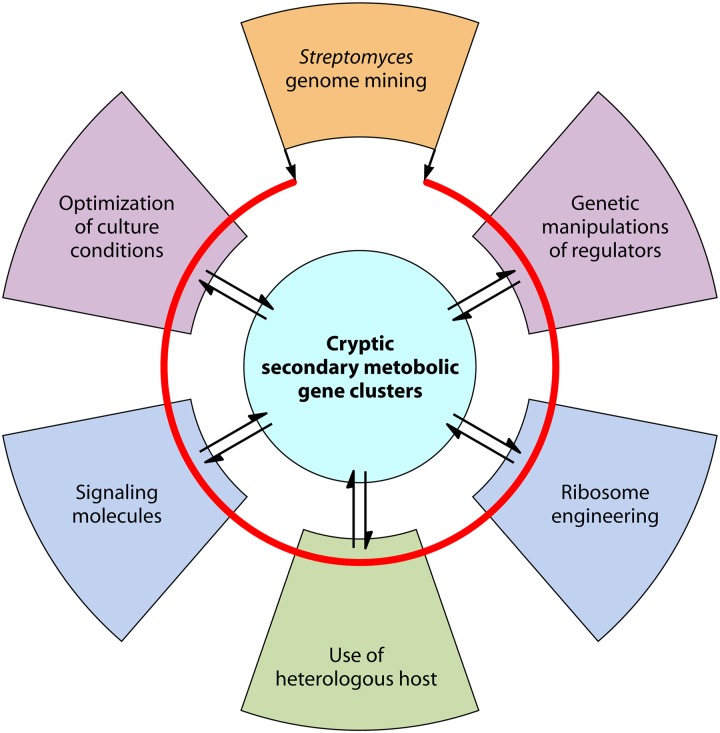
Abstract
SUMMARY
Streptomycetes are the most abundant source of antibiotics. Typically, each species produces several antibiotics, with the profile being species specific. Streptomyces coelicolor, the model species, produces at least five different antibiotics. We review the regulation of antibiotic biosynthesis in S. coelicolor and other, nonmodel streptomycetes in the light of recent studies. The biosynthesis of each antibiotic is specified by a large gene cluster, usually including regulatory genes (cluster-situated regulators [CSRs]). These are the main point of connection with a plethora of generally conserved regulatory systems that monitor the organism's physiology, developmental state, population density, and environment to determine the onset and level of production of each antibiotic. Some CSRs may also be sensitive to the levels of different kinds of ligands, including products of the pathway itself, products of other antibiotic pathways in the same organism, and specialized regulatory small molecules such as gamma-butyrolactones. These interactions can result in self-reinforcing feed-forward circuitry and complex cross talk between pathways. The physiological signals and regulatory mechanisms may be of practical importance for the activation of the many cryptic secondary metabolic gene cluster pathways revealed by recent sequencing of numerous Streptomyces genomes.
INTRODUCTION
Bacteria of the genus Streptomyces are a particularly abundant source of antibiotics and related compounds, providing more than half of medically important antimicrobial and antitumor agents. Early genetic mapping in the model organism Streptomyces coelicolor A3(2) provided the first evidence that the genes for biosynthesis of any particular antibiotic are clustered on the chromosome (1–3) or plasmids (4). Subsequently, molecular analyses revealed such clusters to be large (typically tens of kilobases) and usually to include several operons (5–7). This paradigm has held over the last 2 decades, during which hundreds more such clusters have been characterized. The sequenced gene sets provide an opportunity to study the regulation of antibiotic biosynthesis at the molecular level, illuminating the complex developmental interplay of antibiotic production with morphological differentiation in these mycelial, sporulating bacteria. Such studies may suggest ways of increasing production levels, both at the early stages of characterizing new products and at the level of large-scale industrial production. They may also provide routes to the activation of “silent” gene sets that are revealed by genome-level DNA sequencing.
Mining of genome sequences has revealed at least 29 clusters of likely biosynthetic genes for secondary metabolites in the S. coelicolor A3(2) model organism and 37 in the industrial organism Streptomyces avermitilis. Sequencing of other Streptomyces genomes showed that this was typical and that the majority of clusters are species specific among the genomes analyzed (8). Some of the conserved clusters determine the production of metabolites with roles in the physiology or development of the host, including metal-binding siderophores, spore pigments, and volatile odor compounds (notably geosmin, which is responsible for the earthy smell of streptomycetes). We have generally confined this review to antibiotics—secondary metabolites whose main role appears to be to interfere with neighboring organisms (though we note that roles of many antibiotics in signaling have been proposed) (9–11).
Antibiotic production and morphological differentiation are generally activated when starvation or environmental changes bring about the end of rapid vegetative growth. Under these circumstances, a transient arrest of growth typically ensues, accompanied by a complex series of changes in global gene expression. Antibiotic production and morphological differentiation may then take place (12, 13). Antibiotic production at the industrial level is highly dependent on the fermentation conditions, with the need for a balance between, on one hand, providing nutrients for growth and as precursors for antibiotics and, on the other hand, the repressive effects of some of the most efficient sources of carbon, nitrogen, and phosphate. Numerous metals, such as zinc, iron, and manganese, are essential for bacterial growth, and some of them may also affect antibiotic production (14, 15). Apart from nutrient effects, the pH and dissolved oxygen level are also important for antibiotic production (16, 17). Although the optimization of industrial fermentations is highly specific for particular products and their producing organism, understanding the underlying responses of diverse pathways requires the study of a model organism. Our approach here has therefore been first to provide an up-to-date overview of the regulation of antibiotic production in S. coelicolor A3(2) and then to consider how the concepts developed in the model system are extended, reinforced, or challenged by information coming from biosynthetic gene sets in other streptomycetes. This article builds on three excellent broadly based reviews on the regulation of antibiotic production in Streptomyces (18–20). Other wide-ranging reviews of the topic have also appeared recently (21, 22).
CONTROL OF FIVE EXTENSIVELY STUDIED S. COELICOLOR ANTIBIOTIC PATHWAYS
Streptomyces coelicolor was adopted early as a model streptomycete because of its amenability to genetic analysis, and its status as a model was reinforced by its production of two pigmented antibiotics (actinorhodin [ACT] and undecylprodigiosins [REDs]), as well as a structurally incompletely characterized polyketide (“cryptic polyketide” [CPK]), a calcium-dependent ionophore antibiotic (CDA), and an unusual cyclopentanone antibiotic (methylenomycin [MM]) (Fig. 1). The main architect of S. coelicolor genetic studies, D. A. Hopwood, has written accounts of the organism with much valuable early history (23–25). For the present purposes, it is sufficient to point out that S. coelicolor is a complex mycelial prokaryote that reproduces by the formation of sporulating aerial hyphae. Aerial growth is fueled in part by nutrients from autolysis of the vegetative or substrate mycelium. On agar media, most antibiotic production also takes place at this stage, perhaps providing some protection for the nutrients being released. Under submerged liquid culture conditions (such as are used in industrial antibiotic production fermentations), such morphological differentiation does not usually take place. Nevertheless, antibiotic production is connected to the S. coelicolor life cycle, and this connection has preoccupied many researchers over several decades. With the application of genome-based technologies to Streptomyces biology, there are now signs that the disparate strands of different research programs are beginning to come together.
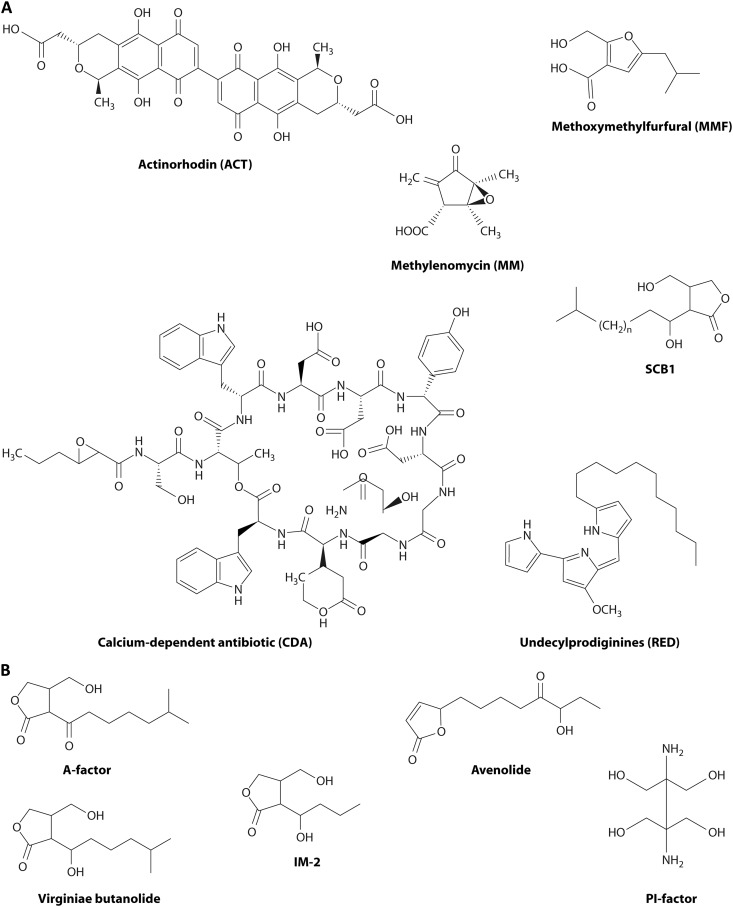
Fig 1.
Diverse antibiotics and autoregulator molecules produced by Streptomyces coelicolor A3(2) and some other streptomycetes. (A) The compounds from S. coelicolor. Actinorhodin (ACT) is a red/blue pH-indicating benzoisochromanequinone made by a type II polyketide synthase-based pathway involving a 22-gene cluster (291). Undecylprodiginines (REDs) are red hydrophobic tripyrroles made by a fatty acid synthase-like pathway involving a 22-gene cluster (292). Methylenomycin A (MM) is an epoxycyclopentenone made by an unusual pathway encoded by 11 genes located on the linear plasmid SCP1 (57). The calcium-dependent antibiotic (CDA) is a lipopeptide made by a route involving nonribosomal peptide synthases and specified by a 48-gene cluster (293). The autoregulator SCB1 (240) is one of several gamma-butyrolactone congeners whose synthesis involves three genes linked to the 19-gene cluster for biosynthesis of a structurally incompletely characterized yellow antibiotic (S. coelicolor polyketide [CPK]) (41, 42, 294). Methylenomycin furans (MMFs) are autoregulators that control MM biosynthesis, and they are made by a pathway resembling that of gamma-butyrolactones that involves three biosynthetic and two regulatory genes next to the MM biosynthetic cluster (58). (B) Autoregulatory molecules from other streptomycetes. A-factor regulates streptomycin biosynthesis in S. griseus (295). Virginiae butanolides (VBs) induce the coordinated production of virginiamycin M and virginiamycin S, synergistically acting but biosynthetically distinct antibiotics in S. virginiae (235). IM-2 controls the production of showdomycin and minimycin in S. lavendulae (237). PI factor enhances pimaricin production in S. natalensis (239). Avenolide is required at nanomolar concentrations for the onset of avermectin biosynthesis in S. avermitilis (238) (avenolide homologs are also found in S. fradiae, Streptomyces ghanaensis, and S. griseoauranticus).
The first molecular analyses of biosynthetic gene clusters showed that they usually contain regulatory genes, often having major effects on the levels of production of the cognate antibiotic. These came to be termed “pathway-specific” regulators, in contrast with “global,” “higher-level,” or “pleiotropic” regulators. One of the first applications of microarray analysis to S. coelicolor revealed that some pathway-specific regulators also had wide-ranging effects on global transcription patterns, leading to the suggestion that the term “cluster-situated regulator” (CSR) would be a more accurate and objective description (26). The value of this term has been reinforced by recent evidence that some CSRs directly control the expression of genes in other clusters (see below). Of course, some of the wide-ranging effects may be indirect, ranging from the consequences of draining pools of precursors to physicochemical properties of the end products such as redox activity (27).
The first parts of this section focus on the five antibiotic biosynthetic pathways of S. coelicolor that have provided paradigms both for pathway-specific regulation and for its dependence on global physiological influences. We particularly emphasize recent work on the complexity of regulation of the CSRs, the roles and diversity of autoregulators, the discovery that end products and late intermediates are sometimes ligands of CSRs, and molecular evidence of regulatory cross talk between apparently unrelated pathways. Global regulators controlling more than one of these pathways are dealt with in the subsequent subsections. In some cases we refer to S. coelicolor genes by their SCO numbers (for further information, see http://strepdb.streptomyces.org.uk).
Activation of Actinorhodin Biosynthesis by Multiple Signal Inputs
Actinorhodin (ACT), a polyketide-derived benzoisochromanequinone (Fig. 1), is a weak antibiotic that is responsible for the pH-sensitive blue/red color from which S. coelicolor gets its name. Its biosynthesis is determined by five transcription units in the act gene cluster, four of which contain more than one gene. These five transcription units are completely dependent on the ActII-ORF4 protein, which binds to sequences in the target promoters via its N-terminal winged helix-turn-helix (HTH) domain and activates transcription through a C-terminal activation domain. Its target motif is repeats of the sequence TCGA at 11-bp intervals, which appear in five promoter regions in the act cluster (28, 29). ActII-ORF4 is a founder member of a protein family called Streptomyces antibiotic regulatory proteins (SARPs) (29). To avoid confusion, we emphasize here that the term SARP does not apply to the many other kinds of cluster-situated regulators of antibiotic biosynthesis, most of which have paralogues in diverse organisms and other physiological contexts. SARPs are a specific family of paralogous proteins that show a high specificity for antibiotic production in actinomycetes. The frequency and properties of SARPs are further surveyed later in this review.
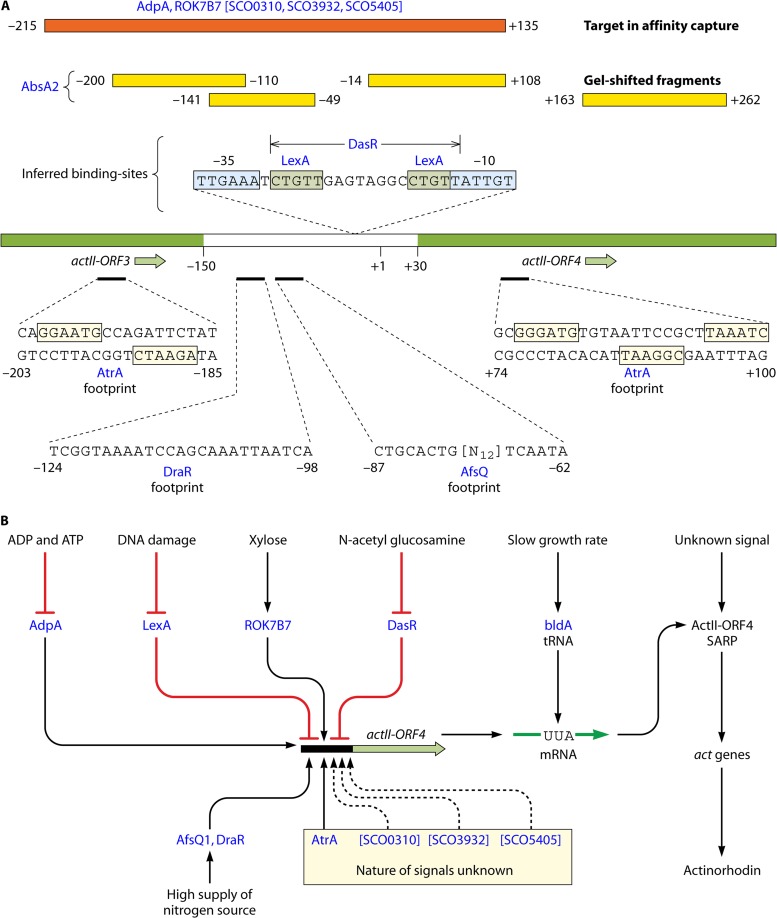
The ActII-ORF4 determinant is embedded in the act cluster. Almost all information from global cellular physiology that affects the timing and level of ACT production is transmitted via ActII-ORF4 and influences either actII-ORF4 transcription or translation or (perhaps) the properties of the protein itself. The promoter region of actII-ORF4 is a direct target for at least eight known regulatory proteins (Fig. 2): AdpA (a pleiotropic regulator of antibiotic production and development) (30), LexA (a global regulator of the DNA damage response) (28), AbsA2 (a global repressor of antibiotic synthesis [see below]) (31, 32), DasR (mediating the global response to N-acetylglucosamine [see below]) (33), DraR and AfsQ1 (activators responding to nitrogen excess) (34, 35), AtrA (a transcriptional activator, which also binds to targets associated with metabolism of acetyl coenzyme A [acetyl-CoA], an ACT precursor) (32, 36), and the xylose operon repressor ROK7B7 (SCO6008) (37) (Fig. 2). In addition, there are indications of binding of the nitrogen regulator GlnR (34) and proteins corresponding to SCO0310, -3932, and -5405 (38). There can be few documented cases of comparably complex regulation of a bacterial promoter, and it will be a challenge for the future to unravel the large number of possible interactions involving regulators binding at separate or overlapping sites. Furthermore, the presence of a particularly rare codon (UUA, one of six leucine codons) in actII-ORF4 mRNA makes its translation dependent on a developmentally significant tRNA encoded by the bldA gene, which is further discussed in a later section (39). A second regulatory gene in the act cluster (actR) encodes a TetR-like protein that represses the adjacent actA operon, encoding the ACT export system (actA also contains a TTA codon) (39). Repression of export is relieved by interaction of ActR with late intermediates in ACT biosynthesis (40). This may ensure that the export system is functional before the potentially toxic end product begins to accumulate in the cytoplasm.
Fig 2.
Complexity of the promoter region of actII-ORF4. (A) Binding of regulatory proteins. The central line represents the actII-ORF3 to actII-ORF4 region, with the noncoding intergenic region given in white and coding sequences in green. Numbers refer to the transcription start site (+1) defined by Gramajo et al. (296). Binding to the diverse regulators (red lettering) was defined by different routes: footprinting results are given below the promoter, while gel-shifted fragments are indicated above as yellow boxes. Brackets indicate proteins defined only by affinity capture to the orange fragment shown. Also shown are binding sites inferred by coupling of experimental evidence of interaction with the promoter region and the presence of matches to known consensus binding sites. References are given in the text. (B) General nature of signal inputs influencing the expression of actII-ORF4. See the text for further details and references.
Regulation of the “Cryptic Polyketide” Gene Cluster
The cpk cluster directs production of a polyketide-derived antibiotic (“cryptic polyketide” [CPK]) and a (presumably related) yellow pigment (yCPK) whose structures are still undetermined (41, 42). Although the difficulty of characterizing CPK has impeded analysis of the regulation of the cluster, it clearly shows significant differences from act regulation: in particular, it involves a gamma-butyrolactone (GBL) signaling molecule, SCB1, and/or its congeners (SCBs) (Fig. 1) (43). Gamma-butyrolactone autoregulators are membrane diffusible and accumulate in cultures until, at above a certain concentration, they bind to a cytoplasmic protein and cause it to dissociate from, and thereby derepress, promoters of target genes (44, 45) (see below). It has been suggested that at least some Streptomyces autoregulators may coordinate physiological changes across the mycelium of a colony (18).
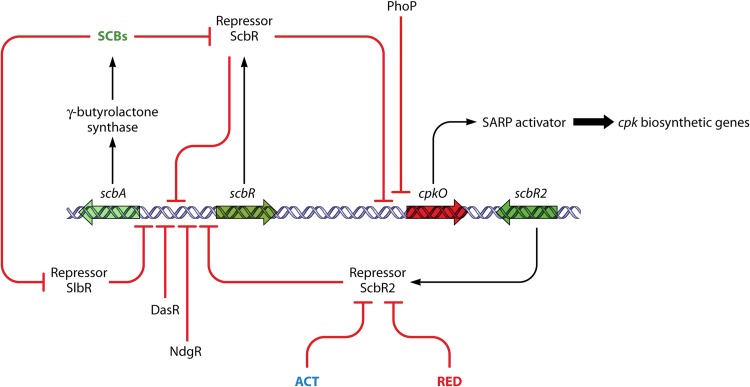
The key SCB biosynthetic gene scbA and a divergent regulatory gene scbR are located at one end of the cpk cluster. ScbR has a TetR-like N-terminal helix-turn-helix domain and belongs to a subfamily of such proteins whose founder member, ArpA, senses the gamma-butyrolactone A-factor in Streptomyces griseus (see below). ScbR binds to operator sites in the bidirectional scbA/scbR promoter and in the promoter for cpkO, a CSR gene essential for CPK biosynthesis (43) (Fig. 3). CpkO has an N-terminal SARP domain and an additional C-terminal domain of unknown function. Under suitable culture conditions, and by an undetermined mechanism, SCBs accumulate to a concentration high enough to bind to ScbR, causing ScbR to dissociate from the bidirectional scbA/scbR promoter. It was suggested that the resulting increase in ScbA and ScbR production has two consequences: a heterodimer of the two proteins is thought to form and then bind to a different site upstream of scbA to activate scbA transcription further, and SCB levels burgeon, sequestering all free ScbR protein and releasing cpkO from repression (41) (Fig. 3). An alternative and perhaps simpler interpretation of the same data is possible, in which the only role of ScbA is to catalyze SCB biosynthesis and the regulatory effects are all mediated by the concentration-dependent interplay of SCBs with ScbR (20). Mathematical modeling of the scbAR/cpk system has shown that its autoamplifying nature may amount to a developmental commitment to CPK production, by acting as a bistable switch (46, 47). Recently, it was reported that a mutation in scbR leading to the amino acid change R120S had occurred independently in two lines of S. coelicolor, one of them being the fairly frequently used strain M600 (48). In some tests, but not others, the mutation appeared to affect the properties of ScbR and the details of timing and levels of expression of cpkO, scbA, and a regulatory gene for RED antibiotic biosynthesis (48). The phenotypic changes may be made difficult to interpret and reproduce if the mutation influences the bistable switch.
Fig 3.
Regulation of CPK biosynthesis involves a gamma-butyrolactone and interplay with other biosynthetic pathways. Genes associated with the cpk cluster are indicated by large open arrows. Regulatory interactions are indicated by bold arrows (activation steps) or bold lines ending with a bar (repressing or inhibitory steps). Small-molecule ligands are indicated in colored letters, with ACT and RED being the products of other pathways encoded by distant gene clusters. For further explanation and references, see the text.
