a source of a variety of bioactive polycyclic alkaloids (10). The prototypical sponge-derived alkaloid manzamine A, which has potent antimalarial activity (11), was found in several geographically and taxonomically unrelated haplosclerid sponges; these observations strongly suggested that manzamine A has a symbiotic origin. A patent has reported the production of manzamine A by an actinobacterium (Micromonospora sp. strain M42) isolated from the sponge Acanthostrongylophora sp.; however, no biosynthetic gene cluster has yet been reported for this actinobacterium (12). The synthesis of structurally complex Haplosclerida alkaloids is putatively derived from the head-to-tail addition of 3-alkylpiridine (or 3-alkylpiperidine) precursors that originate from a modular polyketide synthase (13–15). Until now, these enzymes have been found exclusively in microorganisms.
Polyketide synthase (PKS)-derived secondary metabolites, such as the Haplosclerida alkaloids, are products from the successive condensation of acyl coenzyme A (acyl-CoA) thioesters (16). In bacteria, type I PKSs demonstrate higher complexity as multifunctional and modular enzymes that work in an assembly line fashion to perform the synthesis of complex polyketides. Bacterial modular PKSs can belong to two phylogenetically unrelated groups, which diverge mainly by the presence (cis) or absence (trans) of the acyl-transferase (AT) domain in the extender modules (17).
Phylogenetic analysis of ketosynthase (KS) domains has revealed that modular type I PKSs evolved by duplication but diversified mainly by recombination reprogramming of these and other functionalities (18). KS phylogenetic analysis also showed that the unconventional trans-AT subtype evolved independently of the cis-AT type I PKSs, which possibly implicates horizontal gene transfer and recombination events (19). KS phylogenies have been used to effectively estimate diversity and to precisely dissect type I PKSs from complex metagenome backgrounds (20–24). In a systematic investigation, 150 KS sequences from metagenomic DNA of 20 different demosponges were analyzed, and a sponge-specific PKS group, which is termed symbiont ubiquitous type I PKS (Sup) and is involved in the synthesis of methyl-branched fatty acids, represented 88% of the cloned sequences (23). In addition, modular cis- and trans-AT PKSs, which are typically related to the production of bioactive compounds, were detected at low frequencies. Similarly, both the PKSs in hybrid nonribosomal peptide synthetase (NRPS)-PKS systems and the FAS-like type I PKSs (RkpA and WcbR) related to the synthesis of capsular lipopolysaccharide were detected at low frequencies (23). Orthologs of the RkpA gene and those of rifamycin synthases were also detected by phylogenetic analysis of KSs recovered from alphaproteobacterial (25) and actinomycete (Salinospora) sponge isolates (21).
Efforts to estimate the diversity of PKS systems in sponge holobionts have relied mainly on plasmid cloning strategies, which may lead to underestimations as a result of the cloning process. An exception is the study of the large-scale sequenced PKS amplicons from Cacospongia-associated Poribacteria (26). Moreover, there is a lack of studies on the PKS diversity in sponge species endemic to the southern Atlantic Ocean. One example of an understudied sponge species is Arenosclera brasiliensis, which is the source of cytotoxic and antibacterial complex alkaloid arenosclerins A to C (27, 28). Here, we report an analysis of PKS gene diversity in the A. brasiliensis microbiome conducted by means of massive parallel pyrosequencing and a comprehensive phylogenetic analysis of KS amplicons. We also determined the taxonomic signature of the recovered KS contigs.
MATERIALS AND METHODS
Sample collection and DNA isolation.
Specimens of A. brasiliensis were collected at João Fernandinho's Beach, which is located in the Búzios peninsula in the state of Rio de Janeiro (22°44′49′′/41°52′54′′W), during two expeditions in May 2010 and January 2011. Sponge specimens were transported under controlled temperature (24°C) in aerated seawater for approximately 3 h and then washed and processed for total DNA extraction exactly as previously described (29). Briefly, samples were set on sterile seawater for 5 to 10 min, dissected with a sterile scalpel, dried by gently pressing on sterile paper towels, and ground in liquid nitrogen. Approximately 1 g of ground tissue was used for DNA extraction and purification with 4 M guanidine hydrochloride buffer followed by phenol-chloroform.
KS PCR amplification and pyrosequencing.
