Abstract
We performed a screening of hydroxyurea (HU)-sensitive mutants using a single-gene-deletion mutant collection in Escherichia coli. HU inhibits ribonucleotide reductase (RNR), which leads to arrest of the replication fork. Surprisingly, the wild-type was less resistant to HU than the average for the Keio Collection. Respiration-defective mutants were significantly more resistant to HU, suggesting that the generation of reactive oxygen species (ROS) contributes to cell death. High-throughput screening revealed that 15 mutants were completely sensitive on plates containing 7.5 mM HU. Unexpectedly, translation-related mutants based on COG categorization were the most enriched, and three of them were deletion mutants of nonessential ribosomal proteins (L1, L32, and L36). We found that, in these mutants, an increased membrane stress response was provoked, resulting in increased ROS generation. The addition of OH radical scavenger thiourea rescued the HU sensitivity of these mutants, suggesting that ROS generation is the direct cause of cell death. Conversely, both the deletion of rpsF and the deletion of rimK, which encode S6 and S6 modification enzymes, respectively, showed an HU-resistant phenotype. These mutants increased the copy number of the p15A-based plasmid and exhibited reduced basal levels of SOS response. The data suggest that nonessential proteins indirectly affect the DNA-damaging process.
INTRODUCTION
The Keio Collection is a complete set of precisely defined, nonessential, single-gene knockout mutants of Escherichia coli K-12 (1). The Keio Collection has been used by many groups to observe the effects of gene deletions under specific conditions of interest. These studies have included the determination of mutations affecting antibiotic hypersensitivity (2, 3), swarming motility (4), biofilm formation (5), growth in human blood (6), recipient ability in plasmid conjugation (7), cysteine tolerance and production (8), colicin import and cytotoxicity (9), de-ethylation of 7-ethoxycoumarin (10), and glycogen metabolism (11). In most of these studies, the sets of relevant gene deletions show enrichment for specific cellular functions linked to the conditions of the screen. Although carrying out screens with the Keio Collection is straightforward, the determination of a mechanism explaining why the deletion of certain genes causes a particular effect is rarely obvious. It is often the case with genome-wide screening that unexpected clones are selected, suggesting a lack of our understanding about either gene function or interactions between genes within complex intracellular networks.
Hydroxyurea (HU) is a well-known DNA replication inhibitor that causes replication fork arrest by depleting deoxynucleoside triphosphate (dNTP) pools (12). HU specifically inhibits class 1 ribonucleotide reductase (RNR), the enzyme responsible for the synthesis of dNTPs under aerobic conditions (13, 14). Davies et al. recently clarified how HU induces cell death (15). Unexpectedly, the direct cause of cell death was not inhibition of DNA replication but rather the generation of reactive oxygen species (ROS) in the cytoplasm. In the model of Davies et al., replication fork arrest by HU leads to activation of the MazF/RelE toxins, which generates both incomplete proteins and membrane stress response in the cell. E. coli responds to misfolded or unfolded outer membrane porins in the periplasm by inducing σE-dependent transcription of stress genes (16, 17), which may interfere with the flow of the electron transfer chain, causing an increase in the production of superoxide (17).
In this study, we performed a genome-wide screening with HU. Our data indicated that cells were killed not because of a DNA replication stall but because of ROS generation, followed by DNA damage. Interestingly, we found that nonessential ribosomal proteins were closely related to the process. We show here that most of the deletion strains of a ribosomal protein enhance ROS production, whereas a specific ribosomal protein deletion alleviated DNA damage.
MATERIALS AND METHODS
Strains and growth conditions.
We used mutants from the systematic single-gene knockout mutant collection of the nonessential genes and their parental strain BW25113 (18). Cells were grown at 37°C with vigorous shaking in Luria-Bertani (LB) medium. Where indicated, antibiotics were used at the following concentrations: ampicillin (50 μg ml−1), kanamycin (30 μg ml−1), and chloramphenicol (25 μg ml−1). The gapA-Venus fusion strain was constructed by a one-step recombination method (18) and transferred into a ΔrimK strain by PI transduction.