The initial discovery of SCBs was made possible by their stimulatory effects on pigmented antibiotic production when added exogenously to S. coelicolor cultures, though, paradoxically, eliminating endogenous SCBs by scbA deletion enhanced ACT and RED production (49). This enhanced production is mediated at least partially at the level of regulation of the relevant biosynthetic genes and their CSR regulatory genes. However, since deletion of scbA also caused increased expression of afsS (50), a global activator of antibiotic biosynthesis discussed below, it is possible that the effects on ACT and RED production may be indirect, via AfsS. A further role in cross talk between the CPK, ACT, and RED pathways is played by ScbR2, the gene for which is located close to the SARP gene cpkO. ScbR2 directly represses cpkO and has 35%, near end-to-end, identity to ScbR, but it does not bind SCBs (51) (ScbR2 is therefore considered to be a “pseudo”-gamma-butyrolactone receptor, since there are no other gamma-butyrolactones in S. coelicolor). However, binding of ScbR2 to the cpkO promoter was unexpectedly relieved by adding ACT, leading to the derepression of cpkO and activation of CPK biosynthetic genes (51). Similar results were obtained by adding RED, but the poor solubility of RED and the possibility that it might interact with target DNA rather than the ScbR2 protein hamper unambiguous interpretation of the RED effect (52). Although RED and ACT are very different molecules, ScbR2 structural predictions revealed that its ligand-binding pocket had features reminiscent of QacR and TtgR, other members of the TetR superfamily that possess large helix-rich cavities capable of binding structurally diverse drugs (51, 53, 54). ScbR2 also represses the biosynthesis of SCBs by directly binding to the promoter region of scbA (55). Thus, part of the complex cluster-situated regulatory system for CPK biosynthesis is implicated in cross talk with other antibiotic biosynthetic pathways, and CPK biosynthesis is subject to induction by at least two, and possibly three, entirely different secondary metabolites (SCBs, ACT, and RED) through the action of two ArpA-like repressors.
Other regulators also affect CPK biosynthesis. Thus, cpkO is directly repressed by the phosphate response regulator (RR) PhoP (56), and recent work has shown that another global regulator, AfsQ1, directly activates the divergent promoter between the biosynthetic genes cpkO and cpkD (34).
Cascade Regulation of Methylenomycin Biosynthesis
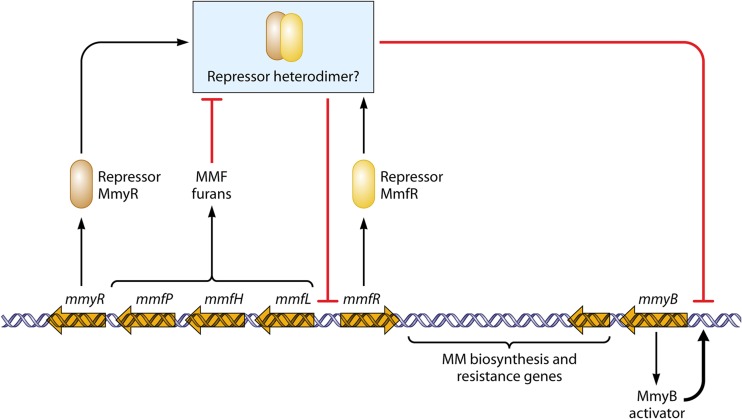
The methylenomycin (MM) biosynthetic pathway of S. coelicolor is encoded by the mmy genes on a large linear plasmid, SCP1, which can remain autonomous or integrate into the chromosome (4). Like the cpk cluster, the mmy genes are regulated by a cascade involving the synthesis of a small autoregulator molecule (57) (Fig. 4). In this case, however, the autoregulator is not a gamma-butyrolactone; instead, it is a mixture of furans (methylenomycin furans [MMFs] such as that shown in Fig. 1), made by a pathway closely similar to that of GBLs (58). The regulatory cascade is complicated by the presence of genes for two ArpA-like proteins (MmyR and MmfR). The three furan biosynthetic genes (mmfL, mmfH, and mmfP) and the two regulatory genes are directly adjacent to the mmy cluster, in the form of divergent transcription units (mmfLHP-mmyR and mmfR). The mode of action of MmfR and MmyR has not been studied in vitro, but genetic evidence suggested that a putative MmyR/MmfR complex represses two targets: the bidirectional promoter between the mmfLHP-mmyR operon and mmfR, to regulate MMF biosynthesis, and the promoter of mmyB, another regulatory gene in the mmy cluster, to regulate the MM biosynthetic genes (57). It is thought that when MMFs reach a concentration that can release repression of the bidirectional promoter, furan biosynthesis accelerates to generate a concentration that relieves MmyR/MmfR-mediated repression of mmyB. MmyB can then activate the MM biosynthetic genes. Early observations that MM production was induced by medium acidification raise the question of whether MmyR or MmfR is itself pH sensitive (59).
Fig 4.
Regulation of methylenomycin biosynthesis, a cascade involving furan autoregulators. Large open arrows indicate genes associated with the gene cluster for MM biosynthesis. Regulatory interactions are indicated by bold arrows (activation steps) or bold lines ending with a bar (repressing or inhibitory steps). See the text for further details and references.
MmyB consists of an N-terminal DNA-binding domain of the Xre type and a C-terminal PAS-like domain such as is involved in many sensory functions (60). The crystal structure of an MmyB-like protein from a thermophilic Gram-negative organism (purified from an Escherichia coli expression system) revealed that its PAS domain was complexed with a saturated fatty acid, prompting speculation that binding of a fatty acid ligand might be required to confer transcription factor activity on MmyB (61). MmyB paralogues are widespread among actinomycetes and particularly among streptomycetes. For example, 13 proteins with near end-to-end similarity to MmyB (expected values of lower than 2e−7) are encoded in the S. coelicolor chromosome (61). About half of them are located very close to genes with homologues in secondary metabolism, and the others are next to diverging genes for medium- or short-chain alcohol dehydrogenases. Only one of the paralogues, SCO4944, is widely conserved in other streptomycetes, and in S. griseus this gene is just one gene away from the biosynthetic gene for A-factor and is regulated by A-factor (61). Thus, MmyB-like proteins may be closely associated with secondary metabolism and fatty acid metabolism.
Like for ACT, MM production is dependent on the translation of regulatory UUA codons, in this case in the mRNAs of mmyB and mmfL. As in the act cluster, there is also a specific regulatory system for MM export/self-resistance: an MM export gene (mmr) is negatively regulated by an adjacent gene, mmyJ, encoding a member of the ArsR family of repressors, and MM or a late biosynthetic intermediate may release this repression, perhaps acting as an MmyJ ligand (62, 63).
Regulation of the Calcium-Dependent Antibiotic Gene Cluster
Biosynthesis of the lipopeptide calcium-dependent antibiotic (CDA) (Fig. 1) involves huge modular nonribosomal peptide synthetases that generate peptide antibiotics, a class that, like polyketides, has received much attention from natural products scientists. The regulation of CDA biosynthesis has not been extensively studied, but, by analogy with other gene clusters, it is likely that the SARP CSR encoded by cdaR activates the cda biosynthetic genes. Several other regulatory genes are associated with the cluster, but the only ones to have been studied are the absA1/absA2 genes, which encode a classical two-component regulatory system of the type shown in Fig. 5. AbsA1 is a membrane-located sensor histidine protein kinase (64), the target of which is AbsA2, a response regulator whose phosphorylation increases its binding efficiency to, and repression of, the promoter of cdaR (31, 32, 65). Probably AbsA1 responds to an unknown signal by changing the ratio of its kinase/phosphatase activity in favor of the phosphatase, with the consequent dephosphorylation of phosphorylated AbsA2 (AbsA2∼P) leading to derepression of cdaR. Remarkably, AbsA2∼P also binds to and represses the promoters of pathway-specific regulatory genes in some other clusters (actII-ORF4 and redZ, but not cpkO) (31, 32, 65). This is a further example of cross talk between pathways to add to the case of ScbR2 (50, 51) (see above). At least one global regulator, AfsQ1, directly activates the cdaR promoter (34).
Fig 5.
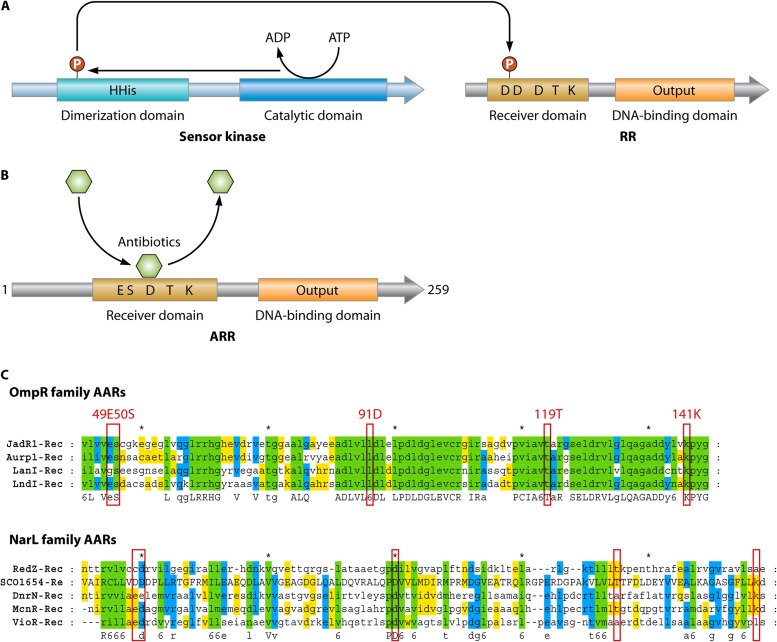
RedZ and other variations on the theme of two-component regulators associated with antibiotic production. (A) The typical two-component system containing a sensor histidine kinase and a cognate response regulator (RR), usually encoded by a pair of adjacent genes. ATP or GTP is used for autophosphorylation by the histidine kinase, and the phosphoryl group is transferred to the RR, which then controls the transcription of target genes. (B) Atypical response regulators (ARRs) such as RedZ (shown here) do not contain all the conserved amino acid residues important for phosphorylation of the receiver domain. Some ARRs can be activated by binding to the end product or late biosynthetic intermediates of secondary metabolites, such as antibiotics (see the text). (C) Alignments of ARRs from Streptomyces. Top, OmpR family: JadR1 (jadomycin, Streptomyces venezuelae [69]), Aur1P (auricin, Streptomyces aureofaciens [297]), LanI (landomycins, Streptomyces cyanogenus [298]), and LndI (landomycin E, Streptomyces globisporus [299]). Bottom, NarI family: RedZ (undecylprodigiosin, S. coelicolor [67, 68]), DnrN (daunorubicin, Streptomyces peucetius [209]), NcnR (naphthocyclinone, Streptomyces arenae [300]), VioR (tuberactinomycin, Streptomyces vinaceus [301]), and SCO1654 (protein of S. coelicolor with unknown function). The red boxes indicate residues corresponding to the conserved residues of conventional response regulators.
A Minicascade Regulating Undecylprodigiosin Biosynthesis
The pyrrole-based undecylprodiginines (REDs) (Fig. 1) are made by a mixed pathway involving a fatty acid synthase-like part and a specialized nonmodular form of nonribosomal peptide synthase (66). The red pathway genes are regulated via a minicascade of two CSRs, with RedZ activating the expression of redD, the direct activator gene for the biosynthetic genes (67). RedZ is an aberrant orphan response regulator that differs from the conventional response regulator component of two-component systems in two respects: its gene is not located next to a cognate histidine protein kinase gene, and its structure, though readily modeled onto known response regulator structures of the NarL-like class, does not have a complete set of conserved residues needed for phosphorylation (Fig. 5). Regulation of redZ has some features in common with that of actII-ORF4: both are direct targets for repression by AbsA2∼P (31, 32), binding of DasR and AfsQ1 to the redZ promoter suggests that the promoter may respond to GlcNAc (33) and a glutamate-related signal (34), and redZ mRNA contains a bldA-dependent UUA codon, which appears to be the reason for the bldA dependence of RED production (67, 68).
In vitro binding of RedZ to the redD promoter is inhibited by the addition of RED antibiotic, implying that a feedback loop may modulate RED biosynthesis (69). RedD, like ActII-ORF4, is a SARP. It is presumed that RedD activates red biosynthetic genes directly, and a putative target site for RedD (repeats of the tetramer TCAG at 11-bp intervals) has been identified upstream of the large red operon that starts with redP (28). Surprisingly, this motif was also found upstream of some genes unconnected with RED biosynthesis (28).
Phosphate Regulation of Antibiotic Production in S. coelicolor
Under laboratory conditions, phosphate limitation of growing cultures activates phosphate scavenging and induces the growth transition that precedes stationary phase and secondary metabolism (70), (71). The two-component system PhoR-PhoP (which is widely distributed across prokaryotes) is the major signal transduction system for phosphate control in S. coelicolor and affects ACT and RED production by (directly or indirectly) influencing the transcription of act and red genes (72, 73). Similar effects on antibiotic production in other streptomycetes, including S. griseus (candicidin) (74, 75), Streptomyces natalensis (pimaricin) (76), and Streptomyces rimosus (oxytetracycline) (75, 77), imply that this role of the PhoR-PhoP system is widespread. A recent extensive survey of the PhoP regulon, using microarray analysis of chromatin immunoprecipitated with anti-PhoP antiserum, concluded that, in addition to activating pathways for phosphate scavenging and for cell wall polymer biosynthesis, PhoP mediates the transient shutdown of central metabolic pathways, some secondary metabolic pathways, and repressors of morphological differentiation (56) (Fig. 6).
Fig 6.
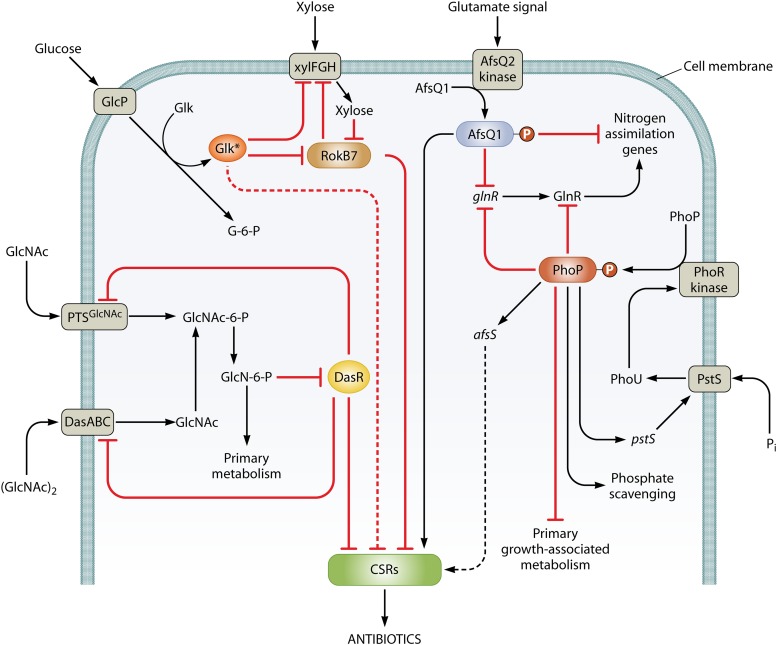
Nutrient-sensing regulators of antibiotic production in S. coelicolor and their cross talk. The diagram summarizes routes by which the availability of sources of carbon, nitrogen, and phosphate influence the expression of the cluster-situated regulators (CSRs) that activate pathways leading to antibiotic biosynthesis. Nutrient availability is sensed by membrane-located sensor kinases or through the transport of nutrients, leading to activation of global regulators (circled). The global regulators control both central metabolic genes and CSR genes, either directly (solid lines) or through unknown routes (dotted lines). Arrows indicate activation, and bars indicate repression. For further information and references, see the text.
PhoR is a membrane-bound sensor kinase that senses phosphate deprivation and then phosphorylates PhoP, its target response regulator, which binds to specific sequences in target promoters (Streptomyces PHO boxes, which are degenerate versions of consensus GTTCACC usually repeated at 11-bp intervals), to influence their expression (56, 78). In other bacteria, signal input to the PhoR sensor kinase is mediated by PhoU, a regulatory protein that is associated with a high-affinity phosphate uptake system (PstABCS) (79). In S. coelicolor, a PhoU-like protein is encoded by SCO4228, which is next to the phoRP genes, but it is not known whether this protein is implicated in phosphate signal transduction. However, elimination of the PstS component of the phosphate uptake system caused actinorhodin overproduction (80), so it is likely that phosphate signal transduction is much the same in streptomycetes as in other bacteria (Fig. 6). In a homeostatic feedback loop, PhoP regulates levels of the PstS transporter, which, in turn, probably influences PhoP phosphorylation (80). Phosphorylation of PhoP presumably enhances DNA binding. Although the overall picture of PhoP-mediated regulation is well established, understanding of the action of PhoP at the molecular level is still limited, apart from the finding that the positions of PHO boxes correlate with positive or negative action (81). Some reported observations are hard to explain. For example, PhoP binds to, and represses, the promoter of afsS, encoding a global activator of antibiotic production (82). This negative effect is counterintuitive since phosphate limitation activates production of ACT and RED, an effect mediated by enhanced transcription of the relevant biosynthetic genes. Likewise, deletion of phoP causes reduced ACT and RED production under phosphate limitation (82). Another PhoP-repressible gene encodes RpoZ, the omega subunit of RNA polymerase, yet RpoZ is required for normal levels of production of ACT and RED (72). It is notable that the effects of phoP deletion on Streptomyces lividans, a very close relative of S. coelicolor, were quite different: in that organism, in which ACT and RED production is turned off under most conditions, phoP deletion caused strongly increased production of the two antibiotics (73). These complications are probably partly caused by the complex constellation of other genes affected by PhoP, which include several pleiotropically acting developmental genes that themselves influence antibiotic production (56).
Although PhoP seems not to target CSR genes in the ACT or RED pathways directly, it does directly repress cdaR, the CSR gene for CDA biosynthesis (56). It also represses the bidirectional scbA-scbR promoter, presumably affecting the biosynthesis and signal transduction of SCB autoregulator molecules. The consequences of this regulation for the bistable switching of CPK biosynthesis mediated by the action of SCBs are not readily predictable (56). Surprisingly, PhoP also binds at exceptionally high levels to three sites internal to cpkB and cpkC, encoding two of the large CPK type I polyketide synthase (PKSs) (56). This binding appears to be necessary for transcription of the cpkBC genes.
PhoP also mediates cross talk between cellular responses to phosphate, carbon, and nitrogen availability (Fig. 6). Thus, PhoP-mediated regulation of ptsS was observed only at very high levels of extracellular carbon sources (80), and PhoP-repressible genes include several involved in nitrogen metabolism, notably glnR, whose product is the major nitrogen regulator of primary metabolism (83, 84), providing a means of adjusting nitrogen metabolism to phosphate availability.
Regulation of Antibiotic Production by N-Acetylglucosamine
Morphological differentiation and secondary metabolite biosynthesis are supported by nutrients released by autolysis of substrate mycelium (9, 85). N-Acetylglucosamine (GlcNAc) released from the cell wall during this process is reimported and phosphorylated by a phosphotransferase-based transport system (PTS) (36, 86). It is also likely that GlcNAc released by the breakdown of fungal chitin may help Streptomyces to sense the presence of fungi, triggering a “competition stress” mechanism to produce secondary metabolites (83), though it has been noted that chitin breakdown yields predominantly chitooligosaccharides rather than GlcNAc (87). After uptake, the subsequent deacetylation of GlcNAc∼P results in the cytoplasmic accumulation of GlcN∼P. This serves as a signaling molecule, binding to the GntR-like regulator DasR and relieving repression of DasR target genes (Fig. 6). These include the genes for the PTS for GlcNAc uptake, forming a self-reinforcing feedback loop reminiscent of that involving PhoP and the phosphate uptake system (see above) (33, 36, 88), while downstream targets include the CSR genes for ACT (actII-ORF4) and RED (redZ) (33, 36, 88). The addition of GlcNAc to solid minimal medium cultures stimulates antibiotic production and development, whereas an opposite, inhibitory effect is observed with rich medium, an observation that is difficult to explain in terms of the known activities of DasR (20). DasR can also induce transcription of SCO6264, encoding a reductase believed to play a role in modification of the SCB gamma-butyrolactone signaling molecules (88).
Chitin is very abundant in soil and is likely to be a major source of carbon and nitrogen for streptomycetes (89). Consistent with this, especially large effects of DasR were observed when S. coelicolor and its dasR mutant were grown in the presence of added chitin on soil (or soil extract rich medium): the dasR mutant exhibited marked differences in expression of 679 genes, including many in the act, red, and cpk clusters (89). This illuminating study should encourage the use of similar conditions for the activation of antibiotic biosynthesis in other streptomycetes.
Carbon Catabolite Repression of Antibiotic Production
It has long been known that extracellular glucose represses the production of many antibiotics, including ACT, but the mechanism has resisted molecular analysis, even though a role for glucose kinase in repression of the utilization of alternative carbon sources was indicated 30 years ago (90). Current evidence suggests that both catalytic activity and a specific interaction of the enzyme with glucose permease are involved in glucose repression (91). A thorough review of this topic is included in reference 20. A recent proteomic study examined the effects of added glucose on exponentially growing cultures of S. coelicolor and its glucose kinase (glkA) mutant in minimal medium supplied with mannitol or fructose (92). Significant effects of glucose on carbon source uptake, central carbon and nitrogen metabolic enzymes (and the nitrogen regulator GlnR), some developmental proteins, and proteins closely or directly involved in RED, CDA, and CPK antibiotic production were found, but there was no effect on DasR. Notably, GlkA was involved in the effects on RED and CDA production but did not affect glucose regulation of SCB1 and CPK biosynthetic enzymes, revealing an uncharacterized second mechanism (92). The central enzyme pyruvate phosphate dikinase, which was particularly highly repressed by glucose in a GlkA-independent manner, was suggested as a possible key player in this second mechanism (92). These complex data sets will be difficult to interpret until samples are also examined during the antibiotic production phase, and progress has been made on the molecular basis of glucose kinase effects.
Another major environmental carbon source is polymers containing xylose, and this is reflected in the ability of the xylose operon repressor (ROK7B7) to activate the DasR-repressible nagE2 gene for GlcNAc permease (20) and to bind to the promoters of the CSR genes actII-ORF4 and redD (38, 54) (Fig. 6). Interestingly, rok7B7 seems to be subject to GlkA-mediated glucose repression (92).
Nitrogen Regulation of Antibiotic Biosynthesis
Mutations in the regulatory genes draR and afsQ1 have effects on antibiotic production that are discernible only under nitrogen excess (35, 64, 65). Both genes are part of two-component regulatory systems (DraRK and AfsQ1Q2) that also affect morphological differentiation (35, 64, 65). In a comprehensive recent paper, AfsQ1 has been revealed as a direct repressor of primary nitrogen assimilation genes and as an activator of several antibiotic biosynthetic regulatory (actII-ORF4, redZ, and cdaR) and structural (cpkA and cpkD) genes, as well as regulating some developmental genes (bldM, whiD, and amfC) (34). It appears to function in the maintenance of C, N, and P balance, as judged by the finding that it also directly represses pstS (phosphate uptake) and binds to promoters of genes for a xylanase (xysA) and a glyceraldehyde 3-phosphate dehydrogenase (gap1) (34) (Fig. 6). AfsQ1 recognizes a moderately conserved pair of 5-bp sequences separated by 6 bp, with the 5-bp sequences similar to that recognized by the nitrogen regulator GlnR. This results in cross-recognition of some sites (especially those in the promoters of nitrogen assimilation genes), leading to actual competition between GlnR activation and AfsQ1 repression (34). It is thought that a possibly glutamate-related signal activates the apparently membrane-associated AfsQ2 kinase and hence the phosphorylation-dependent activation of AfsQ1. This response is somehow enhanced by the AfsQ1-dependent expression of the sigQ gene, which diverges from the afsQ1Q2Q3 operon (34). No such detailed analysis of the DraRK system has been published.