The ketosynthase (KS) gene fragment was amplified from three samples of A. brasiliensis metagenomic DNA (Ab1, Ab2, and Ab5). Reactions were performed as previously described (20, 30). We used 0.1 μg of the template DNA and the degenerated primer pairs KSDPQQF (5′-MGN GAR GCN NWN SMN ATG GAY CCN CAR CAN MG-3′)/KSHGTGR (5′-GGR TCN CCN ARN SWN GTN CCN GTN CCR TG-3′), KSF2.i (5′-GCI ATG GAY CCI CAR CAR MGI VT-3′)/KS5R.i (5′-GTI CCI GTI CCR TGI SCY TCI AC-3′), and KSF2.gc (5′-GCS ATG GAY CCS CAR CAR CGS VT-3′)/KS5R.gc (5′-GTS CCS GTS CCR TGS SCY TCS AC-3′). The products from 10 PCRs with each DNA template were then combined and gel purified using the QIAquick gel extraction kit (Qiagen). Finally, ∼500 ng of the purified KS amplicons (∼750 bp in length) were sequenced by the 454 pyrosequencing methodology (31) using GS-FLX TITANIUM chemistry with the GS-FLX instrument (Roche Applied Science).
KS sequence assembling and comparisons.
The sequences obtained were assembled with Newbler (software package for de novo DNA sequence assembly) using highly stringent conditions, including a minimum alignment length of 100 nucleotides and at least 99% similarity. For querying KS sequences, we built a hidden Markov model (HMM) single profile by aligning 80 KS sequences from type I PKS enzymes that had been retrieved from the Uniprot database (32) using the MEGA5 software (33) and HMMER version 3 software (34). In parallel, A. brasiliensis contigs and the remaining singletons were translated in all six reading frames using the Transeq software available in the European Molecular Biology Open Source Suit (Emboss) (35). Finally, the translated database was queried for the KS HMM profile with an E value of 10−10. We used the special blast tool in order to search for conserved domains (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The recovered sequences were queried against the NCBInr (nonredundant) protein database of the NCBI using the BLASTp for confirmation of KS identity. Additionally, more than 200 Mb of A. brasiliensis-derived shotgun pyrosequencing data (29) was screened for KS sequences. To access the rare KS sequence, we assembled all of the sequencing data (samples Ab1 to Ab7) with Newbler (version 2.6) using stringent parameters, including a minimum alignment of 60 nucleotides and at least 95% similarity. The generated contigs and singletons were then screened using the designed KS single profile at HMMER, as described above. KS sequences before and after assembling were submitted to MG-RAST (36). The potential of these sequences to be associated with known metabolic functions was tested using the Uniprot-TrEMBL database with a maximum E value of 10−5. The functional annotation results were then compared.
Phylogenetic reconstruction and taxonomic assignment.
KS sequences were aligned in MEGA5 (33). The resulting alignment was used for neighbor-joining (NJ) and maximum likelihood (ML) grouping with 1,000 and 100 bootstrap replicates, respectively. All KS sequences were queried using BLASTp (E value of 10−5) against the GenBank Microbe Genomes database (22 September 2011 version) and nonredundant GenBank database. The identity values of the best scores were retained for each bacterial phylum/class.
Nucleotide sequence accession numbers.
The raw input and the assembled sequences of KS amplicon pyrosequencing data are available at the MG-RAST server under project “Arenosclera brasiliensis_PKS diversity.” The sequence data of A. brasiliensis metagenomes is also available under project “Arenosclera brasiliensis_Metagenome” (29). The sequences of KS contigs were deposited into GenBank under accession numbers JX012425 to JX012657 (contigs from amplicon pyrosequencing) and JX945643 (contig from sponge metagenome shotgun pyrosequencing).
RESULTS AND DISCUSSION
To estimate the diversity of PKS genes, ketosynthase genes were amplified from metagenomic DNA of three A. brasiliensis specimens. Thereafter, the amplified products were pyrosequenced, generating approximately 40 Mb of sequence data. The reads from each KS metagenome (MG_KS1-3) had an average length of 269 bp and were assembled to form contigs with a maximum size equal to that of the purified amplicon bands (600 to 800 bp) (see Table S1 in the supplemental material). The number of contigs formed, the amount of reads assembled, and the number of contigs from each metagenome used in phylogenetic analysis are detailed in Table S1 in the supplemental material. Both the assembled and unassembled KS metagenomes were taxonomically and functionally annotated using the MG-RAST server (Fig. 1). The taxonomic annotation assigned more than 90% of data as bacterial sequences, and functional annotation generated a vast majority of hits for the ketosynthase domain or associated nomenclature (i.e., polyketide synthase, β-ketoacyl synthase) (Fig. 1A and B). These results confirmed the specificity of our PCRs and the high quality of the assemblage.
Fig 1.
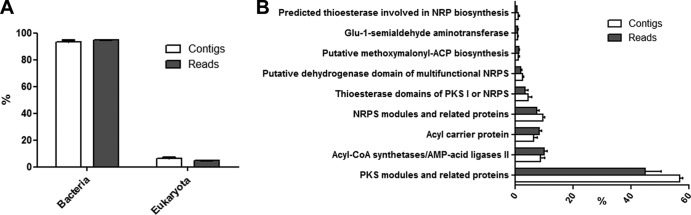
MG-RAST annotation. (A) Taxonomic classification of contigs and reads based on GenBank annotation; (B) function annotation of contigs and reads based on Subsystems annotation. The number of reads was 172,629, and the number of contigs was 3,936.
PKS diversity.
In parallel to the MG-RAST annotation, we screened the metagenomic data for the generated HMM KS profile and recovered a total of 3,936 KS contigs. Of the 314 contigs signed as eukaryote at MG-RAST (Fig. 1A), only a subset of 262 sequences was validated as ketosynthase gene fragments. Interestingly, a blast search of these KS sequences against the nr database revealed that more than 60% of the top 10 best hits obtained were assigned to bacteria (see Fig. S1A in the supplemental material). The prevailing classes were Proteobacteria (61%), Actinobacteria (16%), and Cyanobacteria (14%) (see Fig. S1B in the supplemental material). The differing annotation results of our blast searches might reflect the weak taxonomic signal of the query KS sequences, as well as the bias intrinsically associated with both the MG-RAST and GenBank's nr databases. However, considering that the nr database is bigger, our results suggest that these KS sequences might be derived from bacteria. Nevertheless, such PKS sequences might also be acquired by eukaryotes via lateral gene transfer events with bacteria as the donor. BLAST searches of eukaryotic sequences from A. brasiliensis metagenomics resulted in automatically annotated hits of alveolata and fungi genomes or KS genes of unknown pathways (37).
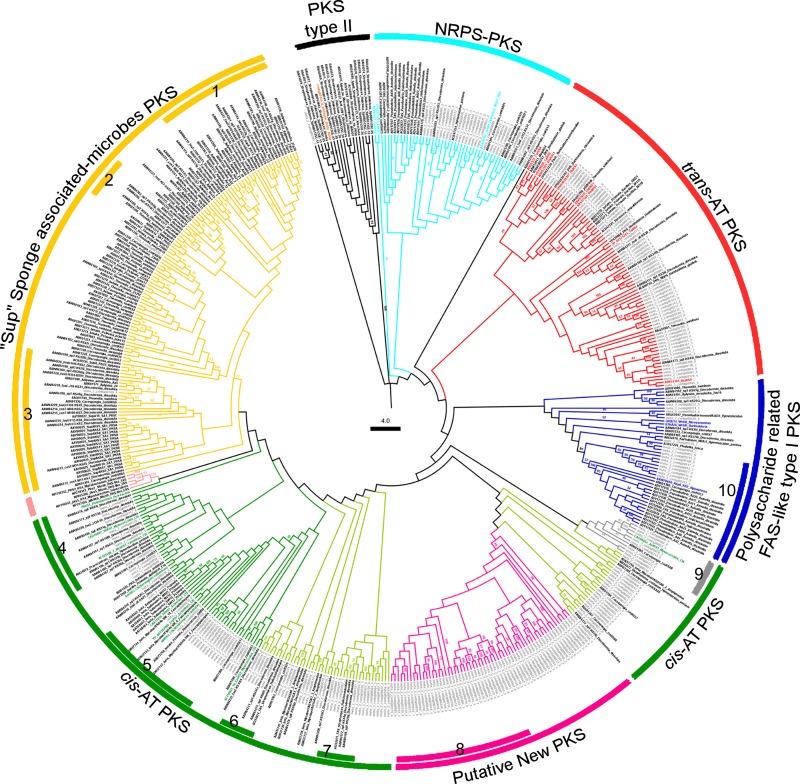
Only the sequences annotated as bacteria were included in our phylogenetic analysis. A total of 235 of these KS contigs encoded uninterrupted protein sequences of at least 150 amino acids; these results demonstrated that the quality of the data was high and sufficient for further phylogenetic analysis. These contigs also revealed the presence of conserved cysteine and histidine residues in the catalytic triad (CHH) of active KS domains from type I PKSs (38, 39) as well as the primer-attaching conserved motifs DPQQR and HGTGT (40, 41) (see Fig. S2 in the supplemental material). The 235 KS contigs were aligned with 47 KS domains from reference PKS systems as well as 226 other KS sequences previously recovered from sponge-microbe association studies (see Table S2 in the supplemental material). The total alignment of 508 sequences was then used for neighbor-joining (NJ) and maximum likelihood (ML) phylogenetic reconstruction. Both methods generated topologically similar trees, although the ML topology was more robust and presented higher bootstrap values (Fig. 2). Based on the reference KS sequences, we detected that our amplicons fell within all six KS groups in the phylogenetic (ML) reconstruction (Fig. 2; the PKS type II outgroup is in black). This result demonstrates the large variety of type I PKS enzymes present in these samples (Fig. 2; sequences from A. brasiliensis are labeled in gray).
Fig 2.