This is certainly not the whole story for nitrogen regulation. It was recently claimed that GlnR binds to the promoters of actII-ORF4, redZ, and cdaR (34), and one of its target genes, glnK, encoding the nitrogen-regulatory PII protein, influences the rates of antibiotic production and sporulation in a medium-dependent manner (93). Also, mutational inactivation of NdgR, an IclR-type regulator for nitrogen source-dependent growth in S. coelicolor, enhanced the production of ACT in minimal media containing certain amino acids (94). This effect seemed to be direct, since NdgR could bind to the promoters of some genes involved in antibiotic biosynthesis, including the scbA-scbR intergenic region in S. coelicolor (see below) and promoters of doxorubicin biosynthetic genes in Streptomyces peucetius (94). Interestingly, the ndgR promoter is a target for the redox stress-induced sigma factor SigR (95), suggesting that redox stress might repress ACT production under certain nutritional conditions.
Amino Acid Limitation and Ribosome-Mediated Effects
When the supply of amino acids becomes rate limiting for protein synthesis, bacteria produce the alarmones guanosine tetraphosphate (ppGpp) and pentaphosphate (pppGpp), which inhibit the synthesis of rRNA and tRNA and activate expression of other genes by altering the RNA polymerase core-binding competitiveness of sigma factors (96). The (p)ppGpp synthase enzyme RelA is activated when a shortage of amino acids leads to an uncharged tRNA molecule entering the A site of the ribosome. In S. coelicolor, a relA disruption mutant did not produce ACT (97). This linking of (p)ppGpp to the onset of secondary metabolism was reinforced by the finding that (p)ppGpp overaccumulation increased ACT and RED production (98). Likewise, bialaphos production in Streptomyces hygroscopicus (99) and production of clavulanic acid and cephalomycin in Streptomyces clavuligerus (100) were reported to be stimulated by (p)ppGpp accumulation. However, in a contradictory report, disruption of relA resulted in the enhanced production of clavulanic acid and cephalomycin C (101), so it is possible that not all antibiotic clusters respond in the same way to (p)ppGpp levels. In a further level of regulation of (p)ppGpp production, the transcription of relA itself responds to nitrogen limitation through the agency of an extracytoplasmic function (ECF) sigma factor, SigT (102). Mutations generating rifampin-resistant RNA polymerase can suppress the antibiotic deficiency of some relA or relC mutants (103, 104). Thus, the available information is consistent with the idea that (p)ppGpp may increase the relative affinity of RNA polymerase for sigma factors that direct the enzyme to genes specific for antibiotic production.
During the transition of S. coelicolor from exponential growth to stationary phase, more than half of the ribosomes are degraded (105), and cleavage of a significant fraction of tRNAs leads to the accumulation of 30- to 35-nucleotide (nt) RNA species (106). Since the onset of secondary metabolite biosynthesis occurs during the transition, it is possible that ribosome and tRNA degradation are related to secondary metabolism, with the further possibility that the 30- to 35-nt RNA species could be signaling molecules for secondary metabolism and morphological differentiation.
Some ribosomal mutants show enhanced production of secondary metabolites. A point mutation in rpsL (encoding ribosomal protein S12) dramatically increased ACT production (107). Overexpression of ribosome recycling factor also causes the overproduction of antibiotics, perhaps reflecting enhanced protein synthesis during stationary phase, probably through increased ribosomal stability under amino acid starvation (108). Mutation of rsmG, which encodes a 16S rRNA methyltransferase, elevates protein synthesis and in turn enhances antibiotic production (109). Although there is not enough evidence to confirm that changes to ribosome structure contribute to the onset of antibiotic production, it has been reported that ribosome structure determines translation accuracy and speed (110). Slowing of translation speed when growth slows may enhance folding of nascent antibiotic biosynthetic proteins, thus enhancing antibiotic production (Y. Pan et al., unpublished data).
Regulation at a Meeting Point of Primary and Secondary Metabolic Pathways
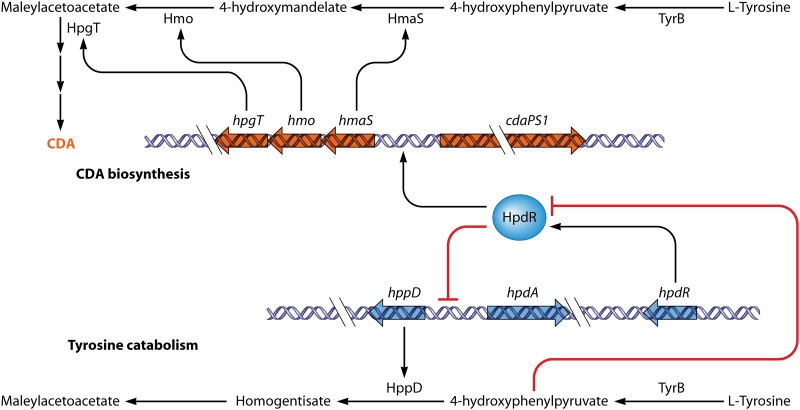
One of the intermediates in CDA biosynthesis is 4-hydroxyphenylpyruvate (4HPP). 4HPP is also the substrate of the product of the hppD gene, which is involved in tyrosine catabolism. This gene is repressed both directly and indirectly by HpdR, an IclR-type protein encoded by a nearby gene (111) (Fig. 7). Repression is relieved by 4HPP (111). Remarkably, HpdR also binds to the promoter of the CDA biosynthetic gene hmaS, which encodes an enzyme converting 4HPP to 4-hydroxymandelate (112). Since hmaS transcription is reduced in an hpdR mutant, HpdR functions to activate hmaS. It seems that 4HPP accumulated from tyrosine catabolism during stationary phase binds to HpdR, relieving autorepression of hpdR. The overexpressed HpdR represses hppD as a repressor but enhances hmaS expression as an activator. As a result, 4HPP is redirected by HmaS to 4-hydroxymandelate, which is a precursor of CDA. Thus, 4HPP acts both as a metabolic intermediate and as a regulatory ligand in the partitioning of tyrosine catabolism between primary and secondary metabolism (Fig. 7) (112).
Fig 7.
Model for the regulation of HpdR during tyrosine catabolism and CDA biosynthesis in S. coelicolor. As an autoregulator, HpdR regulates its own transcription (not shown). It also represses hppD, the product of which catalyzes the conversion of 4-hydroxyphenypyruvate (4HHP) to homogentisate, but activates hmaS, which is involved in CDA biosynthesis. During vegetative growth, l-tyrosine in the medium is catabolized to 4HHP by TyrB. The accumulated 4HHP binds to HpdR and causes it to dissociate from the hppD and hmaS promoters, therefore initiating hppD expression and preventing hmaS expression. During stationary phase, l-tyrosine in the medium is limited, 4HHP is reduced, and expression of hpdD is repressed, while hmaS expression is activated, directing 4HPP into CDA biosynthesis.
AbsC and Zinc Dependency of Antibiotic Production
About 5% of S. coelicolor proteins are predicted to bind zinc (113). It is therefore not surprising that the timing and levels of production of ACT and RED both depend on the amount of zinc supplied (114). A MarR-like repressor, AbsC, was implicated in this zinc response by virtue of the nonproduction of ACT and RED (and CDA) by an absC mutant on medium low in zinc (114). The effect of AbsC on antibiotic production appears to be indirect. AbsC represses a gene cluster for a zinc-binding siderophore, coelibactin, and together with another repressor, Zur, which binds nearby in the same promoter, helps to maintain zinc homeostasis. The influence of AbsC on antibiotic production could possibly be mediated through coelibactin (coelibactin overproduction inhibits sporulation) (115), either in a signaling role or indirectly through changed intracellular zinc levels. Alternatively, some other AbsC-regulated gene(s) may be involved. The influence on ACT and RED biosynthesis is ultimately exerted via the CSRs actII-ORF4 and redD, both of which are underexpressed in the absC mutant (114). Indeed, there is DNA affinity capture evidence (disputed in reference 114) that AbsC may bind directly to the promoters of actII-ORF4 and redD (38).
The AfsK/AfsR/AfsS System and a Possible Interface with Hyphal Growth
Apart from the regulatory systems already described that link antibiotic production to major aspects of nutrition, many other global antibiotic production regulatory genes that are not closely linked to biosynthetic gene clusters have been identified in S. coelicolor (116). For example, AfsR positively controls the biosynthesis of pigmented antibiotics (ACT and RED) in S. coelicolor. The afsR gene is located far from the act and red gene clusters (117, 118), appears to be present in all streptomycetes, and positively regulates the production of diverse antibiotics (119–121). AfsR (993 amino acids [aa]) has an N-terminal SARP domain, a central NB-ARC-like ATPase domain that may act as a molecular switch (122), and a C-terminal possible tetratricopeptide repeat (TPR) domain suggesting protein-protein interaction (SMART database) (123). Similar ATPase domains are also present in CdaR and in two other SARPs of unknown function encoded in the S. coelicolor genome (see below).
In vitro, AfsR can be phosphorylated by AfsK, one of the 34 protein serine/threonine kinases (STKs) encoded in the S. coelicolor genome (124). The autokinase activity of AfsK, and hence its ability to phosphorylate AfsR, is inhibited by direct interaction with the product of the neighboring gene, kbpA (both genes are present in nearly all streptomycetes but absent from other actinomycetes) (125) (Fig. 8). It is interesting that six paralogues of KbpA are encoded in the S. coelicolor genome and that one paralogue, SgaA, in S. griseus has conditional effects on morphology (126) (interestingly, the AfsK-AfsR system is also conditionally required for development in S. griseus [127]). However, it is not known whether any of the KbpA paralogues interact with protein kinases.
Fig 8.
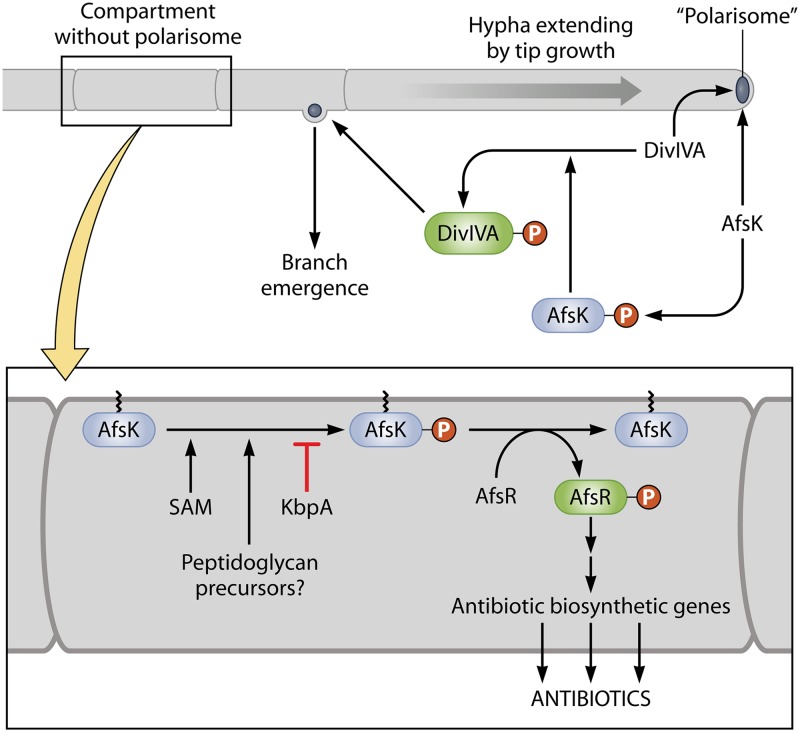
Regulation of antibiotic production by AfsR and its possible interface with hyphal tip growth. The upper part of the figure represents a hypha extending by tip growth, under the control of a tip-located “polarisome.” DivIVA and AfsK are located in the polarisome. As the tip extends, the DivIVA and AfsK content increases, and AfsK action causes polarisome splitting and the nucleation of a new branch point, such as that seen emerging from the subapical compartment. Also shown is a compartment that happens not to have captured a nascent polarisome. This is enlarged in the lower part of the figure. It is suggested that DivIVA-free AfsK in this compartment phosphorylates AfsR. The phosphorylation is potentially modulated by KbpA protein, S-adenosylmethionine, and precursors of cell wall biosynthesis that may accumulate in the absence of a growth point. For further explanation and references, see the text.
Although the close linkage of afsK and afsR suggests that their interaction is meaningful (the two genes are rarely separated by more than 10 genes in Streptomyces genomes), mutation of afsK does not completely eliminate AfsR phosphorylation, and two other STKs (PkaG and AfsL) that can phosphorylate AfsR in vitro have been identified (128). It is therefore possible that AfsR integrates signals from more than one STK-dependent signal transduction system. Remarkably, S. coelicolor AfsK is also implicated in the phosphorylation of a key determinant of mycelial polar growth, DivIVA (129). It was found that AfsK is almost entirely located at hyphal tips, where DivIVA is concentrated, as part of a “polarisome” that controls tip extension (Fig. 8). The activity of AfsK on DivIVA is crucial for the splitting of polarisomes to initiate new hyphal branches and is also stimulated by the inhibition of cell wall synthesis, with AfsK perhaps being responsive to intermediates in peptidoglycan biosynthesis (129). It is not yet possible to relate the two quite different AfsK activities, because the AfsR and DivIVA experiments were done under different culture conditions. However, it may be profitable to investigate the possibility that antibiotic production is initiated mainly in compartments that do not have a tip (i.e., are not apical) and in which any AfsK present could therefore not be tip located and might be able to make contact with alternative substrates such as AfsR. This could provide a mechanism for switching between growth and antibiotic production (Fig. 8).
Nuclear magnetic resonance (NMR) analysis has suggested that AfsK binds S-adenosylmethionine (SAM), though the consequences of this interaction are not known; however, there have been several reports indicating that endogenously increased SAM synthetase or exogenously added SAM increases antibiotic production in different streptomycetes (130–133). There appears to be no information about whether endogenous SAM levels increase as growth comes to a stop (for example, if methionine is shunted to SAM biosynthesis when protein synthesis slows down). In the light of the discovery of the AfsK-DivIVA interaction, the question arises of whether binding of SAM might change the partner choice of AfsK.
AfsR recruits RNA polymerase to the promoter of the adjacent afsS gene (134). AfsS (also called AfsR2) has been studied as an apparent positive regulator of antibiotic production since its discovery (135). It has some homology with the conserved linker domain 3 of sigma factors but is unlikely to function as a sigma factor on its own (136). A suggestion that AfsS might act cooperatively with sigma factors of the ECF subclass, which lack domain 3 (136), seems unlikely given that ECF sigma factors are ubiquitous while AfsS-like proteins are found in only some (not all) streptomycetes. When present, afsS is seldom as closely linked to afsR as in S. coelicolor (though quite often the two genes are within a few tens of genes of each other). AfsS may provide some species-specific beneficial modulation of AfsR function by virtue of its (probably mostly indirect) influences on the transcription of various stress response and stationary-phase genes (136). More than 117 genes were up- or downregulated in an afsS disruption mutant (136). Disruption of afsS also causes diminished actinorhodin production during phosphate starvation, an observation underpinned by the finding that AfsR and PhoP bind to overlapping sequences in the promoter region of afsS (82). Although AfsS is usually thought of as a regulator of antibiotic biosynthetic genes, transcriptome analysis showed that it also has major effects on nutritional starvation genes, which might possibly be responsible for the effects on antibiotic production (136).
AtrA, a TetR-Like Global Regulator of Antibiotic Production
A biochemical approach followed by reverse genetics led to the identification of AtrA, a TetR-like protein that binds to the promoter of the actII-ORF4 CSR gene, as an essential activator of actII-ORF4 expression. Neither RED nor CDA production was affected by an atrA mutation, but in S. griseus AtrA binds to the promoter of strR, the CSR gene for streptomycin production (32). AtrA orthologues are present in all Streptomyces genomes sequenced so far. atrA is regulated by the PhoRP system, providing a link with phosphate availability, and a further nutritional link is provided by the activation by AtrA of nagE2, encoding the GlcNAc permease. nagE2 is also subject to repression by the GlcNAc-sensing pleiotropic regulator DasR (20, 36). Hence, the two regulators act synergistically on GlcNAc-mediated control of the DasR regulon (Fig. 6).
Other Two-Component Systems Influencing Antibiotic Production
We have already mentioned roles in antibiotic production for four two-component systems (AbsA1A2, PhoPR, DraRK, and AfsQ1Q2). These are among 67 two-component systems encoded in the S. coelicolor chromosome (137). Other two-component systems influencing antibiotic production, but not closely linked to antibiotic biosynthetic genes, include the following: CutRS, affecting ACT production (138); EcrA1A2 (SCO2517 and -8), affecting only RED production (139); SCO0203/0204, which interplay with the orphan RR SCO3818 in exerting medium-dependent effects on ACT production (140, 141); and SCO5784 and -5, affecting the timing of ACT and RED production and sporulation, possibly through effects on ppGpp synthesis (142). In two surveys of several two-component systems, the AbrA1A2 system acted negatively on production of ACT, RED, and CDA and the formation of aerial mycelium, while the AbrC1C2C3 system, involving two histidine protein kinases, acted positively on all four aspects (143), and RapA1A2 was shown to stimulate ACT and CPK production (140). Several of the two-component systems implicated in regulating antibiotic production in S. coelicolor are not widely conserved among different species (AbsA1A2, CutRS, AbrA1A2, and SCO5784 and -5).
MalT-Like Regulators
LALs (large ATP-binding regulators of the LuxR family) are paralogues of MalT of E. coli that contain an N-terminal ATP/GTP-binding domain and a C-terminal DNA-binding domain with a LuxR-like helix-turn-helix motif. They have often been found as CSRs in Streptomyces spp. Examples include AmphRI and AmphRIII (amphotericin, Streptomyces nodosus) (144), NysRI and NysRIII (nystatin, Streptomyces noursei) (145), PikD (pikromycin, Streptomyces venezuelae) (146), and RapH (rapamycin, S. hygroscopicus) (147). None of the CSRs identified in S. coelicolor is a LAL, but 14 LAL-encoding genes unlinked to clusters were identified in the S. coelicolor genome (148). Two of these (SCO0877 and SCO7173) were relatively well expressed on a medium favoring antibiotic production, leading to their selection for mutational analysis. Both mutants showed reduced ACT production, attributable to decreased expression of actII-ORF4, along with some effects on global regulatory genes for antibiotic production (148).
RNA Regulation of Antibiotic Production
Several RNases (polynucleotide phosphorylase [PNPase], RNase PH, oligo-RNase, RNase III, and RNase E) are widespread in Streptomyces (149). Of these, RNase III, a double-strand-specific endonuclease involved in the processing of RNA transcripts (150), plays an important role in the regulation of antibiotic biosynthesis. An S. coelicolor RNase III (absB) mutant was deficient in ACT, RED, MM, and CDA because of low expression of the cognate CSR genes (151). RNase III was also reported to cleave the mRNA for AdpA, a pleiotropic transcription factor affecting both differentiation and antibiotic production (see below) (152). Recently, RNA-Seq showed that depletion of RNase III resulted in increased levels of many transcripts, including some (SCO5608, SCO0168, and SCO0864) thought to be involved in antibiotic production (153). An alternative explanation of the effect of RNase III on antibiotic production is that RNase III is responsible for rRNA maturation, which is important for the translation of long mRNA (149, 154).
Polynucleotide phosphorylase (PNPase) catalyzes the 3′-5′-phosphorolysis of RNAs and can also polymerize nucleotide diphosphates to produce ribopolymers. Overexpression of PNPase led to decreased antibiotic production in Streptomyces antibioticus (155). Since the expression of pnp is RNase III dependent in S. coelicolor (156), it will be interesting to investigate the connection of PNPase, RNase III, and antibiotic production. Little is known about the functions of other RNases in Streptomyces.
A Special Regulatory Role for the Rare Leucine Codon UUA in Antibiotic Production
In Streptomyces genes, with an average GC content of more than 70%, triplets using only T and A residues are rare. There is therefore a particularly low frequency in mRNA of UUA codons for leucine, for which there are five other synonymous codons that are all less AU rich. In-frame TTA triplets occur in only 147 chromosomal genes in S. coelicolor, and most of these genes are not widely conserved between species (157). The genomes of other streptomycetes analyzed contain up to 400 TTA codons. Deletion of the gene (bldA) for the cognate tRNA does not impair growth but reduces or eliminates the translation of most TTA-containing genes, so it has been assumed that no TTA-containing genes are essential. However, bldA mutants are defective in development and in the production of many antibiotics, and, consistent with a role in regulating stationary-phase biology, the bldA-specified tRNA increases in abundance when growth slows (158).
In S. coelicolor, a TTA codon is present in actII-ORF4, redZ, and mmyB, as well as in mmfL, one of the structural genes for the MMF autoregulators. Changing the TTA codon of actII-ORF4, or of mmyB and mmfL, to an alternative leucine codon caused the specific restoration of production of the relevant antibiotic in a bldA mutant (39, 57), providing a potentially useful route to the elimination of competing bldA-dependent pathways. It is very common for a TTA codon to be present in cluster-situated regulatory genes: among 143 secondary metabolic biosynthetic gene clusters, 109 included TTA-containing genes, most of which encoded regulators (159). This, and the TTA codon in adpA (see below), explains why disruption of bldA eliminates biosynthesis of a variety of secondary metabolites in diverse streptomycetes.
Unusually, the TTA-containing CSR gene ccaR for clavulanic acid/cephamycin C biosynthesis in S. clavuligerus is fully expressed in a bldA mutant (160, 161). This is thought to indicate a context effect on the extent to which UUA codons are translatable in a bldA mutant, with UUA codons followed by G or A being more likely to be mistranslated in frame (160, 161). Further analysis of this hypothesis is needed in order that the effects of TTA codons in antibiotic gene clusters can be reliably predicted.
The Master Regulator AdpA and Its Interplay with Developmental Genes
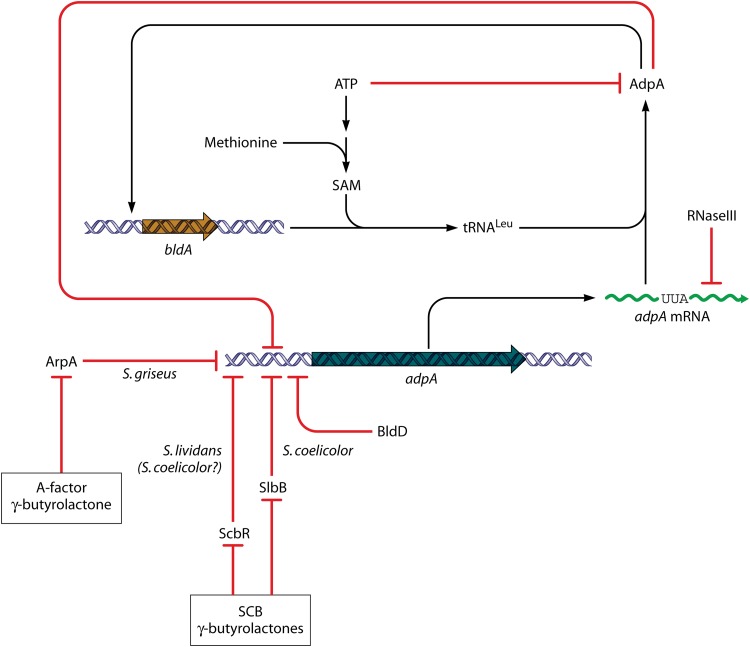
AdpA is a key transcriptional activator that has been studied biochemically mainly in S. griseus, in which it was discovered (30). It is present in all streptomycetes (orthologues are absent from most other actinomycetes), and adpA mutants (originally called bldH in S. coelicolor) have pleiotropic deficiencies in aerial mycelium formation and antibiotic production, which may be medium dependent (162–166). Like most proteins of the AraC/XylS family, to which AdpA belongs, it contains two DNA-binding helix-turn-helix (HTH) motifs in its C-terminal region. Preliminary structural analysis of the DNA-binding domain of the S. griseus protein complexed with DNA has been reported (167). A type 1 glutamine amidotransferase (GATase 1)-like domain in its N-terminal region suggests that AdpA may sense molecular signals.
In S. griseus, the adpA promoter is the sole target for the ArpA repressor, which is the sensor for the pleiotropic S. griseus-specific regulatory gamma-butyrolactone molecule A-factor (30). When the concentration of A-factor reaches a threshold, ArpA is released from the adpA promoter region. The pleiotropic effects of A-factor in S. griseus are manifested entirely via AdpA. Recently, evidence has been obtained pointing to the possibility that even in S. coelicolor adpA may be regulated by the endogenous SCB gamma-butyrolactones (Fig. 9). In DNA affinity capture experiments with S. lividans, which is very closely related to S. coelicolor, proteins binding to the adpA promoter included ScbR (the SCB-sensing paralogue of ArpA) (168). Moreover, in independent DNA affinity capture experiments done with S. coelicolor (169), a quite different protein, the SCO0608 protein (designated SlbR), was found to bind to both the adpA promoter and the scbA-scbR promoter and to be sensitive to SCBs even though it did not resemble any known gamma-butyrolactone-binding protein. Mutation of SCP0608 caused early production of ACT and RED on solid medium and hypersporulation (169). Few other streptomycetes have candidate SlbR orthologues, but proteins with moderate end-to-end similarity are present in about half the species that have been subjected to genome sequencing.
Fig 9.
Regulation of the key pleiotropic regulatory gene adpA. Transcription of adpA has been studied in S. griseus and S. coelicolor, as well as in S. lividans, which is very closely related to S. coelicolor. In each case, repressors that sense species-specific gamma-butyrolactones interact with the adpA promoter; and the pleiotropic regulator BldD also represses expression. AdpA is also autorepressing—circuitry that also implicates cross-regulation with bldA, whose product is the tRNA needed to translate a rare UUA codon that is found in the same place in virtually all streptomycetes analyzed. For further explanation and references, see the text.
Aside from these effects, regulation of adpA is complex in both S. griseus and S. coelicolor (Fig. 9). It is repressed both by its own gene product (170) and, in S. coelicolor, by another pleiotropic regulator, BldD (171). In S. lividans, and therefore probably in S. coelicolor, another protein, ArfA, appears to activate adpA expression (168). AdpA itself may sense adenine nucleotides: it was very recently shown that AdpA competes with the initiator protein DnaA for binding to the S. coelicolor origin of chromosome replication and that the binding of AdpA to this region was relieved by ATP or ADP (172). It is therefore possible that depletion of adenine nucleotides may affect the autoregulation of adpA expression, consistent with the observation that intracellular ATP levels are low at the onset of antibiotic production (173).
The S. coelicolor adpA mRNA is a target for processing by RNase III (152) and contains a bldA-dependent UUA codon (adpA is the only gene present in all streptomycetes that always has a TTA codon). It is not known whether these two posttranscriptional processes interact. The UUA codon in adpA mRNA always falls between the segments encoding the two major AdpA domains and accounts largely (but not entirely) for the aerial mycelium-defective phenotype of bldA mutants of S. coelicolor (164, 166). The abundance of bldA tRNA is important in determining whether AdpA reaches levels sufficient to activate its target genes, which include bldA itself, establishing a remarkable mutual feed-forward cascade (174). At least in S. lividans, there is even a further level of adpA regulation, since it appears that the antibiotic production-stimulatory effect of the addition of SAM (see above) is at least partly mediated through the UUA codon of adpA mRNA, perhaps by modulating the maturation of the bldA tRNA (175). We note in passing that the effects of SAM and adenosine nucleotides on AdpA activity might make their interconversion a pivot in AdpA-mediated activation of secondary metabolism and development.
The binding site for AdpA in DNA targets is rather degenerate (5′-TGGCSNGWWY-3′), and there are about 1,500 direct AdpA-binding sites in the S. griseus chromosome (176). About 75% of these seem not to be involved in gene regulation, but probably ca. 500 of them have regulatory significance (176). In general, when bound to a target site, AdpA recruits RNA polymerase and activates transcription, though some targets (like adpA itself) are repressed by AdpA (176). Targets in S. coelicolor and S. griseus include a considerable number of genes involved in secondary metabolism and morphological differentiation, though the spectrum of target genes is not completely congruent in the two species (30, 177). A notable target in S. griseus is strR, the CSR for streptomycin biosynthesis (the first AdpA target to be defined). The CSR gene (griR) for the biosynthesis of the A-factor-dependent yellow pigment grixazone in S. griseus, on the other hand, is only indirectly dependent on AdpA (178).
AdpA-binding sites have also been found upstream of other CSR genes for antibiotic production, including actII-ORF4 in S. coelicolor (38) and sanG for nikkomycin biosynthesis in Streptomyces ansochromogenes (179). The role of AdpA at such target promoters can be complex. For example, of five AdpA-binding sites located upstream of sanG, two (sites I and V) are used to activate transcription of sanG, while three (sites II, III, and IV) lead to repression. It is speculated that a DNA loop formed via protein-protein interactions between AdpA bound to sites II, III, and IV prevents RNA polymerase from chain elongation and represses the transcription of sanG. Interestingly, nikkomycin production in a site III mutant is much higher (3-fold) than that in the wild type (179).
How Does BldD Regulate Development and Antibiotic Production?
Mutation of bldD results in failure of S. coelicolor to develop aerial mycelium or to produce ACT, RED, MM, or CDA on most media (180, 181). BldD is structurally related to SinR, a protein of Bacillus subtilis, and to lambda repressor, both of which mediate transitions between different cellular states by acting as repressors of some genes and activators of others (182, 183). BldD is autorepressing (184–186). More than 150 other direct targets of BldD have been determined, leading to the delineation of a refined consensus target site, 5′-nTnCnC(A/T)GnGTnAn-3′ (171). Among a relatively high representation of regulatory genes, the target genes include the following: adpA and bldA; bldC, encoding a small protein with a single MerR-like domain that is needed for development and antibiotic production (187); and nsdA, which encodes an apparent repressor of ACT, RED, and CDA production and of differentiation (188, 189). Orthologues of BldD and BldC are found in many actinobacteria, including some that are morphologically simple, have small genomes, and are not known as antibiotic producers, while NsdA orthologues are present in all streptomycetes and their closest relatives but are absent from other actinomycetes. NsdA is not obviously related to proteins of known structure or function, but streptomycetes usually have several paralogues. In Streptomyces bingchengensis, NsdA represses production of the macrolide milbemycin and a polyether, nangchangmycin (189). The effects of NsdA are thus applied to a wide range of antibiotic biosynthetic pathways. In the case of ACT, the effect of NsdA is exerted, directly or indirectly, via actII-ORF4 (188). It is possible that some of the pleiotropic defects of a bldD mutant are attributable to the derepressed expression of nsdA.
Other Developmental Genes with Effects on Antibiotic Production: bldB, abaA and whiJ, and wblA
The small protein BldB is required for antibiotic production and aerial growth, possibly through interaction with another protein, though little more is known about its mode of action (190). BldB appears to universal among, but peculiar to, streptomycetes. S. coelicolor has many small proteins giving end-to-end alignments with BldB, and one of these, SCO4542, has been further studied genetically. Its deletion caused apparent overproduction of ACT and/or RED and loss of aerial mycelium formation (191). Further analysis indicated a complex, medium-sensitive interplay of SCO4542 protein with the products of two neighboring genes, SCO4543 (whiJ) and SCO4544, both of which have predicted regulatory roles (191). It was suggested that the three proteins are involved in transducing an environmental signal that influences developmental progression and that SCO4542 mutants are abnormally rich in a cell type that produces pigmented antibiotics (192). Streptomycetes all have multiple gene clusters comprising paralogues of SCO4542 to -4 (191, 193). One such cluster is the abaA cluster, discovered earlier through its presence in a DNA fragment that stimulated S. coelicolor colony pigmentation at high copy number (194). Some of these clusters are widely conserved among different species, but others, including the whiJ cluster, are species specific. It has been suggested that these clusters may generally determine responses of secondary metabolism and development to environmental signals associated with particular ecological niches (191). Interestingly, there are markedly fewer such clusters in S. venezuelae, an organism that grows and sporulates rapidly and produces at least one antibiotic, chloramphenicol (Cm), during vegetative growth (51).
Another developmental gene, wblA, encodes one of 11 S. coelicolor proteins of the Wbl (WhiB-like) family (195). These small, redox- and nitric oxide-sensitive iron-sulfur proteins are widespread in actinobacteria and are absent from all other bacteria (195). Disruption of wblA dramatically increased antibiotic production in S. coelicolor (195), Streptomyces peucetius (196), and Streptomyces sp. strain C4412 (197), prevented nearly all aerial hyphae from sporulating in S. coelicolor (195) and Streptomyces ghanaensis (198), affected the expression of hundreds of genes in S. coelicolor (195), and slightly increased S. coelicolor oxidative stress responses (199). On the other hand, a high copy number of wblA depressed the production of ACT, RED, and CDA in S. coelicolor (and of doxorubicin in S. peucetius) (200). It was suggested that WblA mediates a switch between antibiotic production and morphological development, such that (as suggested above for WhiJ systems) mutation of wblA causes increased representation in the colony of an antibiotic-producing cell type that is intermediate between growth and a developmentally committed state (195).
BROADENING FROM THE S. COELICOLOR MODEL: EVOLUTION AND GENERALIZATION
Among the hundreds of antibiotic biosynthetic clusters that have been sequenced, there is considerably greater diversity in the numbers, types, and linkage arrangements of CSR genes than has been revealed in S. coelicolor. Comparatively few of the regulatory features of clusters from nonmodel organisms have been analyzed experimentally, but where they have been, new concepts usually arise, as we illustrate in this section with a few examples. Clearly, current knowledge is only the tip of the iceberg.
Antibiotic Biosynthesis Regulation: Cascades, Feedback Control, and Cross Talk
Pathway-specific regulation can be very simple, yet idiosyncratic. For example, although the higher-level regulatory input into streptomycin production is quite complex, only one CSR, StrR, is involved (201). StrR does not resemble other characterized proteins, except in possessing a likely ATPase domain like those of ParA partitioning proteins. Few paralogues of StrR have been found—none are present in S. coelicolor, and we found only four in the seven genomes in StrepDB. Among 236 antibiotic biosynthesis clusters in the sequence database that we collected (Table 1, footnote a), one more strR-like gene was found, at one end of the lankacidin cluster of Streptomyces rochei, but disruption of this gene had no effect on antibiotic production (202).
Table 1.
SARPs encoded in antibiotic biosynthetic gene clusters from diverse streptomycetesa
| Pathway type | Antibiotic | Organism | SARP length |
No. of TTA codons | Gene name | |
|---|---|---|---|---|---|---|
| <400 aa | >400 aa | |||||
| Type II polyketides | Aranciamycin | S. echinatus | 270 | 70 | orf15 | |
| 943 | orf4 | |||||
| Lactonamycin | S. rishiriensis | 626 | lct23 | |||
| 277 | lct15 | |||||
| 221 | lct21 | |||||
| 510 | lct22 | |||||
| FD-594 | Streptomycessp. strain TA-0256 | 636 | 620 | pxnR2 | ||
| Griseorhodin | Streptomyces sp. strain JP95 | 274 | grhR2 | |||
| 274 | grhR1 | |||||
| A-74528 | Streptomyces sp. strain SANK 61196 | 611 | sanR1 | |||
| Aclacinomycin | S. galilaeus | 281 | acl1 | |||
| 273 | aknO | |||||
| Medermycin | Streptomyces sp. strain AM-7161 | 259 | med-ORFII | |||
| Auricin | S. aureofaciens | 274 | sa35 | |||
| 269 | sa23 | |||||
| 293 | sa37 | |||||
| 273 | sa24 | |||||
| Polyketomycin | S. diastatochromogenes | 271 | 8 | pokR2 | ||
| 1,066 | pokR1 | |||||
| Chartreusin | S. chartreusis | 217 | chaR1 | |||
| Lysolipin | S. tendae | 614 | 30 | llpR4 | ||
| Gilvocarcin | S. griseoflavus | 1,090 | gilS | |||
| Hedamycin | S. griseoruber | 273 | orf16 | |||
| Granaticin | S. violaceoruber | 284 | gra-ORF9 | |||
| Polyketide 5 | S. avermitilis | 271 | 129 | ? | ||
| Frenolicin | S. roseofulvus | 283 | frnG | |||
| Fredericamycin | S. griseus | 612 | 191 | fdmR1 | ||
| Alnumycin | Streptomyces sp. strain CM020 | 272 | 5 | alnR3 | ||
| Enterocin | S. maritimus | 284 | encF | |||
| Chromomycin | S. griseus | 301 | 13 | cmmR | ||
| Steffimycin | S. steffisburgensis | 291 | 241 | stfR1 | ||
| Mithramycin | S. argillaceus | 277 | 6, 226 | mtmR | ||
| SF2575 | Streptomyces sp. strain 2575 | 271 | 8 | ssfT1 | ||
| Totals | 22 pathways | 22 organisms | 24 small SARPs | 9 medium or large SARPs | 11 genes with TTA codons | |
| Type I polyketides | Reveromycin | Streptomyces sp. strain SN-593 | 278 | 27, 30 | revQ | |
| AHBA | Kitasatospora putterlickiae | 769 | orf18 | |||
| Tetronasin | S. longisporus | 257 | tsn17 | |||
| Pactamycin | S. pactum | 399 | orf27 | |||
| Furaquinocin A | Streptomyces sp. strain KO-3988 | 344 | 56, 75, 78 | ? | ||
| Oxazolomycin | S. albus | 930 | ozmU | |||
| Chlorothricin | S. antibioticus | 262 | chlF2 | |||
| Natamycin | S. chattanoogensis | 1,177 | 37, 351 | scnR1 | ||
| Piericidin | S. piemogenus | 200 | pieR | |||
| Salinomycin | S. albus | 856 | orf15 | |||
| Bafilomycin | S. lohii | 610 | 172 | bafG | ||
| Nangchangmycin | S. nangchangensis | 254 | 31 | nanR2 | ||
| 242 | nanR1 | |||||
| Polyketide 5 | S. avermitilis | 271 | 129 | ? | ||
| Virginiamycin | S. virginiae | 330 | vrmS | |||
| Rubradirin | S. achromogenes subsp. rubradiris | 291 | rubRg3 | |||
| 267 | rubRg1 | |||||
| Tetronomycin | Streptomyces sp. strain NRRL 11266 | 257 | 31 | trrn18 | ||
| Aureothin | S. thioluteus | 272 | 11 | aurD | ||
| Dorrigocin/migrastatin | S. platensissubsp. rosaceus | 1,218 | 447, 1059 | |||
| Asukamycin | S. nodosussubsp. asukaensis | 255 | asuR5 | |||
| Concanamycin A | S. neyagawaensis | 702 | 5 | orf17 | ||
| Tylosin | S. fradiae | 278 | 68 | tylS | ||
| Totals | 21 pathways | 21 organisms | 16 small SARPs | 8 medium or large SARPs | 12 genes with TTA codons | tylT |
| Nonpolyketides | Nosiheptide (thiopeptide) | S. actuosus | 324 | nosP | ||
| Skyllamycin (nonribosomal peptide) | Streptomyces sp. strain Acta 2897 | 647 | sky44 | |||
| Polyoxin (nucleoside) | S. cacaoisubsp. asoensis | 1,112 | 63 | polR | ||
| 963 | 152, 360 | ? | ||||
| Peptide-2 (nonribosomal peptide) | S. avermitilis | 616 | ? | |||
| Thienamycin (atypical beta-lactam) | S. cattleya | 267 | thnU | |||
| 4-Hydroxy-3-nitrosobenzamide | S. murayamaensis | 323 | 41 | nspR | ||
| Nikkomycin | S. tendae | 1,063 | orfR | |||
| Himastatin | S. himastatinicus | 1,049 | ? | |||
| Virginiamycin (two polyketide/peptide components) | S. virginiae | 330 | vmsS | |||
| Phenalinolactone (terpene) | Streptomyces sp. strain Tu6071 | 266 | plaR1 | |||
| Heboxidiene (?) | S. chromofuscus | 262 | 200 | ? | ||
| Prodigiosin (pyrrole) | S. griseoviridis | 254 | rphD | |||
| Rubrinomycin (?) | S. collinus | 662 | rubS | |||
| Cinnamycin (lantibiotic) | S. cinnamoneus | 262 | 72 | cinR1 | ||
| Totals | 14 pathways | 14 organisms | 8 small SARPs | 7 medium or large SARPs | 5 genes with TTA codons | |
Gene clusters were initially identified by searching the NCBI database for “Streptomyces” under the field “organism” and for “cluster” under the field “title.” Limits of 5,000 to 500,000 nucleotides were set. The exact query submitted to the database was, “Streptomyces [organism] AND cluster [title] AND 5000:5000000 [sequence length].” From 365 clusters initially listed, manual editing was used to eliminate clusters that were not for antibiotic biosynthesis. Further manual editing eliminated clusters that were SARP free but incomplete. From a final total of 236 clusters, 24% (57 clusters) encoded at least one SARP (homologues of ActII-ORF4 using BLASTP analysis). These are listed here in relation to the general type of biosynthetic pathway. “Small” SARPs are up to 400 aa long, “medium” are 401 to 799 bp, and “large” 800 bp or more. AHBA, 3-amino-5-hydroxybenzoic acid.
In contrast to the case for streptomycin, the complex regulation of tylosin production in Streptomyces fradiae involves at least five CSRs, including two SARPs and two ArpA-like proteins (203, 204). TylR, a homologue of CSRs for carbomycin and spiramycin biosynthesis (205), is the direct activator of the tylosin biosynthetic genes. The expression of tylR requires the combined action of two SARPs, TylS and TylU (which is described as a “SARP helper”). In a positive reinforcement circuit, TylS also activates tylU. Two ArpA-like proteins, TylP and TylQ, provide overriding negative control of tylR expression. Thus, TylQ directly represses tylR, and TylP directly represses tylS. Further subtlety is conferred by the ability of TylP to repress both its own gene and that of TylQ. Repression by TylP can be lifted by a (still-unidentified) small-molecule ligand extractable from stationary-phase cultures. Thus, the accumulation of this ligand probably sets the regulatory system off. The logic of the two-step negatively acting part of the overall regulatory system is unclear, but in view of the evidence discussed elsewhere in this review, it seems possible that TylQ might be sensitive to a different ligand, perhaps generated by the tylosin pathway, or possibly the product of another biosynthetic pathway in S. fradiae.
Different regulatory strategies are found in different pathways for biosynthesis of polyether antibiotics, such as nanchangmycin and monensin, low-molecular weight compounds that are widely used in agriculture. The nanchangmycin (nan) biosynthetic gene cluster includes at least six regulatory genes (206). NanR1 and NanR2 are SARPs essential for nanchangmycin biosynthesis, and NanR4 is AraC like and represses nanR1 and nanR2, such that deletion of nanR4 resulted in a 3-fold increase of nanchangmycin (207). The roles of NanR3 (a putative LacI-like repressor) and the NanT5/NanT3 two-component system (206) remain undetermined. Only three putative monensin CSRs have been recognized (109). Overexpression of the SARP MonR1 caused increased monensin production, but the roles of MonRII, which resembles the negative regulator TcmR from Streptomyces glaucescens, and MonH, which resembles CSRs with an N-terminal ATP-binding domain in other clusters, have not been studied (208). Thus, the pathways for different polyethers have different regulatory strategies.
Bearing in mind the binding of the RED antibiotic by RedZ, it was of interest to consider the occurrence and functions of atypical RRs (ARRs) in other antibiotic biosynthetic clusters. RedZ belongs to the NarL family ARRs, which include DnrN, which is required for daunorubicin (DNR) biosynthesis in S. peucetius (209) (Fig. 5). Indeed, mutagenesis of residues in the position of the phosphorylation pocket of DnrN had no effect on daunorubicin production levels (209), strongly suggesting that DnrN is not regulated by conventional phosphorylation (similar conclusions were also reached for some orphan RRs with other physiological roles in Streptomyces [210–212]). Other NarL family ARRs associated with antibiotic clusters are shown in Fig. 5, as well as SCO1645, an S. coelicolor protein with unknown function that is very similar to RedZ.
Other Streptomyces ARRs belong to the OmpR family and are highly conserved (Fig. 5) (213). The most studied of these is JadR1 of S. venezuelae. JadR1 has a winged HTH motif in its C-terminal output domain, and two important aspartic acid residues are replaced by Glu49 and Ser50 in its N-terminal receiver domain. The jadR1 gene is located upstream of jadJ, the first structural gene of the jadomycin biosynthetic cluster (69). As JadR1 could not be phosphorylated, it should sense some signaling molecules to start the antibiotic biosynthesis. Recent studies found that JadR1 activates the expression of jadomycin B (JdB) biosynthetic genes in the presence of a low concentration of JdB and its homologues, but high JdB concentrations led to the dissociation of JadR1 from its target promoters. Thus, JdB interacts with JadR1 directly in a dose-dependent manner (Fig. 10) (69). This was the first report that any ARR could be activated by binding to the end product or late biosynthetic intermediates of secondary metabolites. Clearly there is a need to investigate both the molecular basis of ARR-ligand interactions and their broader significance in relation to antibiotic production.
Fig 10.
Cross-coordination of different antibiotic biosynthetic pathways by the pseudo-GBL receptors in S. venezuelae. The pseudo-gamma-butyrolactone receptor JadR2 directly represses the transcription of jadR1 in the absence of ethanol and also binds chloramphenicol (Cm) and jadomycins. The cluster-situated regulator JadR1 activates the biosynthesis of jadomycin B by activating the transcription of biosynthetic structural genes. JadR1 also represses the production of Cm by binding to the promoters of the structural genes (51).
End product-mediated feedback regulation has also been found with CSRs of a different type. In the three-step CSR cascade controlling daunorubicin production in S. peucetius, the TetR-like product of dnrO activates the diverging gene dnrN, encoding an ARR which then initiates the transcription of dnrI, the SARP product of which turns on daunorubicin (DNR) biosynthesis. dnrO is repressed by its own product, DnrO, but once daunorubicin or its glycosylated late precursors are produced, they bind to DnrO, derepressing dnrO and apparently activating dnrN (214).