Maximum likelihood tree of partial PKS sequences (approximately 229 amino acids). Ketosynthase sequences from (i) the metagenome of sponge A. brasiliensis (gray labels), (ii) other sponge metagenomes (black labels), (iii) reference sequences of type I PKS subtypes (labeled in the color of the respective PKS subtype), and (iv) other specific taxonomic groups to help with the identification of the KS subtypes amplified from the A. brasiliensis metagenomes. Bootstrap values greater than 40% are shown at the nodes. The sequences marked in salmon are Mycobacterium fatty-acid synthases (FAS). The subtypes of PKSs are indicated. The internal bars indicate taxonomic clades. 1, 2, 3, and 5, Actinobacteria; 4, 6, and 7, Cyanobacteria; 8 and 9, Deltaproteobacteria; 10, Alphaproteobacteria.
In order to determine the closest taxonomic neighbors of the bacterial KS contigs recovered, each aligned KS sequence was blasted against GenBank's microbial genome database. The values of identity that were obtained varied between 46 and 96%, and a high level of similarity was obtained (82%) for the A. brasiliensis KS contig (meta_2_contig00168_4, meta_2_contig00109_1) toward Mycobacterium marinum and Agrobacterium vitis (see Table S2 in the supplemental material). The average best-identity scores were approximately 60% (see Table S2), which enabled identification at the level of family or a higher taxonomic rank.
Sup group.
Approximately 54% of the publicly available sponge-derived KS amplicons (122 sequences) are grouped into the symbiont ubiquitous PKS group (Sup) (Fig. 2, marked in yellow). Only two (0.8%) of the KS sequences (meta_2_contig00100_1 and meta_2_contig00196) amplified from the A. brasiliensis metagenomes fell within this clade. These observations were in opposition to the previously observed dominance of Sup-like PKS clones (84%), which was reported for the metagenomes of 20 different sponge species (23). Such discrepancies may reflect the relative abundance of the microbial community containing the Sup PKS genes. Once we excluded plasmid-cloning artifacts and performed our analyses in triplicate (pyrosequencing KS amplicons from three A. brasiliensis specimens), the possibility that any KS amplicon had been favored during our sequencing protocol was low. Therefore, a more convincing explanation would be that A. brasiliensis belongs to a group of Demospongiae in which the Sup PKS-containing microbes, such as Poribacteria (42, 43), are not as abundant. Finally, taxonomic assignment of the sequences revealed that three internal groups of Sup contained KS domains that showed higher similarities to regions within Mycobacterium genomes (Fig. 2, internal bars 1 to 3).
Cis-AT-PKS group.
Following the ML topology counterclockwise, we found that 26 (11%) of the A. brasiliensis KS sequences grouped within the KS domains from the typical cis-AT type I PKSs (Fig. 2, marked in dark green). Additionally, 46 recovered KSs (19.5%) formed minor clades with other sponge-derived KSs (20, 23) that were also phylogenetically related to the cis-AT PKS systems (Fig. 2, clades in light green; see also Table S2 in the supplemental material).
Approximately 19.6% of such A. brasiliensis-derived KS contigs were closely related to actinobacterial KS sequences (Fig. 2, internal bar 5; see Table S2 in the supplemental material). Actinobacteria, particularly the species belonging to the order Actinomycetales, are known to be a source of cis-AT PKS systems, including those involved in the synthesis of clinically validated antibiotics as well as of several cis-AT PKS systems that remain unexplored (44). The production of manzamine A, an alkaloid found in several haplosclerid sponges, has been assigned to an Actinomycetales strain of the genus Micromonospora (12). Furthermore, our previous metagenomic analysis indicated that Actinobacteria was one of the most abundant bacterial phyla in the microbiome associated with A. brasiliensis (29). Therefore, it is reasonable to suggest that A. brasiliensis-associated Actinobacteria communities might be responsible for the production of secondary metabolites as well as of bioactive polyketides.
A subgroup of A. brasiliensis-derived KS contigs showed the greatest similarity to deltaproteobacteria genomes (Fig. 2, internal bar 9). This subgroup also formed a well-supported (bootstrap of 98%) monophyletic group with KS domains from the myxobacterial NRPS-PKS hybrid routes, which have specificities for unusual starter units (Fig. 2, highlighted in gray, and Fig. 3). Three additional internal clusters were related to Cyanobacteria (Fig. 2, internal bars 4, 6, and 7); together, a total of five clusters were assigned to a specific bacterial phylum.
Fig 3.
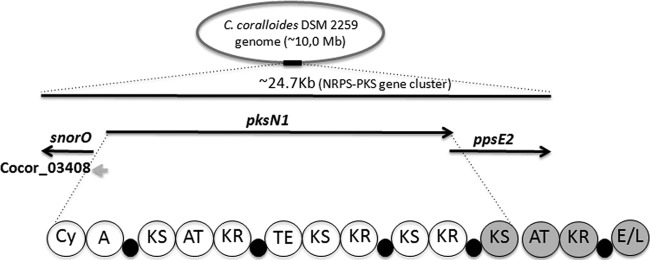
Scheme of the Corallococcus coralloides DSM 2259 orphan NRPS-PKS gene group. The catalytic domains encoded by pksN1 are shown as bubbles. Abbreviations: Cy, cyclization; A, adenylation; KS, ketosynthase; KR, ketoreductase; TE, thioesterase; and E/L, esterase-lipase. The thiolation domains are shown as small black bubbles. In gray, the PKS module split between the putative PksN1 and PpsE2 PKSs is illustrated. Microbial-BLAST searches querying the KS contigs from the new PKS group, consisting only of A. brasiliensis, returned the sequence of the PksN1 KS4 domain (the only one in gray) as the best hit.
Novel PKS cluster.
Our phylogenetic reconstructions also consistently grouped 59 of the recovered KSs from A. brasiliensis (25%) into a monophyletic group (Fig. 2, group colored in pink). This group, consisting only of A. brasiliensis KSs, was maintained in spite of the inclusion of KSs from other known cis- and trans-AT PKS and NRPS-PKS hybrid systems, as well as the FAS-like PKSs involved in the biosynthesis of polyunsaturated fatty acids (PUFA) (45), enediynes (46), and cyanobacterial heterocysts (47) (see Fig. S3 in the supplemental material).
A substantial portion (69%) of the KS contigs that formed this novel PKS cluster showed great similarity to deltaproteobacteria, particularly the fourth KS domain encoded by the gene pksN1 (YP_005369384) from the myxobacteria Corallococcus coralloides DSM 2259 (Fig. 2, internal bar 8; see also Table S2 in the supplemental material) (48). The pksN1 encodes an unusual NRPS-PKS hybrid protein with at least 15 identifiable catalytic domains that could be organized into four active modules (Fig. 3). The fourth KS (KS4) domain of the putative PksN1 megasynthase stands alone at the protein's carboxyl terminus (Fig. 3, KS in gray). Downstream of pksN1, the gene ppsE2 encodes a PKS-bearing protein that contains four catalytic domains and hypothetically creates a module with PksN1 KS4 (Fig. 3, hypothetical module in gray). In silico analysis suggested that the PksN1 NRPS module loads and cyclizes a cysteine residue, leading to a thiazoline-bearing hypothetical metabolite that has yet to be isolated from any Haplosclerida sponge.
FAS-like type I PKS cluster.
Another 6% of the recovered KS contigs were closely related to either the KS domain in the RkpA and WcbR reference PKSs or to the RkpA-orthologous environmental KS sequences previously discovered (23, 25, 43) (Fig. 2, dark blue cluster; see Table S2 in the supplemental material). In fact, eight KS contigs that were more similar to the alphaproteobacterial genomes grouped corroboratively with RkpA-like sequences from Pseudovibrio sp. with a bootstrap support value of 89 (Fig. 2, internal bar 10; see also Table S2).
The capsular polysaccharide (KPS) synthesized by the rkpABCDE operon has been shown to be a determining factor in both symbiotic (49) and pathogenic (50) bacteria-host interactions. The repetitive isolation of rkpA homologues from sponge metagenomes supports the long-standing hypothesis that capsule production may function as a critical factor for sponge-associated bacteria to escape recognition and phagocytosis by host cells (51).
Trans-AT PKS group.
A total of 58 contigs recovered from A. brasiliensis (approximately 24%) grouped with the KS domains from characterized trans-AT type I PKS routes as well as other sponge-derived sequences that had previously been related to these PKS systems (Fig. 2, group marked in red; see Table S2 in the supplemental material). Twenty-four of such contigs (40%) formed robust clusters (bootstrap values above 60) with the KS domains from Burkholderia, including RhiKS4 and TaiKS2; RhiKS4 and TaiKS2 are known to be involved in the production of the anticancer compound rhizoxin (52) and the synthesis of thailandamides (19), respectively (Fig. 2). Interestingly, we previously showed that A. brasiliensis metagenomes are enriched for sequences that are taxonomically assigned to Burkholderiales (29). Most of the taxonomic assignments for the trans-AT type I PKS cluster were assigned to the Firmicutes (44%) and betaproteobacteria (24%) groups (see Table S2). However, even the internal, robust clusters within this group could not be assigned to a specific bacterial taxon.