In S. venezuelae, the key jadomycin CSR JadR1 not only activates jadomycin biosynthesis directly but also represses the chloramphenicol biosynthetic genes (51) (Fig. 10). JadR1 binds directly to an extended sequence in the cml cluster, located downstream of the transcription start site of cmlJ. The mechanism by which binding at this site represses transcription has not been investigated, and it is also unclear how this binding sequence is related to those used in the jad cluster to activate jadomycin biosynthesis, but the discovery of this kind of CSR-mediated cross talk implies yet a further layer of complexity in the regulation of antibiotic biosynthesis.
The jadR1 gene is itself directly repressed by an ScbR2-like pseudo-gamma-butyrolactone receptor, JadR2, encoded by the adjacent diverging gene. Thus, during JadR2-mediated repression of jadR1, Cm biosynthesis is derepressed. Repression of jadR1 is relieved by binding of either jadomycins or Cm to JadR2 (51) (Fig. 10). The resulting accumulation of JadR1 represses Cm biosynthesis and activates jadomycin biosynthesis. This resonates with similar observations with ScbR2 in S. coelicolor (see “Regulation of the ‘Cryptic Polyketide’ Gene Cluster”). Streptomyces genomes often encode several putative ArpA homologues, including many that group together with ScbR2 and JadR2 pseudo-gamma-butyrolactone receptors in the phylogenetic tree of ArpA-like proteins (215). Examples include Aur1R from Streptomyces aureofaciens and BarB from Streptomyces virginiae. This suggests that the coordination of the biosynthesis of disparate antibiotics by receptors of this kind may be widespread in Streptomyces.
A Survey of SARPs
As shown above, SARPs are the most frequently encountered CSRs in S. coelicolor and are associated with antibiotic biosynthetic clusters in many other streptomycetes. The most well known SARP, ActII-ORF4, contains 255 amino acid residues, and activates actinorhodin biosynthesis in S. coelicolor (39). Some pathways involve more than one SARP, forming parts of cascades of pathway-specific regulatory steps (e.g., tylosin and nangchangmycin [see above]) (203, 206).
The N-terminal winged helix-turn-helix (HTH) DNA-binding motif of SARPs binds repeated motifs (often heptamers) at an 11-nt spacing, and the adjacent transcriptional activation domain switches on the expression of target antibiotic production genes (29). The specific sequences recognized by SARPs generally overlap the −35 regions of their targets, but sometimes the sequences are far from the transcriptional start point (tsp) of their target genes. Some SARP-binding sites contain three discernible heptamers, such as those upstream of actII-ORF1 and actVI-ORF1 in S. coelicolor (29), claR, cefD, and cefF in S. clavuligerus (216), vlmJ and vlmA-vlmH in Streptomyces viridifaciens (217), dnrD in S. peucetius (218), fdmD in S. griseus (219), pimM in S. natalensis (220), and so on. Other sites contain only two obvious heptamers, such as sanO-sanN in S. ansochromogenes (221), polB and polC in Streptomyces cacaoi subsp. asoensis (222), cmcI and ceaS2-II in S. clavuligerus (223), and fdmR2 in S. griseus (219). Degeneracy of the repeats may make the structure of the targets difficult to characterize, perhaps explaining why the spacers appear to vary from 4 to 15 nt in different targets (216, 217, 219). Alternatively, these differences could perhaps be related to variations in the structure of SARPs.
Like ActII-ORF4 and RedD, many other SARPs are less than 300 residues long, and some of these have been shown, mainly by mutant phenotypes, to function as pathway-specific activators, including CcaR (cephamycin-clavulanic acid supercluster in S. clavuligerus) (216), DnrI (daunorubicin biosynthesis in S. peucetius) (218, 224), Aur1PR2 and Aur1PR3 (auricin biosynthesis in S. aureofaciens) (225), TylS and TylT (which activate the transcription of the tylosin CSR gene tylR in S. fradiae) (226), FdmR1 (fredericamycin biosynthesis in S. griseus) (219), ThnU (cephamycin C biosynthesis in Streptomyces cattleya) (227), SrrY and SrrZ (lankamycin biosynthetic regulatory cascade in Streptomyces rochei) (228), and VlmI (valanimycin biosynthesis in Streptomyces viridifaciens) (217). These ActII-ORF4-like SARPs are referred to here as “small SARPs.” Members of a second class of SARP CSRs (“medium SARPs”) resemble CdaR in being around 600 aa long, and these all show end-to-end similarity to each other (229). Still other SARP CSRs are much larger, containing about 1,000 residues (“large SARPs”). All of these have the same three major functional domains: an N-terminal SARP domain, a central AAA domain (ATPase associated with diverse cellular activities) (230), and a conserved C-terminal domain of unknown function.
Several large SARPs have been shown to activate their cognate pathways. For example, SanG positively regulates nikkomycin biosynthesis in S. ansochromogenes (221, 231), with its helix-turn-helix domain directly binding to the bidirectional sanN-sanO promoter region and activating the biosynthetic genes, and PolR controls polyoxin biosynthesis in S. cacaoi subsp. asoensis (222) through binding to the promoter regions of polC and polB. polR itself is activated by another large SARP, PolY, the product of the adjacent gene (232). Binding of PolY at a typical SARP-binding sequence in the promoter region of polR is enhanced by binding of ADP/ATPγS to the ATPase domain of PolY, which triggers PolY oligomerization (232) (Fig. 11). Since changes of ADP/ATP concentrations significantly affect the binding activity of PolY in vivo, the ATPase domain may sense endogenous ADP/ATP levels. Other large SARPs studied genetically include PteR (filipin, S. avermitilis) (233) and PimR (pimaricin, S. natalensis) (220, 234). It has been pointed out that all the examples of large SARP CSRs are in pathways for antifungal antibiotics, leading to the suggestion that such SARPs might recognize a common signal related to this antifungal activity (220).
Fig 11.
PolY controls the transcription of polR by sensing the ATP/ADP in the cells of S. cacaoi. Binding of ADP/ATPγS to the ATPase domain of PolY triggers the oligomerization of PolY and enhances its DNA binding affinity in vitro. Changes of ADP/ATP concentrations significantly affect the binding activity of PolY in vivo (232).
Using our database of 236 antibiotic biosynthetic clusters from streptomycetes other than S. coelicolor, we found 57 with at least one SARP (Table 1). We evaluated the relative frequencies of the three SARP classes and their association with different classes of antibiotics (Table 1). Small SARPs were the most abundant (48 examples were found in 40 clusters), while medium (13 were present in 12 clusters) and large (11 were present in 10 clusters) SARPs were both moderately common. Although SARPs were associated with the biosynthetic genes for diverse classes of antibiotics, there was a strong association of all three classes with pathways involving polyketide biosynthesis (43 of 57 SARP-containing clusters were for polyketides). Perhaps SARPs sense some physiological parameter that is particularly relevant to polyketide biosynthesis, such as the availability of precursors or the balance between growth-associated fatty acid consumption and starvation-associated lipid degradation. Of the 72 SARP genes listed in Table 1, 28 (39%) contain TTA codons, about 10 times more often than the average for all Streptomyces genes, consistent with the notion that UUA codons are significant in regulating antibiotic biosynthesis (see above).
Although SARP genes are most often located within antibiotic production clusters, a BLAST survey of the S. coelicolor genome shows that in addition to AfsR, there are three others that are not CSRs: one medium and two large CSRs (Table 2). Unlike CSR SARPs, these are all present in a significant fraction of streptomycetes. It will be of considerable interest to investigate whether these SARPs influence production of different antibiotics in different species.
Table 2.
Predicted SARPs encoded in the genome of S. coelicolor
| SCO no. | Length (aa) | Domain(s)a | Functionb | Species with orthologuec |
|---|---|---|---|---|
| 5085 | 255 | SARP | ActII-ORF4 (UCSR for actinorhodin) | |
| 6288 | 276 | SARP | CSR in Cpk cluster | |
| 4116 | 1,113 | SARP, ATPase, X | ? | S. avermitilis, S. venezuelae |
| 2259 | 1,334 | SARP, Y, ATPase, X | ? | S. avermitilis, S. clavuligerus, S. griseus, S. scabiei, S. venezuelae |
| 4426 | 993 | SARP, NB-ARC, TPR | AfsR (global regulator of antibiotic biosynthesis) | S. avermitilis, S. clavuligerus, S. griseus, S. scabiei, S. venezuelae |
| 6280 | 543 | SARP, Z | KasO (= CpkO) (UCSR for CPK) | |
| 5877 | 265d | SARP | RedD (UCSR for RED) | |
| 3217 | 638 | SARP, NB-ARC | CdaR (UCSR for CDA) | |
| 0898 | 660 | SARP, W | ? | S. avermitilis, S. clavuligerus, S. griseus |
Domains are abbreviated as follows: SARP, Streptomyces antibiotic regulatory protein; NB-ARC, pfam00931, signaling motif shared by plant resistance gene products and regulators of cell death in animals; TPR, tetratricopeptide domain; W, X, Y, and Z, domains of unknown function.
UCSR, ultimate cluster-situated regulators” (CSRs that directly activate biosynthetic genes).
Orthologues were determined as reciprocal best hits in BLASTP analysis. These genomes are presented in annotated form at http://strepdb.streptomyces.org.uk.
An alternative translational start to that usually allocated has been chosen, to take account of the known transcription start site.
Hormone-Like Autoregulators
Following the S. griseus A-factor paradigm (177), A-factor-like autoregulatory signal molecules have been found in many different Streptomyces species, with such autoregulatory systems often being associated with antibiotic biosynthetic gene clusters. For example, virginiae butanolides (VBs) (Fig. 1B) induce the coordinated production of virginiamycin M and virginiamycin S, synergistically acting but biosynthetically distinct antibiotics in S. virginiae (235), and IM-2 (Fig. 1B) controls the production of showdomycin and minimycin in Streptomyces lavendulae (236, 237). Three further chemical classes of autoregulator have been identified (45) (Fig. 1): furans such as the MMFs; avenolide, which is required at nanomolar concentrations for the onset of avermectin biosynthesis in S. avermitilis (238) (avenolide homologues are also found in S. fradiae, S. ghanaensis, and Streptomyces griseoauranticus); and PI factor [2,3-diamino-2,3-bis(hydroxymethyl)-1,4-butanediol], which enhances pimaricin production in S. natalensis (239). Like A-factor, MMFs and avenolide appear to bind to specific ArpA-like binding proteins (240). The nature of the PI factor effect is not understood, since the PI factor requirement could be bypassed by adding A-factor, glycerol, ethylene glycol, or propanediol (76, 239).
Typically, the afsA-like biosynthetic gene (often with additional cotranscribed factor biosynthetic genes) is located next to a diverging, arpA-like gene, whose product regulates both itself and the biosynthetic operon. This autoregulatory organization presumably leads to a sudden accumulation of autoregulator. Unusually, the A-factor autoregulatory system in S. griseus is determined not by genes close to the streptomycin biosynthetic gene cluster but by scattered genes (afsA and brpA for A-factor biosynthesis and arpA for the A-factor receptor protein ArpA) (177). The separation of the arpA and afsA genes from each other may be responsible for the loss of the kind of gamma-butyrolactone autoinduction found in most other systems, so that A-factor is accumulated more gradually in a constitutive growth-dependent manner, eventually giving concentrations high enough to relieve repression of adpA by ArpA (177).
Knocking out afsA-like genes not only causes reduced production of particular antibiotics but also often impairs differentiation, while mutating ArpA-like binding proteins can accelerate it. For example, SabR of S. ansochromogenes, a regulatory protein encoded away from the san (nikkomycin biosynthesis) cluster, not only activates the CSR sanG, but also represses sporulation. No ligand for SabR has been found, but deletion of sanG causes morphological deficiency and loss of the brown pigment, raising the possibility that a nikkomycin-related metabolite might bind to SabR (231). Likewise, elimination of a similar protein, SpbR, caused defects in growth, pristinamycin biosynthesis, and morphological differentiation in Streptomyces pristinaespiralis (241). The possibility that these effects involve activity of the autoregulatory systems on the expression of adpA (see above) would be worth investigation.
Global Regulators
The S. coelicolor studies have shown that numerous global regulators transmit various signal inputs, including nutrient availability, translation rates, developmental state, diverse stresses, and other environmental information, to the CSR genes. Such layered regulation has been called “pyramidal” or “cascade” regulation, though neither term seems apposite for systems in which multiple inputs drive a process forward (“confluent regulation” might be more accurate). Comparative genomic analysis of 14 sequenced Streptomyces genomes (G. Chandra and K. F. Chater, unpublished data) has shown that most of the global regulators described in the preceding sections are universal among streptomycetes, with an exception being the whiJ cluster and some of its paralogous clusters. Probably, therefore, the endogenous regulatory systems and their interplay are broadly the same in all streptomycetes, with species-specific responses to specialized environmental variables being conferred in part by constellations of WhiJ-like complexes. The acquisition or loss of sequences in promoters, increasing or decreasing the affinity of regulatory proteins, causes modest species-specific variation in the regulons controlled by conserved regulators, and some such changes may account for important species-specific differences in the phenotypes of regulatory mutants (242).
In this comparative genomic analysis, apparent orthologues of many of the global regulatory genes were also found in actinomycetes other than streptomycetes, often including morphologically and metabolically simple genera with small genomes. This information is summarized in Fig. 12 in the form of likely points in the phylogeny of actinomycetes at which each gene was acquired. Probably, the genes acquired earliest preceded the evolution of a complex secondary metabolism and have (or had) functions related to growth limitation. This fits with the broad range of attributes regulated by BldC, BldD, and WblA. From this point of view, relatively simple regulatory interfaces of gene clusters for antibiotic production with the ubiquitous global regulatory systems might be sufficient to ensure appropriate timing of antibiotic production in any host that the gene cluster enters through horizontal gene transfer, at least under some conditions in which production has a selective advantage. This would provide selective pressure for the retention of the gene cluster, which would presumably subsequently acquire regulatory adaptations increasing its value to the host.
Fig 12.
Acquisition of global regulators during the evolution of streptomycetes. The multilocus phylogeny on which the diagram is based was derived from a catenated set of seven conserved proteins (AtpD, DnaA, DnaG, DnaK, GyrB, RecA, and RpoB) (G. Chandra, unpublished data). Points of acquisition of global regulators are based on the presence or absence of orthologues encoded in the genomes of organisms before or after branch points.
ACTIVATION OF CRYPTIC SECONDARY METABOLITE GENE CLUSTERS
Sequencing of several Streptomyces genomes revealed the presence of a large number of secondary metabolic gene clusters for previously unsuspected products and thus the potential to produce many more natural products than had previously been recognized (233, 243, 244). It has therefore become necessary to devise methods and strategies to identify physiological signals and regulatory mechanisms that can activate these “cryptic” pathways, thus unleashing the full biosynthetic potential of these prodigious producers of valuable natural products (245, 246). Currently used or proposed methods include manipulation of fermentation conditions, the use of ecologically based tricks, genome mining, ribosome engineering, genetic manipulation of the regulators controlling gene clusters, regulation of signaling molecules, and heterologous expression of gene clusters in different host strains (Fig. 13).
Fig 13.
Strategies for the activation of cryptic secondary metabolic gene clusters in Streptomyces. The red line indicates the sequenced Streptomyces linear chromosome. Trapezoidal blocks with different colors represent different methods based on gene clusters to activate the possible expression of clusters.
Fermentation Conditions and Antibiotic Production
Traditionally, modifying fermentation conditions has been crucial for industrial yield improvement. Some of the molecular information described in this review may help to make such explorations increasingly knowledge led and may permit greater interplay between fermentation development and targeted genetic manipulations. However, empirical observations may continue to be useful; for example, it was recently reported that addition of the rare earth scandium to the fermentation medium significantly stimulated the production of actinorhodin in S. coelicolor, of actinomycin in Streptomyces antibioticus, and of streptomycin in S. griseus (247). Genomic techniques may also be useful: based on transcriptome analysis, several novel compounds were isolated from Streptomyces flaveolus through screening six different media (248). This suggests that combining changes of fermentation conditions with -omic studies may identify ways of awakening cryptic gene clusters for novel compounds.
Coculture with Organisms Sharing Common Ecology
Specific environmental signals or nutritional components required for the activation of cryptic gene clusters may in some cases reflect the presence of interacting microorganisms in the natural environment. Coculture can be an effective method for activating the production of cryptic metabolites. For example, the coculture of a Streptomyces strain with mycolic acid-containing Tsukamurella pulmonis from soil samples induced the Streptomyces strain to produce a novel antibiotic (249). It is also noteworthy that Streptomyces rapamycinicus can specifically induce expression of silent biosynthetic gene clusters in Aspergillus nidulans, which shares the same habitat (250). The activation is achieved by Streptomyces-triggered fungal histone acetylation modifications (251). The growing understanding of cross talk between Streptomyces and other species may provide more opportunities for the discovery of novel natural products. However, even though this approach is effective, it is still laborious and obscure. Therefore, miniaturized high-throughput screening methods need to be used. A recent paper (252) describes how the small molecules in and around a living bacterial colony can be identified and quantified over time and space by nanoscale mass spectrometry. The method rapidly generated profiles of complex interorganism signal responses at the adjacent edges of pairs of different organisms, such as B. subtilis and S. coelicolor. The procedure involves establishing a dynamic droplet (liquid bridge) held between a solvent input capillary and an outlet capillary leading to a nanospray mass spectrometry (MS) system (nano-DESI). A colony is brought into contact with the droplet, and the solvent-extractable molecules are drawn off and subjected to MS-MS. Covarying fragments are used to identify particular molecular species. The same colony can be analyzed repeatedly to generate a time lapse sequence. The method is able to generate new structural information that had been unobtainable by previous approaches. This approach to the discovery of new bioactive molecules through molecular ecology is exceptionally promising.
Ribosome Engineering and Related Strategies
On the basis of findings that certain mutations of ribosomal components activated antibiotic biosynthesis, Ochi proposed ribosome engineering as a means to activate Streptomyces cryptic gene clusters (253). With this strategy, strains were cultured on sublethal concentrations of antiribosomal (and some other) antibiotics to select antibiotic-resistant mutants, which were tested for secondary metabolite profiles. The feasibility of this concept was first exemplified by the activation of ACT production in S. lividans, in which the act genes are normally not expressed (107). Other examples are summarized elsewhere (253). This strategy was also successful with unknown compounds. Hosaka et al. (254) subjected 1,068 nonproducing actinomycetes to sublethal concentrations of rifampin, gentamicin, and streptomycin. Antibacterial activity against Staphylococcus aureus 209P was detected in 43% of Streptomyces strains. Eight novel piperidamycins were identified alone in derivatives of one strain carrying mutations in rpsL and rpoB. Mutations in rpoB increased affinities of mutant RNA polymerase for promoters of certain genes involved in piperidamycin biosynthesis, which in turn enhanced the transcription of these genes. Mutations in rpsL enhanced protein synthesis during late growth phase (254). Further understanding of the mechanisms will facilitate the discovery of novel bioactive compounds. It has proved useful to introduce rpsL and rpoB mutations into strains of S. coelicolor designed to maximize the expression of introduced heterologous sets of biosynthetic genes (255, 256).
Discovery of New Signaling Molecules
A lack of specific signaling molecules such as gamma-butyrolactones (see above) may be a factor causing the silence of gene clusters. Other substances promote secondary metabolism and morphogenesis at low concentrations in various actinomycetes. Examples include goadsporin, a 19-aa peptide containing four oxazole and two thiazole residues produced by Streptomyces sp. strain TP-A0584 (257, 258), and desferrioxamine E, a siderophore secreted by S. griseus (257, 258). The discovery of more signaling molecules and their use against genome-sequenced Streptomyces strains will speed up the process of cryptic gene cluster activation and deepen our understanding of the roles of signaling molecules (45).
Genetic Manipulation of CSR Genes
It is possible to activate the expression of cryptic gene cluster by genetic manipulation of CSR genes (activators and repressors) (192). A straightforward strategy, simply to overexpress activators or delete repressors (259), has proved effective. For example, amplification of sanG significantly increased nikkomycin production in S. ansochromogenes (222, 231), overexpression of polR resulted in a 2-fold increase of polyoxin in S. cacaoi subsp. asoensis (222, 231), and a giant type I modular PKS gene cluster of Streptomyces ambofaciens ATCC 23877, spanning almost 150 kb, was activated by the overexpression of a LAL family regulatory gene, leading to the discovery of stambomycins, unusual glycosylated macrolides with unique chemical structures and promising antiproliferative activity against human cancer cell lines (260). Similarly, the activation of the cryptic cpk gene cluster and production of a novel antibiotic followed the deletion of scbR2 in S. coelicolor (41), and transcription of cpk genes was also markedly increased in a mutant lacking the master regulator DasR (33). Deletion of the scbR2 homologue jadR2 in S. venezuelae activated jadomycin B production in the absence of the normally required stress challenges (51). The inactivation of a putative repressor gene, pgaY, in Streptomyces sp. strain PGA64 resulted in the production of two major angucycline metabolites, UWM6 and rabelomycin, which had not been detected in the parental wild-type strain, and many other metabolites were also produced in the mutant (261). Deletion of the negatively acting CSR gene alpW improved kinamycin production in S. ambofaciens from very low to workable levels (262).
Heterologous Expression in Engineered Hosts
Knowledge of regulatory factors has informed another method of activating cryptic gene clusters, i.e., heterologous expression of the cluster in different host strains, which is also often used to confirm the integrity of gene clusters synthesizing secondary metabolites (263–265) and for combinatorial biosynthesis to produce novel derivatives of bioactive secondary metabolites. In an early example, Martin et al. identified a cryptic PKS gene cluster in the genome of Streptomyces collinus by heterologous expression of the cluster in the first engineered host, S. coelicolor CH999 (266). More well-developed surrogate hosts have been derived in S. coelicolor and the industrial strain S. avermitilis. For example, Gomez-Escribano and Bibb (255) constructed a strain of S. coelicolor from which the gene clusters for ACT, RED, CDA, and CPK had been deleted and production-promoting mutations in rpoB and rpsL introduced. Using this strain, they were able to obtain high production levels of ACT, chloramphenicol, and congocidine (255). Another group also demonstrated the effectiveness of these strains by heterologous expression of novobiocin (267). An extensively engineered S. avermitilis host has also proved useful in the expression of heterologous clusters (268).
DEVELOPING ASPECTS
As is clear from this article, transcriptional regulation has been investigated extensively in the production of antibiotics and other secondary metabolites. However, little is known about the regulation of antibiotic biosynthesis at other levels. This regulation can occur both at the chromatin level (pretranscriptional) and the posttranscriptional level, via small noncoding RNAs (sRNAs) and the protein degradation machinery.
sRNAs
Small noncoding RNAs (sRNAs) control a wide variety of cellular processes in bacteria by modulating the stability and/or translation of mRNA targets or by modifying protein activity (269). Combinations of bioinformatic predictions and experimental approaches have facilitated the identification of many sRNAs in S. coelicolor and other species (52, 269–271), and what little is known about their functions points to a boost in the perceived importance of sRNA-mediated regulation of antibiotic production in the near future. Thus, the overexpression of scr5239, an sRNA recently identified in S. coelicolor, decreased ACT production, whereas depletion of scr5239 increased production (272), and overexpression of cnc2198.1as significantly decreased RED production (273). The mechanisms of these effects remain to be understood.