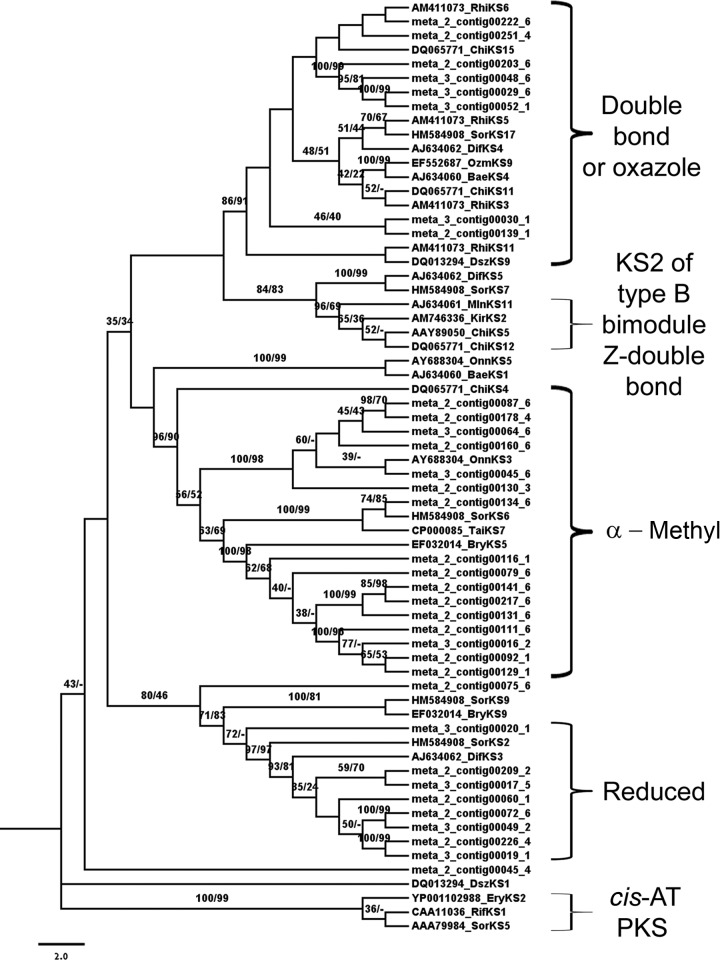
KS domains from the trans-AT PKSs have been shown to group according to their substrate type, independently of being a membership belonging to a specific bacterial taxa (19). Therefore, the contigs forming the trans-AT PKS cluster were realigned with additional reference KSs of known substrate specificity (Fig. 4). The new phylogenies revealed the putative specificity of several contigs for different substrates, including double-bonded, reduced β-methylated and pyran moieties (Fig. 4). Although the characterization of an entire trans-AT PKS route would be necessary for an accurate prediction of the chemistry of these compounds, this is another indication that A. brasiliensis microbiome might produce complex polyketides.
Fig 4.
Detailed phylogenetic reconstitution of trans-AT PKS sequences. NJ (1,000 replicates) and ML (100 replicates) bootstrap values are shown on the branch nodes. Scale bar represents substitutions/site.
NRPS-PKS group.
The remaining 21 KS contigs that were recovered (9%) grouped with KS domains that recognized amino acid substrates in the typical NRPS-PKS hybrid systems (Fig. 2, light-blue cluster). No internal robust clade of this group could be assigned to a particular bacterial phylum.
Finally, a unique KS contig was recovered from approximately 150 Mb of data from the A. brasiliensis shotgun pyrosequencing. This KS-like region clustered with a type II PKS outgroup (Fig. 2, sequence in orange). In agreement with these observations, the sequence of this putative protein returned greater BLAST hits for fatty-acid biosynthesis-related beta-ketoacyl-acyl-carrier-protein synthase (KAS).
Concluding remarks.
The microbiome of the marine sponge Arenosclera brasiliensis harbors a large diversity of type I PKS subtypes. Actinobacteria, Cyanobacteria, Alphaproteobacteria, and Deltaproteobacteria (Burkholderia) were related to subgroups of the PKS subtypes, which implies that these bacteria exchange PKS genes in the sponge holobiont. We also identified a novel type I PKS group containing KS sequences homologous to a KS domain from a putative PKS-NRPS hybrid enzyme in the myxobacterial strain DSM 2259. Therefore, our results enabled the study of a promising biosynthetic gene group from this strain. Our findings contribute to the discovery of new genes for drug production, and they may open new avenues for polyketide production via fermentation processes using culturable strains along with heterologous expression of specific PKS gene cassettes.
Supplementary Material
ACKNOWLEDGMENTS
We thank FAPESP (2010/50190-2 to R.G.S.B. and 2009/11612-1 to A.E.T.-S.), FAPERJ (F.L.T.), CNPq (R.G.S.B. and F.L.T.), and CAPES for financial support.
The authors have no conflicts of interests to declare.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03354-12.
REFERENCES
- 1. Vacelet J, Donadey C. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Marine Biol. Ecol. 30:301–314 [Google Scholar]
- 2. Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, Perez T, Rodrigo A, Schupp PJ, Vacelet J, Webster N, Hentschel U, Taylor MW. 2011. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 6:564–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simister RL, Deines P, Botté ES, Webster NS, Taylor MW. 2012. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 14:517–524 [DOI] [PubMed] [Google Scholar]
- 5. Webster NS, Taylor MW, Behnam F, Lucker S, Rattei T, Whalan S, Horn M, Wagner M. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12:2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitt S, Hentschel U, Taylor MW. 2011. Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687:341–351 [Google Scholar]
- 7. Pawlik JR. 2011. The chemical ecology of sponges on Caribbean reefs: natural products shape natural systems. Bioscience 61:888–898 [Google Scholar]
- 8. Amoutzias GD, Van de Peer Y, Mossialos D. 2008. Evolution and taxonomic distribution of nonribosomal peptide and polyketide synthases. Future Microbiol. 3:361–370 [DOI] [PubMed] [Google Scholar]
- 9. Schmidt EW. 2008. Trading molecules and tracking targets in symbiotic interactions. Nat. Chem. Biol. 4:466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Oliveira JHHL, Nascimento AM, Kossuga MH, Cavalcanti BC, Pessoa CO, Moraes MO, Macedo ML, Ferreira AG, Hajdu E, Pinheiro US, Berlinck RG. 2007. Cytotoxic alkylpiperidine alkaloids from the Brazilian marine sponge Pachychalina alcaloidifera. J. Nat. Prod. 70:538–543 [DOI] [PubMed] [Google Scholar]
- 11. Ang KKH, Holmes MJ, Higa T, Hamann MT, Kara UA. 2000. In vivo antimalarial activity of the beta-carboline alkaloid manzamine A. Antimicrob. Agents Chemother. 44:1645–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hill RT, Peraud O, Hamann MT, Kamke J. November 2005 Manzamine-producing actinomycetes. US patent 20050244938A1
- 13. Fontana A. 2006. Biogenetic proposals and biosynthetic studies on secondary metabolites of opisthobranch molluscs, p 304–332 In Cimino G, Gavagnin M. (ed), Molluscs: from chemo-ecological study to biotechnological application. Springer-Verlag, Berlin, Germany: [DOI] [PubMed] [Google Scholar]
- 14. Almeida AM, Hajdu E, Berlinck RGS. 1997. Alcalóides alquilpiridínicos de esponjas marinhas. Quím. Nova 20:170–185 [Google Scholar]
- 15. Andersen RJ, van Soest RWM, Kong F. 1996. 3-Alkylpyridine alkaloids isolated from marine sponges in the order Haposclerida, p 301–355 In Pelletier SW. (ed), Alkaloids: chemical and biological perspectives, vol 10 Pergamon, London, United Kingdom. [Google Scholar]
- 16. Fischbach MA, Walsh CT. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468–3496 [DOI] [PubMed] [Google Scholar]
- 17. Piel J. 2010. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 27:996–1047 [DOI] [PubMed] [Google Scholar]
- 18. Jenke-Kodama H, Borner T, Dittmann E. 2006. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLoS Comput. Biol. 2:1210–1218 doi:10.1371/journal.pcbi.0020132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. 2008. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26:225–233 [DOI] [PubMed] [Google Scholar]
- 20. Schirmer A, Gadkari R, Reeves CD, Ibrahim F, DeLong EF, Hutchinson CR. 2005. Metagenomic analysis reveals diverse polyketide synthase gene clusters in microorganisms associated with the marine sponge Discodermia dissoluta. Appl. Environ. Microbiol. 71:4840–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim TK, Fuerst JA. 2006. Diversity of polyketide synthase genes from bacteria associated with the marine sponge Pseudoceratina clavata: culture-dependent and culture-independent approaches. Environ. Microbiol. 8:1460–1470 [DOI] [PubMed] [Google Scholar]
- 22. Kennedy J, Baker P, Piper C, Cotter PD, Walsh M, Mooij MJ, Bourke MB, Rea MC, O'Connor PM, Ross RP, Hill C, O'Gara F, Marchesi JR, Dobson AD. 2008. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish waters. Mar. Biotechnol. 11:384–396 [DOI] [PubMed] [Google Scholar]
- 23. Fieseler L, Hentschel U, Grozdanov L, Schirmer A, Wen G, Platzer M, Hrvatin S, Butzke D, Zimmermann K, Piel J. 2007. Widespread occurrence and genomic context of unusually small polyketide synthase genes in microbial consortia associated with marine sponges. Appl. Environ. Microbiol. 73:2144–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. 2009. Polyketide assembly lines of uncultivated sponge symbionts from structure-based gene targeting. Nat. Chem. Biol. 5:494–501 [DOI] [PubMed] [Google Scholar]
- 25. O'Halloran JA, Barbosa TM, Morrissey JP, Kennedy J, O'Gara F, Dobson AD. 2011. Diversity and antimicrobial activity of Pseudovibrio spp. from Irish marine sponges. J. Appl. Microbiol. 110:1495–1508 [DOI] [PubMed] [Google Scholar]