Protein Degradation Machinery
Transcriptional regulators or response proteins in antibiotic regulatory cascades may have to be removed after the fulfillment of their responsibilities, by target-specific or nonspecific proteases. Specificity of intracellular proteases is important for the efficient degradation of targeted proteins and the avoidance of undesired destruction of essential cellular proteins. Bacteria have different types of ATP-dependent proteases, including ClpAP, ClpCP, ClpEP, ClpXP, Lon, FtsH, HslUV, and the recently identified 20S proteasome (274, 275). The Clp complex consists of the proteolytic ClpP subunit and the ATPase regulatory subunit, ClpA, ClpC, ClpE, or ClpX. In contrast to only one clpP gene being present in most bacteria, five clpP-like (clpP1, clpP2, clpP3, clpP4, and clpP5) genes were identified in S. lividans (276), and at least three are present in all the streptomycetes represented in StrepDB. The Clp complex is a regulatory component of Streptomyces secondary metabolism, as demonstrated by increased ACT production in S. coelicolor and the activation of ACT production in S. lividans in clpX overexpression strains. Activation of ACT production in S. lividans is clpP1 dependent (276). It would be interesting to identify the ClpX targets implicated in antibiotic production. Involvement of other proteases in antibiotic production in other bacteria has been reported (277).
Proteasomes are large self-compartmentalized proteases found in archaea, eukaryotes, and certain actinobacteria, including Streptomyces, Frankia, and Mycobacterium (275, 278–280). The structure of proteasomes and their roles in intracellular protein degradation were summarized recently (281). The functions of the proteasome and its individual components have been investigated in Mycobacterium tuberculosis. The targeting of proteins to proteasomes requires the conjugation of a small “Pup” (prokaryotic ubiquitin-like protein) tag to substrates (275). The Streptomyces 20S proteasome was first identified in 1998, and its functions were initially investigated in 2007 (278, 282). A different protein profile was noticed in proteasome mutants of S. coelicolor, though no specific proteasome target was identified. Our preliminary data indicate that a Pup-like protein is a regulator of antibiotic production in S. coelicolor (G. Niu et al., unpublished data).
Epigenetic Regulation
Bacterial chromosomes are organized as nucleoids that, unlike the eukaryotic nucleus, are not surrounded by a nuclear membrane. Both the nucleoid and nucleus are compacted in ways that are compatible with DNA replication, chromosome segregation, and gene expression. It is well known that histones play important roles in nucleus packing, chromatin structure, and gene transcription in eukaryotes, and analogous roles are played by bacterial nucleoid-associated proteins (NAPs), which are significant regulators of nucleoid physical structure and gene transcription (283).
Manipulations of genes encoding heterochromatin protein 1 (HepA), histone methyltransferase (ClrD), and histone deacetylase (HDAC) resulted in modified chemical diversity in A. nidulans (284, 285). Application of DNA methyltransferase and HDAC inhibitors with different fungal genera resulted in enhanced chemical diversity of natural product profiles (285–287). These studies demonstrated that small-molecule epigenetic modifiers are effective tools for modifying biosynthetic pathways and for generating new compounds in fungi. In Streptomyces, both nucleoid-associated HU and Dps proteins affect nucleoid structure in spores of S. coelicolor (288), but little is known about the functions of NAPs in the regulation of gene transcription, especially in antibiotic biosynthesis. A more-detailed understanding of NAPs and their modifications is needed to know whether epigenetic modification exists in Streptomyces as prokaryotes. McArthur and Bibb developed an in vivo DNase I sensitivity method to monitor the correlation between the physical nucleoid and the transcriptional profile of Streptomyces. This approach provides an opportunity to characterize and compare the NAPs associated with the active and repressive portions of the nucleoid (289). If coupled with the decoy oligonucleotide technique developed by the same group (290), this approach can be used to identify histone-like proteins associated with antibiotic regulatory genes and to examine the role of these proteins in antibiotic production.
CONCLUDING REMARKS
The importance of understanding the regulation of antibiotic production has encouraged intensive international research, and the emerging picture is extremely complex. Basic cellular physiology (including nutrition, stress, developmental stage, and population density) plays a leading part in determining whether antibiotic production can go ahead, and the transmission of signals to specific sets of pathway genes involves conserved positively and negatively acting regulators, some of which are subject to regulation by small-molecule ligands or by phosphorylation. An emerging concept is that of cross talk between pathways, often mediated by unexpected promiscuity in the ligands and targets of regulatory proteins. Signal input and cross talk are coordinated in part by regulatory cascades and in part by the convergence of regulators upon common target promoters, especially those of cluster-situated regulators. Very often, there are multiple pathway-associated genes concerned with regulation, permitting signal input at more than one regulatory step. Both the nature of the regulators and the “wiring” of the cascades are diverse, but some features recur, such as the frequent involvement of SARPs, autoregulatory molecules such as gamma-butyrolactones and their receptor proteins, and TTA codons in regulatory genes. Other nascent areas expected to generate rapid advances in the near future include chromatin immunoprecipitation (ChIP)-Seq for the analysis of the targets of regulators and nano-DESI analysis of small molecules at the level of individual colonies. The complexity of these data will mean that system-level approaches will be needed to maximize information retrieval. These will greatly increase our understanding of the biology surrounding the production of antibiotics.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministry of Science and Technology of China (grant numbers 2009CB118905 and 2013CB734001) and the National Natural Science Foundation of China (grant number 31030003). K.F.C was supported by a John Innes Foundation Emeritus Fellowship.
Biographies

Gang Liu obtained his Ph.D. in microbial genetics in 1998 at the Institute of Microbiology, Chinese Academy of Sciences (CAS). From 1998 to 2001, he worked on fungal secondary metabolic biosynthesis and regulation at Leon University and Texas A&M University as a postdoctoral fellow. In 2001, he joined Professor Huarong Tan's group and started to work on the molecular regulation of secondary metabolic biosynthesis in Streptomyces. In 2007, he was appointed Full Professor in molecular microbiology at the Institute of Microbiology, CAS. His research focuses mainly on the regulation of microbial antibiotic production. So far, he has published more than 30 papers in this research field.

Keith F. Chater obtained his Ph.D. in Salmonella genetics in 1969 at the University of Birmingham, United Kingdom. Since then he has worked at the John Innes Centre, Norwich, United Kingdom, focusing on genetic approaches to the biological complexity of Streptomyces. He has published more than 200 primary research papers. He has been the Head of the Departments of Genetics and Molecular Microbiology and is now a John Innes Foundation Emeritus Fellow, an Honorary Professor at the Institute of Microbiology, Chinese Academy of Sciences (CAS), and a Fellow of The Royal Society.

Govind Chandra obtained a Ph.D. in botany at Kanpur University, Kanpur, India, before focusing on plant virology, with two postdoctoral periods at The National Botanical Research Institute in Lucknow. He joined the John Innes Centre, Norwich, United Kingdom, in 2000, initially as a visiting worker in the Department of Disease and Stress Biology before being appointed in 2001 as a bioinformaticist focusing on Streptomyces genomics in the Department of Molecular Microbiology. He has published 22 papers in this field. He maintains the StrepDB website (with on average 13,000 hits per week), thus providing genome-based bioinformatic support to the worldwide actinomycete community.

Guoqing Niu obtained his Ph.D. in microbial genetics in 2006 at the Institute of Microbiology, Chinese Academy of Sciences (CAS). After graduation, he worked for 4 years as Associate Research Scholar at The University of Oklahoma Health Sciences Center. In 2010, he was appointed Associate Professor in molecular microbiology at the Institute of Microbiology, CAS. His research focuses on the molecular regulation of secondary metabolites in Streptomyces. In the past few years, he has published 10 research papers.

Huarong Tan obtained his Ph.D. in microbial genetics in 1991 at the University of East Anglia, England. In 1992, he started to work on the molecular regulation of differentiation and secondary metabolites in Streptomyces. In 1994, he was appointed as Full Professor in molecular microbiology at the Institute of Microbiology, Chinese Academy of Sciences (CAS), and in 1999 to 2008, he was the Deputy Director of the Institute of Microbiology, CAS. In 2001 to 2011, he was a Vice President of the Chinese Society for Microbiology. From 2006 to the present, he has been an Editor in Chief for Acta Microbiologica Sinica. He has published more than 120 papers in his research field.
REFERENCES
- 1. Rudd BA. 1978. Genetics of pigmented secondary metabolites in Streptomyces coelicolor. University of East Anglia, Norwich, United Kingdom [Google Scholar]
- 2. Rudd BA, Hopwood DA. 1979. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J. Gen. Microbiol. 114: 35– 43 [DOI] [PubMed] [Google Scholar]
- 3. Rudd BA, Hopwood DA. 1980. A pigmented mycelial antibiotic in Streptomyces coelicolor: control by a chromosomal gene cluster. J. Gen. Microbiol. 119: 333– 340 [DOI] [PubMed] [Google Scholar]
- 4. Kirby R, Wright LF, Hopwood DA. 1975. Plasmid-determined antibiotic synthesis and resistance in Streptomyces coelicolor. Nature 254: 265– 267 [DOI] [PubMed] [Google Scholar]
- 5. Chater KF, Bruton CJ. 1985. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 4: 1893– 1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Malpartida F, Hopwood DA. 1986. Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 205: 66– 73 [DOI] [PubMed] [Google Scholar]
- 7. Malpartida F, Niemi J, Navarrete R, Hopwood DA. 1990. Cloning and expression in a heterologous host of the complete set of genes for biosynthesis of the Streptomyces coelicolor antibiotic undecylprodigiosin. Gene 93: 91– 99 [DOI] [PubMed] [Google Scholar]
- 8. Nett M, Ikeda H, Moore BS. 2009. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 26: 1362– 1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34: 171– 198 [DOI] [PubMed] [Google Scholar]
- 10. Hofer I, Crusemann M, Radzom M, Geers B, Flachshaar D, Cai X, Zeeck A, Piel J. 2011. Insights into the biosynthesis of hormaomycin, an exceptionally complex bacterial signaling metabolite. Chem. Biol. 18: 381– 391 [DOI] [PubMed] [Google Scholar]
- 11. Yim G, Wang HH, Davies J. 2007. Antibiotics as signalling molecules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362: 1195– 1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCormick JR, Flärdh K. 2012. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36: 206– 231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen OM, Sletta H, Alam MT, Merlo ME, Moore J, Omara WA, Morrissey ER, Juarez-Hermosillo MA, Rodriguez-Garcia A, Nentwich M, Thomas L, Iqbal M, Legaie R, Gaze WH, Challis GL, Jansen RC, Dijkhuizen L, Rand DA, Wild DL, Bonin M, Reuther J, Wohlleben W, Smith MC, Burroughs NJ, Martin JF, Hodgson DA, Takano E, Breitling R, Ellingsen TE, Wellington EM. 2010. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11: 10 doi:10.1186/1471-2164-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coisne S, Bechet M, Blondeau R. 1999. Actinorhodin production by Streptomyces coelicolor A3(2) in iron-restricted media. Lett. Appl. Microbiol. 28:199– 202 [DOI] [PubMed] [Google Scholar]
- 15. Owen GA, Pascoe B, Kallifidas D, Paget MS. 2007. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and sigmaR. J. Bacteriol. 189: 4078– 4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai RP, Leaf T, Woo E, Licari P. 2002. Enhanced production of heterologous macrolide aglycones by fed-batch cultivation of Streptomyces coelicolor. J. Ind. Microbiol. Biotechnol. 28: 297– 301 [DOI] [PubMed] [Google Scholar]
- 17. Ronnest NP, Stocks SM, Eliasson Lantz A, Gernaey KV. 2011. Introducing process analytical technology (PAT) in filamentous cultivation process development: comparison of advanced online sensors for biomass measurement. J. Ind. Microbiol. Biotechnol. 38: 1679– 1690 [DOI] [PubMed] [Google Scholar]
- 18. Bibb MJ. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8: 208– 215 [DOI] [PubMed] [Google Scholar]
- 19. Martin JF, Liras P. 2010. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13: 263– 273 [DOI] [PubMed] [Google Scholar]
- 20. van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28: 1311– 1333 [DOI] [PubMed] [Google Scholar]
- 21. Martín JF, Liras P. 2012. Cascades and networks of regulatory genes that control antibiotic biosynthesis. Subcell. Biochem. 64: 115– 138 [DOI] [PubMed] [Google Scholar]
- 22. Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Avalos M, Guzman-Trampe S, Rodriguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J. Antibiot. (Tokyo) 63: 442– 459 [DOI] [PubMed] [Google Scholar]
- 23. Chater K. 1999. David Hopwood and the emergence of Streptomyces genetics. Int. Microbiol. 2: 61– 68 [PubMed] [Google Scholar]
- 24. Hopwood DA. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40: 1– 23 [DOI] [PubMed] [Google Scholar]
- 25. Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. Oxford University Press, New York, NY [Google Scholar]
- 26. Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih C- J, Kao CM, Buttner MJ, Cohen SN. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol. Microbiol. 58: 1276– 1287 [DOI] [PubMed] [Google Scholar]
- 27. Shin JH, Singh AK, Cheon DJ, Roe JH. 2011. Activation of the SoxR regulon in Streptomyces coelicolor by the extracellular form of the pigmented antibiotic actinorhodin. J. Bacteriol. 193: 75– 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iqbal M, Mast Y, Amin R, Hodgson DA, Wohlleben W, Burroughs NJ. 2012. Extracting regulator activity profiles by integration of de novo motifs and expression data: characterizing key regulators of nutrient depletion responses in Streptomyces coelicolor. Nucleic Acids Res. 40: 5227– 5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wietzorrek A, Bibb M. 1997. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25: 1181– 1184 [DOI] [PubMed] [Google Scholar]
- 30. Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69: 431– 439 [DOI] [PubMed] [Google Scholar]
- 31. Sheeler NL, MacMillan SV, Nodwell JR. 2005. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J. Bacteriol. 187: 687– 696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uguru GC, Stephens KE, Stead JA, Towle JE, Baumberg S, McDowall KJ. 2005. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol. Microbiol. 58: 131– 150 [DOI] [PubMed] [Google Scholar]
- 33. Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9: 670– 675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang R, Mast Y, Wang J, Zhang W, Zhao G, Wohlleben W, Lu Y, Jiang W. 26 October 2012. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol. Microbiol doi:10.1111/mmi.12080 [DOI] [PubMed] [Google Scholar]
- 35. Yu Z, Zhu H, Dang F, Zhang W, Qin Z, Yang S, Tan H, Lu Y, Jiang W. 2012. Differential regulation of antibiotic biosynthesis by DraR-K, a novel two-component system in Streptomyces coelicolor. Mol. Microbiol. 85: 535– 556 [DOI] [PubMed] [Google Scholar]
- 36. Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 75: 1133– 1144 [DOI] [PubMed] [Google Scholar]
- 37. Heo GY, Kim WC, Joo GJ, Kwak YY, Shin JH, Roh DH, Park HD, Rhee IK. 2008. Deletion of xylR gene enhances expression of xylose isomerase in Streptomyces lividans TK24. J. Microbiol. Biotechnol. 18: 837– 844 [PubMed] [Google Scholar]
- 38. Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 36: 1073– 1083 [DOI] [PubMed] [Google Scholar]
- 39. Fernandez-Moreno MA, Caballero JL, Hopwood DA, Malpartida F. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66: 769– 780 [DOI] [PubMed] [Google Scholar]
- 40. Tahlan K, Yu Z, Xu Y, Davidson AR, Nodwell JR. 2008. Ligand recognition by ActR, a TetR-like regulator of actinorhodin export. J. Mol. Biol. 383: 753– 761 [DOI] [PubMed] [Google Scholar]
- 41. Gottelt M, Kol S, Gomez-Escribano JP, Bibb M, Takano E. 2010. Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156: 2343– 2353 [DOI] [PubMed] [Google Scholar]
- 42. Pawlik K, Kotowska M, Kolesinski P. 2010. Streptomyces coelicolor A3(2) produces a new yellow pigment associated with the polyketide synthase Cpk. J. Mol. Microbiol. Biotechnol. 19: 147– 151 [DOI] [PubMed] [Google Scholar]
- 43. Takano E, Kinoshita H, Mersinias V, Bucca G, Hotchkiss G, Nihira T, Smith CP, Bibb M, Wohlleben W, Chater K. 2005. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56: 465– 479 [DOI] [PubMed] [Google Scholar]
- 44. Hsiao NH, Soding J, Linke D, Lange C, Hertweck C, Wohlleben W, Takano E. 2007. ScbA from Streptomyces coelicolor A3(2) has homology to fatty acid synthases and is able to synthesize gamma-butyrolactones. Microbiology 153: 1394– 1404 [DOI] [PubMed] [Google Scholar]
- 45. Sidda JD, Corre C. 2012. Gamma-butyrolactone and furan signaling systems in streptomyces. Methods Enzymol. 517: 71– 87 [DOI] [PubMed] [Google Scholar]
- 46. Chatterjee A, Drews L, Mehra S, Takano E, Kaznessis YN, Hu WS. 2011. Convergent transcription in the butyrolactone regulon in Streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS One 6: e21974 doi:10.1371/journal.pone.0021974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mehra S, Charaniya S, Takano E, Hu WS. 2008. A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS One 3:e2724 doi:10.1371/journal.pone.0002724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gottelt M, Hesketh A, Bunet R, Puri P, Takano E. 2012. Characterisation of a natural variant of the gamma-butyrolactone signalling receptor. BMC Res. Notes 5:379 doi:10.1186/1756-0500-5-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Butler MJ, Takano E, Bruheim P, Jovetic S, Marinelli F, Bibb MJ. 2003. Deletion of scbA enhances antibiotic production in Streptomyces lividans. Appl. Microbiol. Biotechnol. 61:512– 516 [DOI] [PubMed] [Google Scholar]
- 50. D'Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E. 2011. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2). Microb. Biotechnol. 4: 239– 251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu G, Wang J, Wang L, Tian X, Yang H, Fan K, Yang K, Tan H. 2010. “Pseudo” gamma-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285: 27440– 27448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vockenhuber MP, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B. 2011. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 8: 468– 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daniels C, Daddaoua A, Lu D, Zhang X, Ramos JL. 2010. Domain cross-talk during effector binding to the multidrug binding TTGR regulator. J. Biol. Chem. 285: 21372– 21381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peters KM, Sharbeen G, Theis T, Skurray RA, Brown MH. 2009. Biochemical characterization of the multidrug regulator QacR distinguishes residues that are crucial to multidrug binding and induction of qacA transcription. Biochemistry 48: 9794– 9800 [DOI] [PubMed] [Google Scholar]
- 55. Wang J, Wang W, Wang L, Zhang G, Fan K, Tan H, Yang K. 2011. A novel role of ‘pseudo'gamma-butyrolactone receptors in controlling gamma-butyrolactone biosynthesis in Streptomyces. Mol. Microbiol. 82: 236– 250 [DOI] [PubMed] [Google Scholar]
- 56. Allenby NE, Laing E, Bucca G, Kierzek AM, Smith CP. 2012. Diverse control of metabolism and other cellular processes in Streptomyces coelicolor by the PhoP transcription factor: genome-wide identification of in vivo targets. Nucleic Acids Res. 40: 9543– 9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O'Rourke S, Wietzorrek A, Fowler K, Corre C, Challis GL, Chater KF. 2009. Extracellular signalling, translational control, two repressors and an activator all contribute to the regulation of methylenomycin production in Streptomyces coelicolor. Mol. Microbiol. 71: 763– 778 [DOI] [PubMed] [Google Scholar]
- 58. Corre C, Song L, O'Rourke S, Chater KF, Challis GL. 2008. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. U. S. A. 105: 17510– 17515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hayes A, Hobbs G, Smith CP, Oliver SG, Butler PR. 1997. Environmental signals triggering methylenomycin production by Streptomyces coelicolor A3(2). J. Bacteriol. 179: 5511– 5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479– 506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu Q, van Wezel GP, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, Deacon AM, Wilson IA. 2012. Structure of an MmyB-Like regulator from C. aurantiacus, member of a new transcription factor family linked to antibiotic metabolism in Actinomycetes. PLoS One 7: e41359 doi:10.1371/journal.pone.0041359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hobbs G, Obanye AI, Petty J, Mason JC, Barratt E, Gardner DC, Flett F, Smith CP, Broda P, Oliver SG. 1992. An integrated approach to studying regulation of production of the antibiotic methylenomycin by Streptomyces coelicolor A3(2). J. Bacteriol. 174:1487– 1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Neal RJ, Chater KF. 1991. Bidirectional promoter and terminator regions bracket mmr, a resistance gene embedded in the Streptomyces coelicolor A3(2) gene cluster encoding methylenomycin production. Gene 100: 75– 83 [DOI] [PubMed] [Google Scholar]
- 64. Anderson TB, Brian P, Champness WC. 2001. Genetic and transcriptional analysis of absA, an antibiotic gene cluster-linked two-component system that regulates multiple antibiotics in Streptomyces coelicolor. Mol. Microbiol. 39: 553– 566 [DOI] [PubMed] [Google Scholar]
- 65. Shu D, Chen L, Wang W, Yu Z, Ren C, Zhang W, Yang S, Lu Y, Jiang W. 2009. afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 81: 1149– 1160 [DOI] [PubMed] [Google Scholar]
- 66. Cerdeno AM, Bibb MJ, Challis GL. 2001. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 8: 817– 829 [DOI] [PubMed] [Google Scholar]
- 67. White J, Bibb M. 1997. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179: 627– 633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guthrie EP, Flaxman CS, White J, Hodgson DA, Bibb MJ, Chater KF. 1998. A response-regulator-like activator of antibiotic synthesis from Streptomyces coelicolor A3(2) with an amino-terminal domain that lacks a phosphorylation pocket. Microbiology 144: 727– 738 [DOI] [PubMed] [Google Scholar]
- 69. Wang L, Tian X, Wang J, Yang H, Fan K, Xu G, Yang K, Tan H. 2009. Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U. S. A. 106: 8617– 8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Apel AK, Sola-Landa A, Rodriguez-Garcia A, Martin JF. 2007. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology. 153: 3527– 3537 [DOI] [PubMed] [Google Scholar]
- 71. Martin JF. 2004. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J. Bacteriol. 186: 5197– 5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Santos-Beneit F, Barriuso-Iglesias M, Fernandez-Martinez LT, Martinez-Castro M, Sola-Landa A, Rodriguez-Garcia A, Martin JF. 2011. The RNA polymerase omega factor RpoZ is regulated by PhoP and has an important role in antibiotic biosynthesis and morphological differentiation in Streptomyces coelicolor. Appl. Environ. Microbiol. 77: 7586– 7594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sola-Landa A, Moura RS, Martin JF. 2003. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl. Acad. Sci. U. S. A. 100: 6133– 6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Asturias JA, Martin JF, Liras P. 1994. Biosynthesis and phosphate control of candicidin by Streptomyces acrimycini JI2236: effect of amplification of the pabAB gene. J. Ind. Microbiol. 13: 183– 189 [DOI] [PubMed] [Google Scholar]
- 75. McDowall KJ, Thamchaipenet A, Hunter IS. 1999. Phosphate control of oxytetracycline production by Streptomyces rimosus is at the level of transcription from promoters overlapped by tandem repeats similar to those of the DNA-binding sites of the OmpR family. J. Bacteriol. 181: 3025– 3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mendes MV, Tunca S, Anton N, Recio E, Sola-Landa A, Aparicio JF, Martin JF. 2007. The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab. Eng. 9: 217– 227 [DOI] [PubMed] [Google Scholar]
- 77. Tang Z, Xiao C, Zhuang Y, Chu J, Zhang S, Herron PR, Hunter IS, Guo M. 2011. Improved oxytetracycline production in Streptomyces rimosus M4018 by metabolic engineering of the G6PDH gene in the pentose phosphate pathway. Enzyme Microb. Technol. 49: 17– 24 [DOI] [PubMed] [Google Scholar]
- 78. Sola-Landa A, Rodriguez-Garcia A, Franco-Dominguez E, Martin JF. 2005. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 56: 1373– 1385 [DOI] [PubMed] [Google Scholar]
- 79. Oganesyan V, Oganesyan N, Adams PD, Jancarik J, Yokota HA, Kim R, Kim SH. 2005. Crystal structure of the “PhoU-like” phosphate uptake regulator from Aquifex aeolicus. J. Bacteriol. 187: 4238– 4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Diaz M, Esteban A, Fernandez-Abalos JM, Santamaria RI. 2005. The high-affinity phosphate-binding protein PstS is accumulated under high fructose concentrations and mutation of the corresponding gene affects differentiation in Streptomyces lividans. Microbiology 151: 2583– 2592 [DOI] [PubMed] [Google Scholar]
- 81. Santos-Beneit F, Rodriguez-Garcia A, Franco-Dominguez E, Martin JF. 2008. Phosphate-dependent regulation of the low- and high-affinity transport systems in the model actinomycete Streptomyces coelicolor. Microbiology 154: 2356– 2370 [DOI] [PubMed] [Google Scholar]
- 82. Santos-Beneit F, Rodriguez-Garcia A, Sola-Landa A, Martin JF. 2009. Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72: 53– 68 [DOI] [PubMed] [Google Scholar]
- 83. Martin JF, Sola-Landa A, Santos-Beneit F, Fernandez-Martinez LT, Prieto C, Rodriguez-Garcia A. 2011. Cross-talk of global nutritional regulators in the control of primary and secondary metabolism in Streptomyces. Microb. Biotechnol. 4: 165– 174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rodriguez-Garcia A, Sola-Landa A, Apel K, Santos-Beneit F, Martin JF. 2009. Phosphate control over nitrogen metabolism in Streptomyces coelicolor: direct and indirect negative control of glnR, glnA, glnII and amtB expression by the response regulator PhoP. Nucleic Acids Res. 37: 3230– 3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chater KF. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361: 761– 768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chi WJ, Lee SY, Lee J. 2011. Functional analysis of SGR4635-induced enhancement of pigmented antibiotic production in Streptomyces lividans. J. Microbiol. 49: 828– 833 [DOI] [PubMed] [Google Scholar]
- 87. Usuki H, Yamamoto Y, Kumagai Y, Nitoda T, Kanzaki H, Hatanaka T. 2011. MS/MS fragmentation-guided search of TMG-chitooligomycins and their structure-activity relationship in specific beta-N-acetylglucosaminidase inhibition. Org. Biomol. Chem. 9: 2943– 2951 [DOI] [PubMed] [Google Scholar]
- 88. Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Muller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61: 1237– 1251 [DOI] [PubMed] [Google Scholar]
- 89. Nazari B, Kobayashi M, Saito A, Hassaninasab A, Miyashita K, Fujii T. 2 November 2012. Chitin-induced gene expression involved in secondary metabolic pathways in Streptomyces coelicolor A3(2) grown in soil. Appl. Environ. Microbiol. doi:10.1128/AEM.02217-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hodgson DA. 1982. Glucose repression of carbon source uptake and metabolism in Streptomyces coelicolor A3(2) and its perturbation in mutants resistant to 2-deoxyglucose. J. Gen. Microbiol. 128: 2417– 2430 [Google Scholar]
- 91. van Wezel GP, Konig M, Mahr K, Nothaft H, Thomae AW, Bibb M, Titgemeyer F. 2007. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in streptomyces coelicolor A3(2). J. Mol. Microbiol. Biotechnol. 12: 67– 74 [DOI] [PubMed] [Google Scholar]
- 92. Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP. 18 October 2012. Identification of glucose kinase-dependent and -independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol. Microbiol. doi:10.1111/mmi.12072 [DOI] [PubMed] [Google Scholar]
- 93. Waldvogel E, Herbig A, Battke F, Amin R, Nentwich M, Nieselt K, Ellingsen TE, Wentzel A, Hodgson DA, Wohlleben W, Mast Y. 2011. The PII protein GlnK is a pleiotropic regulator for morphological differentiation and secondary metabolism in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 92: 1219– 1236 [DOI] [PubMed] [Google Scholar]
- 94. Yang YH, Song E, Kim EJ, Lee K, Kim WS, Park SS, Hahn JS, Kim BG. 2009. NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 82: 501– 511 [DOI] [PubMed] [Google Scholar]
- 95. Kim YJ, Moon MH, Song JY, Smith CP, Hong SK, Chang YK. 2008. Acidic pH shock induces the expressions of a wide range of stress-response genes. BMC Genomics 9: 604 doi:10.1186/1471-2164-9-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sharma UK, Chatterji D. 2010. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34:646– 657 [DOI] [PubMed] [Google Scholar]
- 97. Hesketh A, Sun J, Bibb M. 2001. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 39: 136– 144 [DOI] [PubMed] [Google Scholar]
- 98. Kang SG, Jin W, Bibb M, Lee KJ. 1998. Actinorhodin and undecylprodigiosin production in wild-type and relA mutant strains of Streptomyces coelicolor A3(2) grown in continuous culture. FEMS Microbiol. Lett. 168: 221– 226 [DOI] [PubMed] [Google Scholar]
- 99. Holt TG, Chang C, Laurent-Winter C, Murakami T, Garrels JI, Davies JE, Thompson CJ. 1992. Global changes in gene expression related to antibiotic synthesis in Streptomyces hygroscopicus. Mol. Microbiol. 6: 969– 980 [DOI] [PubMed] [Google Scholar]
- 100. Jin W, Ryu YG, Kang SG, Kim SK, Saito N, Ochi K, Lee SH, Lee KJ. 2004. Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology 150: 1485– 1493 [DOI] [PubMed] [Google Scholar]
- 101. Gomez-Escribano JP, Martin JF, Hesketh A, Bibb MJ, Liras P. 2008. Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154: 744– 755 [DOI] [PubMed] [Google Scholar]
- 102. Feng WH, Mao XM, Liu ZH, Li YQ. 2011. The ECF sigma factor SigT regulates actinorhodin production in response to nitrogen stress in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 92: 1009– 1021 [DOI] [PubMed] [Google Scholar]
- 103. Lai C, Xu J, Tozawa Y, Okamoto-Hosoya Y, Yao X, Ochi K. 2002. Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology 148: 3365– 3373 [DOI] [PubMed] [Google Scholar]
- 104. Xu J, Tozawa Y, Lai C, Hayashi H, Ochi K. 2002. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Genet. Genomics 268: 179– 189 [DOI] [PubMed] [Google Scholar]
- 105. Piir K, Paier A, Liiv A, Tenson T, Maivali U. 2011. Ribosome degradation in growing bacteria. EMBO Rep. 12: 458– 462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. 2008. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 36: 732– 741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shima J, Hesketh A, Okamoto S, Kawamoto S, Ochi K. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 178: 7276– 7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hosaka T, Xu J, Ochi K. 2006. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol. Microbiol. 61: 883– 897 [DOI] [PubMed] [Google Scholar]
- 109. Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. 2007. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 189: 3876– 3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Agarwal D, Gregory ST, O'Connor M. 2011. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J. Mol. Biol. 410: 1– 9 [DOI] [PubMed] [Google Scholar]
- 111. Yang H, Wang L, Xie Z, Tian Y, Liu G, Tan H. 2007. The tyrosine degradation gene hppD is transcriptionally activated by HpdA and repressed by HpdR in Streptomyces coelicolor, while hpdA is negatively autoregulated and repressed by HpdR. Mol. Microbiol. 65: 1064– 1077 [DOI] [PubMed] [Google Scholar]
- 112. Yang H, An Y, Wang L, Zhang S, Zhang Y, Tian Y, Liu G, Tan H. 2010. Autoregulation of hpdR and its effect on CDA biosynthesis in Streptomyces coelicolor. Microbiology 156: 2641– 2648 [DOI] [PubMed] [Google Scholar]
- 113. Andreini C, Banci L, Bertini I, Rosato A. 2006. Zinc through the three domains of life. J. Proteome Res. 5: 3173– 3178 [DOI] [PubMed] [Google Scholar]
- 114. Hesketh A, Kock H, Mootien S, Bibb M. 2009. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 74: 1427– 1444 [DOI] [PubMed] [Google Scholar]
- 115. Kallifidas D, Pascoe B, Owen GA, Strain-Damerell CM, Hong HJ, Paget MS. 2010. The zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor. J. Bacteriol. 192: 608– 611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Champness W, Riggle P, Adamidis T, Vandervere P. 1992. Identification of Streptomyces coelicolor genes involved in regulation of antibiotic synthesis. Gene 115: 55– 60 [DOI] [PubMed] [Google Scholar]
- 117. Floriano B, Bibb M. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21: 385– 396 [DOI] [PubMed] [Google Scholar]
- 118. Horinouchi S, Kito M, Nishiyama M, Furuya K, Hong SK, Miyake K, Beppu T. 1990. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2). Gene 95: 49– 56 [DOI] [PubMed] [Google Scholar]
- 119. Horinouchi S. 2003. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 30: 462– 467 [DOI] [PubMed] [Google Scholar]
- 120. Maharjan S, Oh TJ, Lee HC, Sohng JK. 2009. Identification and functional characterization of an afsR homolog regulatory gene from Streptomyces venezuelae ATCC 15439. J. Microbiol. Biotechnol. 19: 121– 127 [DOI] [PubMed] [Google Scholar]
- 121. Parajuli N, Viet HT, Ishida K, Tong HT, Lee HC, Liou K, Sohng JK. 2005. Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res. Microbiol. 156: 707– 712 [DOI] [PubMed] [Google Scholar]
- 122. van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. 2008. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59: 1383– 1397 [DOI] [PubMed] [Google Scholar]
- 123. Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302– 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Parker JL, Jones AM, Serazetdinova L, Saalbach G, Bibb MJ, Naldrett MJ. 2010. Analysis of the phosphoproteome of the multicellular bacterium Streptomyces coelicolor A3(2) by protein/peptide fractionation, phosphopeptide enrichment and high-accuracy mass spectrometry. Proteomics 10: 2486– 2497 [DOI] [PubMed] [Google Scholar]
- 125. Umeyama T, Horinouchi S. 2001. Autophosphorylation of a bacterial serine/threonine kinase, AfsK, is inhibited by KbpA, an AfsK-binding protein. J. Bacteriol. 183: 5506– 5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ando N, Ueda K, Horinouchi S. 1997. A Streptomyces griseus gene (sgaA) suppresses the growth disturbance caused by high osmolality and a high concentration of A-factor during early growth. Microbiology 143: 2715– 2723 [DOI] [PubMed] [Google Scholar]
- 127. Umeyama T, Lee PC, Ueda K, Horinouchi S. 1999. An AfsK/AfsR system involved in the response of aerial mycelium formation to glucose in Streptomyces griseus. Microbiology 145: 2281– 2292 [DOI] [PubMed] [Google Scholar]
- 128. Sawai R, Suzuki A, Takano Y, Lee PC, Horinouchi S. 2004. Phosphorylation of AfsR by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). Gene 334: 53– 61 [DOI] [PubMed] [Google Scholar]
- 129. Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, Parker JL, Richards DM, Jung YG, Buttner MJ, Flardh K. 2012. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 109: E2371– 2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim DJ, Huh JH, Yang YY, Kang CM, Lee IH, Hyun CG, Hong SK, Suh JW. 2003. Accumulation of S-adenosyl-l-methionine enhances production of actinorhodin but inhibits sporulation in Streptomyces lividans TK23. J. Bacteriol. 185: 592– 600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Okamoto S, Lezhava A, Hosaka T, Okamoto-Hosoya Y, Ochi K. 2003. Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3(2). J. Bacteriol. 185: 601– 609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang X, Fen M, Shi X, Bai L, Zhou P. 2008. Overexpression of yeast S-adenosylmethionine synthetase metK in Streptomyces actuosus leads to increased production of nosiheptide. Appl. Microbiol. Biotechnol. 78: 991– 995 [DOI] [PubMed] [Google Scholar]
- 133. Zhao XQ, Gust B, Heide L. 2010. S-Adenosylmethionine (SAM) and antibiotic biosynthesis: effect of external addition of SAM and of overexpression of SAM biosynthesis genes on novobiocin production in Streptomyces. Arch. Microbiol. 192: 289– 297 [DOI] [PubMed] [Google Scholar]
- 134. Tanaka A, Takano Y, Ohnishi Y, Horinouchi S. 2007. AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J. Mol. Biol. 369: 322– 333 [DOI] [PubMed] [Google Scholar]
- 135. Vogtli M, Chang PC, Cohen SN. 1994. afsR2: a previously undetected gene encoding a 63-amino-acid protein that stimulates antibiotic production in Streptomyces lividans. Mol. Microbiol. 14: 643– 653 [DOI] [PubMed] [Google Scholar]
- 136. Lian W, Jayapal KP, Charaniya S, Mehra S, Glod F, Kyung YS, Sherman DH, Hu WS. 2008. Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2). BMC Genomics 9: 56 doi:10.1186/1471-2164-9-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. 2004. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2). Microbiology 150:2795– 2806 [DOI] [PubMed] [Google Scholar]
- 138. Chang HM, Chen MY, Shieh YT, Bibb MJ, Chen CW. 1996. The cutRS signal transduction system of Streptomyces lividans represses the biosynthesis of the polyketide antibiotic actinorhodin. Mol. Microbiol. 21: 1075– 1085 [PubMed] [Google Scholar]
- 139. Li YQ, Chen PL, Chen SF, Wu D, Zheng J. 2004. A pair of two-component regulatory genes ecrA1/A2 in Streptomyces coelicolor. J. Zhejiang Univ. Sci. 5: 173– 179 [DOI] [PubMed] [Google Scholar]
- 140. Lu Y, Wang W, Shu D, Zhang W, Chen L, Qin Z, Yang S, Jiang W. 2007. Characterization of a novel two-component regulatory system involved in the regulation of both actinorhodin and a type I polyketide in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 77: 625– 635 [DOI] [PubMed] [Google Scholar]
- 141. Wang W, Shu D, Chen L, Jiang W, Lu Y. 2009. Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol. Lett. 294: 150– 156 [DOI] [PubMed] [Google Scholar]
- 142. Rozas D, Gullon S, Mellado RP. 2012. A novel two-component system involved in the transition to secondary metabolism in Streptomyces coelicolor. PLoS One 7: e31760 doi:10.1371/journal.pone.0031760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Yepes A, Rico S, Rodriguez-Garcia A, Santamaria RI, Diaz M. 2011. Novel two-component systems implied in antibiotic production in Streptomyces coelicolor. PLoS One 6:e19980 doi:10.1371/journal.pone.0019980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Carmody M, Byrne B, Murphy B, Breen C, Lynch S, Flood E, Finnan S, Caffrey P. 2004. Analysis and manipulation of amphotericin biosynthetic genes by means of modified phage KC515 transduction techniques. Gene 343:107– 115 [DOI] [PubMed] [Google Scholar]
- 145. Sekurova ON, Brautaset T, Sletta H, Borgos SE, Jakobsen MO, Ellingsen TE, Strom AR, Valla S, Zotchev SB. 2004. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 186: 1345– 1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wilson DJ, Xue Y, Reynolds KA, Sherman DH. 2001. Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J. Bacteriol. 183: 3468– 3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Aparicio JF, Molnar I, Schwecke T, Konig A, Haydock SF, Khaw LE, Staunton J, Leadlay PF. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169: 9– 16 [DOI] [PubMed] [Google Scholar]
- 148. Guerra SM, Rodriguez-Garcia A, Santos-Aberturas J, Vicente CM, Payero TD, Martin JF, Aparicio JF. 2012. LAL regulators SCO0877 and SCO7173 as pleiotropic modulators of phosphate starvation response and actinorhodin biosynthesis in Streptomyces coelicolor. PLoS One 7: e31475 doi:10.1371/journal.pone.0031475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Jones GH. 2010. RNA degradation and the regulation of antibiotic synthesis in Streptomyces. Future Microbiol. 5:419– 429 [DOI] [PubMed] [Google Scholar]
- 150. Chang SA, Bralley P, Jones GH. 2005. The absB gene encodes a double strand-specific endoribonuclease that cleaves the read-through transcript of the rpsO-pnp operon in Streptomyces coelicolor. J. Biol. Chem. 280: 33213– 33219 [DOI] [PubMed] [Google Scholar]
- 151. Aceti DJ, Champness WC. 1998. Transcriptional regulation of Streptomyces coelicolor pathway-specific antibiotic regulators by the absA and absB loci. J. Bacteriol. 180: 3100– 3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Xu W, Huang J, Lin R, Shi J, Cohen SN. 2010. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75: 781– 791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Gatewood ML, Bralley P, Weil MR, Jones GH. 2012. RNA-Seq and RNA immunoprecipitation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J. Bacteriol. 194: 2228– 2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sello JK, Buttner MJ. 2008. The gene encoding RNase III in Streptomyces coelicolor is transcribed during exponential phase and is required for antibiotic production and for proper sporulation. J. Bacteriol. 190: 4079– 4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bralley P, Jones GH. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149: 2173– 2182 [DOI] [PubMed] [Google Scholar]
- 156. Gatewood ML, Bralley P, Jones GH. 2011. RNase III-dependent expression of the rpsO-pnp operon of Streptomyces coelicolor. J. Bacteriol. 193: 4371– 4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Li W, Wu J, Tao W, Zhao C, Wang Y, He X, Chandra G, Zhou X, Deng Z, Chater KF, Tao M. 2007. A genetic and bioinformatic analysis of Streptomyces coelicolor genes containing TTA codons, possible targets for regulation by a developmentally significant tRNA. FEMS Microbiol. Lett. 266: 20– 28 [DOI] [PubMed] [Google Scholar]
- 158. Chater KF, Chandra G. 2008. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J. Microbiol. 46: 1– 11 [DOI] [PubMed] [Google Scholar]
- 159. Chandra G, Chater KF. 2008. Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie Van Leeuwenhoek 94: 111– 126 [DOI] [PubMed] [Google Scholar]
- 160. Jensen SE. 5 September 2012. Biosynthesis of clavam metabolites. J. Ind. Microbiol. Biotechnol. doi:10.1007/s10295-012-1191-0 [DOI] [PubMed] [Google Scholar]
- 161. Trepanier NK, Jensen SE, Alexander DC, Leskiw BK. 2002. The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology 148: 643– 656 [DOI] [PubMed] [Google Scholar]
- 162. Champness WC. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170: 1168– 1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lopez-Garcia MT, Santamarta I, Liras P. 2010. Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology 156: 2354– 2365 [DOI] [PubMed] [Google Scholar]
- 164. Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185: 7291– 7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34: 102– 111 [DOI] [PubMed] [Google Scholar]
- 166. Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50: 475– 486 [DOI] [PubMed] [Google Scholar]
- 167. Yao MD, Miyazono KI, Ohtsuka J, Hirano S, Nagata K, Horinouchi S, Ohnishi Y, Tanokura M. 2012. Purification, crystallization and preliminary X-ray analysis of the DNA-binding domain of AdpA, the central transcription factor in the A-factor regulatory cascade in the filamentous bacterium Streptomyces griseus, in complex with a duplex DNA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68: 946– 949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Xu D, Kim TJ, Park ZY, Lee SK, Yang SH, Kwon HJ, Suh JW. 2009. A DNA-binding factor, ArfA, interacts with the bldH promoter and affects undecylprodigiosin production in Streptomyces lividans. Biochem. Biophys. Res. Commun. 379: 319– 323 [DOI] [PubMed] [Google Scholar]
- 169. Yang YH, Song E, Kim JN, Lee BR, Kim EJ, Park SH, Kim WS, Park HY, Jeon JM, Rajesh T, Kim YG, Kim BG. 2012. Characterization of a new ScbR-like gamma-butyrolactone binding regulator (SlbR) in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 96: 113– 121 [DOI] [PubMed] [Google Scholar]
- 170. Kato JY, Ohnishi Y, Horinouchi S. 2005. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350: 12– 26 [DOI] [PubMed] [Google Scholar]
- 171. den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78: 361– 379 [DOI] [PubMed] [Google Scholar]
- 172. Wolanski M., Jakimowicz D., Zakrzewska-Czerwinska J. 2012. AdpA, key regulator for morphological differentiation regulates bacterial chromosome replication. Open Biol. 2: 120097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Meng L, Li M, Yang SH, Kim TJ, Suh JW. 2011. Intracellular ATP levels affect secondary metabolite production in Streptomyces spp. Biosci. Biotechnol. Biochem. 75:1576– 1581 [DOI] [PubMed] [Google Scholar]
- 174. Higo A, Horinouchi S, Ohnishi Y. 2011. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 81: 1607– 1622 [DOI] [PubMed] [Google Scholar]
- 175. Xu D, Kwon HJ, Suh JW. 2008. S-Adenosylmethionine induces BldH and activates secondary metabolism by involving the TTA-codon control of bldH expression in Streptomyces lividans. Arch. Microbiol. 189: 419– 426 [DOI] [PubMed] [Google Scholar]
- 176. Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19: 259– 274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Horinouchi S. 2007. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71: 283– 299 [DOI] [PubMed] [Google Scholar]
- 178. Higashi T, Iwasaki Y, Ohnishi Y, Horinouchi S. 2007. A-factor and phosphate depletion signals are transmitted to the grixazone biosynthesis genes via the pathway-specific transcriptional activator GriR. J. Bacteriol. 189: 3515– 3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Pan Y, Liu G, Yang H, Tian Y, Tan H. 2009. The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogenes. Mol. Microbiol. 72: 710– 723 [DOI] [PubMed] [Google Scholar]
- 180. Elliot M, Damji F, Passantino R, Chater K, Leskiw B. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180: 1549– 1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Merrick MJ. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96: 299– 315 [DOI] [PubMed] [Google Scholar]