- 26. Hochmuth T, Niederkruger H, Gernert C, Siegl A, Taudien S, Platzer M, Crews P, Hentschel U, Piel J. 2010. Linking chemical and microbial diversity in marine sponges: possible role for poribacteria as producers of methyl-branched fatty acids. Chembiochem 11:2572–2578 [DOI] [PubMed] [Google Scholar]
- 27. Torres YR, Berlinck RGS, Magalhaes A, Schefer AB, Ferreira AG, Hajdu E, Muricy G. 2000. Arenosclerins A–C and haliclonacyclamine E, new tetracyclic alkaloids from a Brazilian endemic haplosclerid sponge Arenosclera brasiliensis. J. Nat. Prod. 63:1098–1105 [DOI] [PubMed] [Google Scholar]
- 28. Torres YR, Berlinck RGS, Nascimento GGF, Fortier SC, Pessoa C, de Moraes MO. 2002. Antibacterial activity against resistant bacteria and cytotoxicity of four alkaloid toxins isolated from the marine sponge Arenosclera brasiliensis. Toxicon 40:885–891 [DOI] [PubMed] [Google Scholar]
- 29. Trindade-Silva AE, Rua C, Silva GG, Dutilh BE, Moreira AP, Edwards RA, Hajdu E, Lobo-Hajdu G, Vasconcelos AT, Berlinck RG, Thompson FL. 2012. Taxonomic and functional microbial signatures of the endemic marine sponge Arenosclera brasiliensis. PLoS One 7:e39905 doi:10.1371/journal.pone.0039905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Piel J, Hui DQ, Fusetani N, Matsunaga S. 2004. Targeting modular polyketide synthases with iteratively acting acyltransferases from metagenomes of uncultured bacterial consortia. Environ. Microbiol. 6:921–927 [DOI] [PubMed] [Google Scholar]
- 31. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40:D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics Rev. 14:755–763 [DOI] [PubMed] [Google Scholar]
- 35. Olson S. 2002. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Brief. Bioinformatics 3:87–91 [DOI] [PubMed] [Google Scholar]
- 36. Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kroken S, Glass L, Taylor JW, Yoder OC, Turgeon BG. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 100:15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Hurlbert J, White SW, Rock CO. 2006. Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae. J. Biol. Chem. 281:17390–17399 [DOI] [PubMed] [Google Scholar]
- 39. Qiu X, Choudhry AE, Janson CA, Grooms M, Daines RA, Lonsdale JT, Khandekar SS. 2005. Crystal structure and substrate specificity of the β-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Protein Sci. 14:2087–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beyer S, Kunze B, Silakowski B, Müller R. 1999. Metabolic diversity in myxobacteria: identification of the myxalamid and the stigmatellin biosynthetic gene cluster of Stigmatella aurantiaca Sg a15 and a combined polyketide-(poly)peptide gene cluster from the epothilone producing strain Sorangium cellulosum So ce90. Biochim. Biophys. Acta 1445:185–195 [DOI] [PubMed] [Google Scholar]
- 41. Piel J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. U. S. A. 99:14002–14007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siegl A, Hentschel U. 2010. PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ. Microbiol. 2:507–513 [DOI] [PubMed] [Google Scholar]
- 43. Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C, Dandekar T, Hentschel U. 2011. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 5:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26:1362–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Metz JG. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293 [DOI] [PubMed] [Google Scholar]
- 46. Ahlert J. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173–1176 [DOI] [PubMed] [Google Scholar]
- 47. Campbell EL, Cohen MF, Meeks JC. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 167:251–258 [DOI] [PubMed] [Google Scholar]
- 48. Huntley S, Zhang Y, Treuner-Lange A, Kneip S, Sensen CW, Søgaard-Andersen L. 2012. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 194:3012–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parada M, Vinardell JM, Ollero FJ, Hidalgo A, Gutierrez R, Buendía-Clavería AM, Lei W, Margaret I, López-Baena FJ, Gil-Serrano AM, Rodríguez-Carvajal MA, Moreno J, Ruiz-Sainz JE. 2006. Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajan. Mol. Plant Microbe Interact. 19:43–52 [DOI] [PubMed] [Google Scholar]
- 50. DeShazer D, Waag DM, Fritz DL, Woods DE. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269 [DOI] [PubMed] [Google Scholar]
- 51. Wilkinson CR, Garrone R, Vacelet J. 1984. Marine sponges discriminate between food bacteria and bacterial symbionts: electron microscope radioautography and in situ evidence. Proc. R. Soc. B Biol. Sci. 220:519–528 [Google Scholar]
- 52. Partida-Martinez LP, Hertweck C. 2007. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem 8:41–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.