- 182. Kim IK, Lee CJ, Kim MK, Kim JM, Kim JH, Yim HS, Cha SS, Kang SO. 2006. Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2). Mol. Microbiol. 60: 1179– 1193 [DOI] [PubMed] [Google Scholar]
- 183. Lee CJ, Won HS, Kim JM, Lee BJ, Kang SO. 2007. Molecular domain organization of BldD, an essential transcriptional regulator for developmental process of Streptomyces coelicolor A3(2). Proteins 68: 344– 352 [DOI] [PubMed] [Google Scholar]
- 184. Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40: 257– 269 [DOI] [PubMed] [Google Scholar]
- 185. Elliot MA, Leskiw BK. 1999. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J. Bacteriol. 181: 6832– 6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Elliot MA, Locke TR, Galibois CM, Leskiw BK. 2003. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol. Lett. 225: 35– 40 [DOI] [PubMed] [Google Scholar]
- 187. Hunt AC, Servin-Gonzalez L, Kelemen GH, Buttner MJ. 2005. The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family. J. Bacteriol. 187: 716– 728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Li W, Ying X, Guo Y, Yu Z, Zhou X, Deng Z, Kieser H, Chater KF, Tao M. 2006. Identification of a gene negatively affecting antibiotic production and morphological differentiation in Streptomyces coelicolor A3(2). J. Bacteriol. 188: 8368– 8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Wang XJ, Guo SL, Guo WQ, Xi D, Xiang WS. 2009. Role of nsdA in negative regulation of antibiotic production and morphological differentiation in Streptomyces bingchengensis. J. Antibiot. (Tokyo) 62: 309– 313 [DOI] [PubMed] [Google Scholar]
- 190. Eccleston M, Willems A, Beveridge A, Nodwell JR. 2006. Critical residues and novel effects of overexpression of the Streptomyces coelicolor developmental protein BldB: evidence for a critical interacting partner. J. Bacteriol. 188: 8189– 8195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Ainsa JA, Bird N, Ryding NJ, Findlay KC, Chater KF. 2010. The complex whiJ locus mediates environmentally sensitive repression of development of Streptomyces coelicolor A3(2). Antonie Van Leeuwenhoek 98: 225– 236 [DOI] [PubMed] [Google Scholar]
- 192. Aigle B, Corre C. 2012. Waking up Streptomyces secondary metabolism by constitutive expression of activators or genetic disruption of repressors. Methods Enzymol. 517: 343– 366 [DOI] [PubMed] [Google Scholar]
- 193. Gehring AM, Wang ST, Kearns DB, Storer NY, Losick R. 2004. Novel genes that influence development in Streptomyces coelicolor. J. Bacteriol. 186: 3570– 3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Fernandez-Moreno MA, Martin-Triana AJ, Martinez E, Niemi J, Kieser HM, Hopwood DA, Malpartida F. 1992. abaA, a new pleiotropic regulatory locus for antibiotic production in Streptomyces coelicolor. J. Bacteriol. 174: 2958– 2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Chater KF. 2011. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157: 1312– 1328 [DOI] [PubMed] [Google Scholar]
- 196. Noh JH, Kim SH, Lee HN, Lee SY, Kim ES. 2010. Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Appl. Microbiol. Biotechnol. 86: 1145– 1153 [DOI] [PubMed] [Google Scholar]
- 197. Nah JH, Park SH, Yoon HM, Choi SS, Lee CH, Kim ES. 2012. Identification and characterization of wblA-dependent tmcT regulation during tautomycetin biosynthesis in Streptomyces sp. CK4412. Biotechnol. Adv. 30: 202– 209 [DOI] [PubMed] [Google Scholar]
- 198. Rabyk M, Ostash B, Rebets Y, Walker S, Fedorenko V. 2011. Streptomyces ghanaensis pleiotropic regulatory gene wblA(gh) influences morphogenesis and moenomycin production. Biotechnol. Lett. 33: 2481– 2486 [DOI] [PubMed] [Google Scholar]
- 199. Kim JS, Lee HN, Kim P, Lee HS, Kim ES. 2012. Negative role of wblA in response to oxidative stress in Streptomyces coelicolor. J. Microbiol. Biotechnol. 22: 736– 741 [DOI] [PubMed] [Google Scholar]
- 200. Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES. 2007. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J. Bacteriol. 189: 4315– 4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Retzlaff L, Distler J. 1995. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 18: 151– 162 [DOI] [PubMed] [Google Scholar]
- 202. Mochizuki S, Hiratsu K, Suwa M, Ishii T, Sugino F, Yamada K, Kinashi H. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48: 1501– 1510 [DOI] [PubMed] [Google Scholar]
- 203. Bate N, Butler AR, Gandecha AR, Cundliffe E. 1999. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6: 617– 624 [DOI] [PubMed] [Google Scholar]
- 204. Cundliffe E. 2008. Control of tylosin biosynthesis in Streptomyces fradiae. J. Microbiol. Biotechnol. 18: 1485– 1491 [PubMed] [Google Scholar]
- 205. Karray F, Darbon E, Nguyen HC, Gagnat J, Pernodet JL. 2010. Regulation of the biosynthesis of the macrolide antibiotic spiramycin in Streptomyces ambofaciens. J. Bacteriol. 192: 5813– 5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Sun Y, Zhou X, Dong H, Tu G, Wang M, Wang B, Deng Z. 2003. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 10: 431– 441 [DOI] [PubMed] [Google Scholar]
- 207. Yu Q, Du A, Liu T, Deng Z, He X. 2012. The biosynthesis of the polyether antibiotic nanchangmycin is controlled by two pathway-specific transcriptional activators. Arch. Microbiol. 194: 415– 426 [DOI] [PubMed] [Google Scholar]
- 208. Oliynyk M, Stark CB, Bhatt A, Jones MA, Hughes-Thomas ZA, Wilkinson C, Oliynyk Z, Demydchuk Y, Staunton J, Leadlay PF. 2003. Analysis of the biosynthetic gene cluster for the polyether antibiotic monensin in Streptomyces cinnamonensis and evidence for the role of monB and monC genes in oxidative cyclization. Mol. Microbiol. 49: 1179– 1190 [DOI] [PubMed] [Google Scholar]
- 209. Furuya K, Hutchinson CR. 1996. The DnrN protein of Streptomyces peucetius, a pseudo-response regulator, is a DNA-binding protein involved in the regulation of daunorubicin biosynthesis. J. Bacteriol. 178: 6310– 6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Molle V, Buttner MJ. 2000. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol. Microbiol. 36: 1265– 1278 [DOI] [PubMed] [Google Scholar]
- 211. O'Connor TJ, Nodwell JR. 2005. Pivotal roles for the receiver domain in the mechanism of action of the response regulator RamR of Streptomyces coelicolor. J. Mol. Biol. 351: 1030– 1047 [DOI] [PubMed] [Google Scholar]
- 212. Tian Y, Fowler K, Findlay K, Tan H, Chater KF. 2007. An unusual response regulator influences sporulation at early and late stages in Streptomyces coelicolor. J. Bacteriol. 189: 2873– 2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. de Been M, Bart MJ, Abee T, Siezen RJ, Francke C. 2008. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ. Microbiol. 10: 2796– 2809 [DOI] [PubMed] [Google Scholar]
- 214. Jiang H, Hutchinson CR. 2006. Feedback regulation of doxorubicin biosynthesis in Streptomyces peucetius. Res. Microbiol. 157: 666– 674 [DOI] [PubMed] [Google Scholar]
- 215. Nishida H, Ohnishi Y, Beppu T, Horinouchi S. 2007. Evolution of gamma-butyrolactone synthases and receptors in Streptomyces. Environ. Microbiol. 9: 1986– 1994 [DOI] [PubMed] [Google Scholar]
- 216. Santamarta I, Lopez-Garcia MT, Kurt A, Nardiz N, Alvarez-Alvarez R, Perez-Redondo R, Martin JF, Liras P. 2011. Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Mol. Microbiol. 81: 968– 981 [DOI] [PubMed] [Google Scholar]
- 217. Garg RP, Parry RJ. 2010. Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP). Microbiology 156: 472– 483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Tang L, Grimm A, Zhang YX, Hutchinson CR. 1996. Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol. Microbiol. 22: 801– 813 [DOI] [PubMed] [Google Scholar]
- 219. Chen Y, Wendt-Pienkowski E, Shen B. 2008. Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 190: 5587– 5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Santos-Aberturas J, Vicente CM, Payero TD, Martin-Sanchez L, Canibano C, Martin JF, Aparicio JF. 2012. Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS One 7: e38536 doi:10.1371/journal.pone.0038536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. He X, Li R, Pan Y, Liu G, Tan H. 2010. SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology 156:828– 837 [DOI] [PubMed] [Google Scholar]
- 222. Li R, Xie Z, Tian Y, Yang H, Chen W, You D, Liu G, Deng Z, Tan H. 2009. polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology 155: 1819– 1831 [DOI] [PubMed] [Google Scholar]
- 223. Tahlan K, Anders C, Jensen SE. 2004. The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. J. Bacteriol. 186: 6286– 6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Madduri K, Hutchinson CR. 1995. Functional characterization and transcriptional analysis of the dnrR1 locus, which controls daunorubicin biosynthesis in Streptomyces peucetius. J. Bacteriol. 177: 1208– 1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Novakova R, Rehakova A, Kutas P, Feckova L, Kormanec J. 2011. The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 157: 1629– 1639 [DOI] [PubMed] [Google Scholar]
- 226. Bate N, Stratigopoulos G, Cundliffe E. 2002. Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 43: 449– 458 [DOI] [PubMed] [Google Scholar]
- 227. Rodriguez M, Nunez LE, Brana AF, Mendez C, Salas JA, Blanco G. 2008. Identification of transcriptional activators for thienamycin and cephamycin C biosynthetic genes within the thienamycin gene cluster from Streptomyces cattleya. Mol. Microbiol. 69: 633– 645 [DOI] [PubMed] [Google Scholar]
- 228. Suzuki T, Mochizuki S, Yamamoto S, Arakawa K, Kinashi H. 2010. Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci. Biotechnol. Biochem. 74: 819– 827 [DOI] [PubMed] [Google Scholar]
- 229. Ryding NJ, Anderson TB, Champness WC. 2002. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184: 794– 805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Leipe DD, Koonin EV, Aravind L. 2004. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343: 1– 28 [DOI] [PubMed] [Google Scholar]
- 231. Liu G, Tian Y, Yang H, Tan H. 2005. A pathway-specific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Mol. Microbiol. 55: 1855– 1866 [DOI] [PubMed] [Google Scholar]
- 232. Li R, Liu G, Xie Z, He X, Chen W, Deng Z, Tan H. 2010. PolY, a transcriptional regulator with ATPase activity, directly activates transcription of polR in polyoxin biosynthesis in Streptomyces cacaoi. Mol. Microbiol. 75: 349– 364 [DOI] [PubMed] [Google Scholar]
- 233. Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21: 526– 531 [DOI] [PubMed] [Google Scholar]
- 234. Anton N, Mendes MV, Martin JF, Aparicio JF. 2004. Identification of PimR as a positive regulator of pimaricin biosynthesis in Streptomyces natalensis. J. Bacteriol. 186: 2567– 2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. 1987. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. (Tokyo) 40: 496– 504 [DOI] [PubMed] [Google Scholar]
- 236. Kitani S, Iida A, Izumi TA, Maeda A, Yamada Y, Nihira T. 2008. Identification of genes involved in the butyrolactone autoregulator cascade that modulates secondary metabolism in Streptomyces lavendulae FRI-5. Gene 425: 9– 16 [DOI] [PubMed] [Google Scholar]
- 237. Kitani S, Kinoshita H, Nihira T, Yamada Y. 1999. In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J. Bacteriol. 181: 5081– 5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Kitani S, Miyamoto KT, Takamatsu S, Herawati E, Iguchi H, Nishitomi K, Uchida M, Nagamitsu T, Omura S, Ikeda H, Nihira T. 2011. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 108: 16410– 16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Recio E, Colinas A, Rumbero A, Aparicio JF, Martin JF. 2004. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 279: 41586– 41593 [DOI] [PubMed] [Google Scholar]
- 240. Takano E. 2006. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9: 287– 294 [DOI] [PubMed] [Google Scholar]
- 241. Folcher M, Gaillard H, Nguyen LT, Nguyen KT, Lacroix P, Bamas-Jacques N, Rinkel M, Thompson CJ. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276: 44297– 44306 [DOI] [PubMed] [Google Scholar]
- 242. Chater KF, Horinouchi S. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48: 9– 15 [DOI] [PubMed] [Google Scholar]
- 243. Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141– 147 [DOI] [PubMed] [Google Scholar]
- 244. Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190: 4050– 4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Medema MH, Breitling R, Bovenberg R, Takano E. 2011. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat. Rev. Microbiol. 9: 131– 137 [DOI] [PubMed] [Google Scholar]
- 246. Zerikly M, Challis GL. 2009. Strategies for the discovery of new natural products by genome mining. Chembiochem 10: 625– 633 [DOI] [PubMed] [Google Scholar]
- 247. Kawai K, Wang G, Okamoto S, Ochi K. 2007. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 274: 311– 315 [DOI] [PubMed] [Google Scholar]
- 248. Qu X, Lei C, Liu W. 2011. Transcriptome mining of active biosynthetic pathways and their associated products in Streptomyces flaveolus. Angew. Chem. Int. Ed. Engl. 50: 9651– 9654 [DOI] [PubMed] [Google Scholar]
- 249. Onaka H, Mori Y, Igarashi Y, Furumai T. 2011. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77: 400– 406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 106: 14558– 14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Nutzmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA. 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. U. S. A. 108: 14282– 14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. 2012. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U. S. A. 109: E1743– 1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Ochi K. 2007. From microbial differentiation to ribosome engineering. Biosci. Biotechnol. Biochem. 71: 1373– 1386 [DOI] [PubMed] [Google Scholar]
- 254. Hosaka T, Ohnishi-Kameyama M, Muramatsu H, Murakami K, Tsurumi Y, Kodani S, Yoshida M, Fujie A, Ochi K. 2009. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 27: 462– 464 [DOI] [PubMed] [Google Scholar]
- 255. Gomez-Escribano JP, Bibb MJ. 2011. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4: 207– 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Gomez-Escribano JP, Bibb MJ. 2012. Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol. 517: 279– 300 [DOI] [PubMed] [Google Scholar]
- 257. Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. 2001. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and characterization. J. Antibiot. (Tokyo) 54: 1036– 1044 [DOI] [PubMed] [Google Scholar]
- 258. Yamanaka K, Oikawa H, Ogawa HO, Hosono K, Shinmachi F, Takano H, Sakuda S, Beppu T, Ueda K. 2005. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151: 2899– 2905 [DOI] [PubMed] [Google Scholar]
- 259. Hopwood DA, Chater KF, Bibb MJ. 1995. Genetics of antibiotic production in Streptomyces coelicolor A3(2), a model streptomycete. Biotechnology 28: 65– 102 [DOI] [PubMed] [Google Scholar]
- 260. Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. 2011. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. U. S. A. 108: 6258– 6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Metsa-Ketela M, Ylihonko K, Mantsala P. 2004. Partial activation of a silent angucycline-type gene cluster from a rubromycin beta producing Streptomyces sp. PGA64. J. Antibiot. (Tokyo) 57: 502– 510 [DOI] [PubMed] [Google Scholar]
- 262. Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B. 2011. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of Kinamycins. J. Bacteriol. 193: 1142– 1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Chen W, Huang T, He X, Meng Q, You D, Bai L, Li J, Wu M, Li R, Xie Z, Zhou H, Zhou X, Tan H, Deng Z. 2009. Characterization of the polyoxin biosynthetic gene cluster from Streptomyces cacaoi and engineered production of polyoxin H. J. Biol. Chem. 284: 10627– 10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Young TS, Walsh CT. 2011. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc. Natl. Acad. Sci. U. S. A. 108: 13053– 13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Zaleta-Rivera K, Charkoudian LK, Ridley CP, Khosla C. 2010. Cloning, sequencing, heterologous expression, and mechanistic analysis of A-74528 biosynthesis. J. Am. Chem. Soc. 132: 9122– 9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Martin R, Sterner O, Alvarez MA, de Clercq E, Bailey JE, Minas W. 2001. Collinone, a new recombinant angular polyketide antibiotic made by an engineered Streptomyces strain. J. Antibiot. (Tokyo) 54: 239– 249 [DOI] [PubMed] [Google Scholar]
- 267. Dangel V, Westrich L, Smith MC, Heide L, Gust B. 2010. Use of an inducible promoter for antibiotic production in a heterologous host. Appl. Microbiol. Biotechnol. 87: 261– 269 [DOI] [PubMed] [Google Scholar]
- 268. Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. 2010. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 107: 2646– 2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Swiercz JP, Hindra Bobek J, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA. 2008. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res. 36: 7240– 7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Panek J, Bobek J, Mikulik K, Basler M, Vohradsky J. 2008. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics 9: 217 doi:10.1186/1471-2164-9-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Tezuka T, Hara H, Ohnishi Y, Horinouchi S. 2009. Identification and gene disruption of small noncoding RNAs in Streptomyces griseus. J. Bacteriol. 191:4896– 4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Vockenhuber MP, Suess B. 2012. Streptomyces coelicolor sRNA scr5239 inhibits agarase expression by direct base pairing to dagA coding region. Microbiology 158: 424– 435 [DOI] [PubMed] [Google Scholar]
- 273. D'Alia D, Nieselt K, Steigele S, Muller J, Verburg I, Takano E. 2010. Noncoding RNA of glutamine synthetase I modulates antibiotic production in Streptomyces coelicolor A3(2). J. Bacteriol. 192: 1160– 1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Gur E, Biran D, Ron EZ. 2011. Regulated proteolysis in Gram-negative bacteria—how and when? Nat. Rev. Microbiol. 9: 839– 848 [DOI] [PubMed] [Google Scholar]
- 275. Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. 2008. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322: 1104– 1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Viala J, Mazodier P. 2002. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol. Microbiol. 44: 633– 643 [DOI] [PubMed] [Google Scholar]
- 277. Whistler CA, Stockwell VO, Loper JE. 2000. Lon protease influences antibiotic production and UV tolerance of Pseudomonas fluorescens Pf-5. Appl. Environ. Microbiol. 66: 2718– 2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. 1998. The 20S proteasome of Streptomyces coelicolor. J. Bacteriol. 180: 5448– 5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Pouch MN, Cournoyer B, Baumeister W. 2000. Characterization of the 20S proteasome from the actinomycete Frankia. Mol. Microbiol. 35: 368– 377 [DOI] [PubMed] [Google Scholar]
- 280. Volker C, Lupas AN. 2002. Molecular evolution of proteasomes. Curr. Top. Microbiol. Immunol. 268: 1– 22 [DOI] [PubMed] [Google Scholar]
- 281. Maupin-Furlow J. 2012. Proteasomes and protein conjugation across domains of life. Nat. Rev. Microbiol. 10: 100– 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. De Mot R, Schoofs G, Nagy I. 2007. Proteome analysis of Streptomyces coelicolor mutants affected in the proteasome system reveals changes in stress-responsive proteins. Arch. Microbiol. 188: 257– 271 [DOI] [PubMed] [Google Scholar]
- 283. Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8: 185– 195 [DOI] [PubMed] [Google Scholar]
- 284. Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76: 1376– 1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6: 1656– 1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Fisch KM, Gillaspy AF, Gipson M, Henrikson JC, Hoover AR, Jackson L, Najar FZ, Wagele H, Cichewicz RH. 2009. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J. Ind. Microbiol. Biotechnol. 36: 1199– 1213 [DOI] [PubMed] [Google Scholar]
- 287. Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. 2008. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 6: 1895– 1897 [DOI] [PubMed] [Google Scholar]
- 288. Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flardh K. 2009. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J. Bacteriol. 191: 6489– 6500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. McArthur M, Bibb M. 2006. In vivo DNase I sensitivity of the Streptomyces coelicolor chromosome correlates with gene expression: implications for bacterial chromosome structure. Nucleic Acids Res. 34: 5395– 5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. McArthur M, Bibb MJ. 2008. Manipulating and understanding antibiotic production in Streptomyces coelicolor A3(2) with decoy oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 105: 1020– 1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Okamoto S, Taguchi T, Ochi K, Ichinose K. 2009. Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 16: 226– 236 [DOI] [PubMed] [Google Scholar]
- 292. Mo S, Sydor PK, Corre C, Alhamadsheh MM, Stanley AE, Haynes SW, Song L, Reynolds KA, Challis GL. 2008. Elucidation of the Streptomyces coelicolor pathway to 2-undecylpyrrole, a key intermediate in undecylprodiginine and streptorubin B biosynthesis. Chem. Biol. 15: 137– 148 [DOI] [PubMed] [Google Scholar]
- 293. Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BA, Hayes MA, Smith CP, Micklefield J. 2002. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 9: 1175– 1187 [DOI] [PubMed] [Google Scholar]
- 294. Pawlik K, Kotowska M, Chater KF, Kuczek K, Takano E. 2007. A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Arch. Microbiol. 187: 87– 99 [DOI] [PubMed] [Google Scholar]
- 295. Horinouchi S, Beppu T. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12: 859– 864 [DOI] [PubMed] [Google Scholar]
- 296. Gramajo HC, Takano E, Bibb MJ. 1993. Stationary-phase production of the antibiotic actinorhodin in Streptomyces coelicolor A3(2) is transcriptionally regulated. Mol. Microbiol. 7: 837– 845 [DOI] [PubMed] [Google Scholar]
- 297. Novakova R, Homerova D, Feckova L, Kormanec J. 2005. Characterization of a regulatory gene essential for the production of the angucycline-like polyketide antibiotic auricin in Streptomyces aureofaciens CCM 3239. Microbiology 151: 2693– 2706 [DOI] [PubMed] [Google Scholar]
- 298. Rebets Y, Dutko L, Ostash B, Luzhetskyy A, Kulachkovskyy O, Yamaguchi T, Nakamura T, Bechthold A, Fedorenko V. 2008. Function of lanI in regulation of landomycin A biosynthesis in Streptomyces cyanogenus S136 and cross-complementation studies with Streptomyces antibiotic regulatory proteins encoding genes. Arch. Microbiol. 189: 111– 120 [DOI] [PubMed] [Google Scholar]
- 299. Rebets Y, Ostash B, Luzhetskyy A, Kushnir S, Fukuhara M, Bechthold A, Nashimoto M, Nakamura T, Fedorenko V. 2005. DNA-binding activity of LndI protein and temporal expression of the gene that upregulates landomycin E production in Streptomyces globisporus 1912. Microbiology 151: 281– 290 [DOI] [PubMed] [Google Scholar]
- 300. Brunker P, McKinney K, Sterner O, Minas W, Bailey JE. 1999. Isolation and characterization of the naphthocyclinone gene cluster from Streptomyces arenae DSM 40737 and heterologous expression of the polyketide synthase genes. Gene 227: 125– 135 [DOI] [PubMed] [Google Scholar]
- 301. Yin X, O'Hare T, Gould SJ, Zabriskie TM. 2003. Identification and cloning of genes encoding viomycin biosynthesis from Streptomyces vinaceus and evidence for involvement of a rare oxygenase. Gene 312: 215– 224 [DOI] [PubMed] [Google Scholar]