Plasmid constructions.
The pKDN31 plasmid (a gift from Barry Wanner) was used to amplify a Venus green fluorescent protein (GFP) fragment by PCR with the primers TNP143 and TNP193. The amplified fragment was cloned into the SalI-HindIII site of pSTV29 (TaKaRa Bio), resulting in pTN242. The promoters and short (30 to 60 nucleotides [nt]) N-terminal coding regions of nrdAB, nrdEF, bamA, soxS, and ahpC were amplified with the following primer sets: nrdAB, TNP194 and TNP195; nrdEF, TNP223 and TNP224; bamA, TNP201 and TNP218; soxS, TNP203 and TNP204; and ahpC, TNP214 and TNP215. The amplified fragments were cloned into the BamHI-SalI site of pTN242, creating pTN244, pTN250, pTN246, pTN247, and pTN249, respectively. For construction of the sulA-GFP plasmid, a PCR fragment encompassing the sulA promoter and a short N-terminal portion of the coding region (primers TNP126 and TNP151) was cloned into the BamHI-SalI site of pHSG399 (TaKaRa Bio), resulting in pTN163. The pKDN31 plasmid was used to amplify a Venus GFP fragment by PCR with the primer set TNP143 and NYP273. This PCR fragment was cloned successively into pSTBlue-1 (Novagen) and the SalI-SphI site of pTN163, creating pTN167. The sulA-GFP fragment of pTN167 was recloned into the BamHI-EcoRI site of the low-copy vector pHSG575 (19). The resultant plasmid pTN175 was used for monitoring the SOS response. The frameshift assay plasmids pTN196 and pTN197 were constructed as follows. The pKDN31 plasmid was used to amplify the Venus GFP fragment by PCR with the primer set NYP243 and NYP273, and this PCR fragment was cloned into pSTBlue-1 (pTN155). The long oligonucleotide TNP146, where the termination codon (TGA) within dnaX frameshift region was changed to a cysteine codon (TGC), was prepared and used as a PCR template. PCR was performed with the primers TNP147 and TNP148 for amplification of the −1 frame fusion fragment, and the primer set TNP147 and TNP149 was used for amplification of the in-frame fusion fragment. These fragments were cloned into the BamHI-SphI site of pTN155, creating pTN179 and pTN180, respectively. Both fusion fragments were recloned into the BamHI-HindIII site of pHSG398 (19), and the gapA promoter region amplified with primer set TNP154 and TNP155 was inserted into the SacI-BamHI site of both plasmids, resulting in pTN189 and pTN190. The SacI-HindIII fragments of both plasmids were then transferred into pSTV29, yielding pTN196 and pTN197, respectively. The suppression assay plasmids pTN251 and pTN252 were constructed as follows. The pKDN31 plasmid was used to amplify a Venus GFP fragment by PCR with the primers TNP143 and TNP193. The amplified fragment was cloned into the SalI-HindIII site of pSTV29 (TaKaRa Bio), resulting in pTN242. The gapA promoter regions amplified with primer set TNP205 and TNP206 or TNP207 were inserted into the BamHI-SalI site of pTN242, resulting in pTN251 and pTN252. In order to monitor rpsF promoter expression, pTN174 was constructed as follows. The rpsF promoter region was amplified by PCR with the primers TPN122 and TPN123, and the amplified fragment was cloned into the BamHI-SalI site of pHSG399, resulting in pTN164. The Venus GFP fragment was inserted into the SalI-SphI site of pTN164 (pTN171). The rpsF-GFP fragment was transferred into the low-copy-number vector pHSG575 (pTN174).
Screening of HU-sensitive clones, colony quantification, normalization, and clustering analysis.
The medium used for primary screening was LB medium containing kanamycin (30 μg ml−1), 1.0% agar, and 5 to 15 mM HU. The mutants from the Keio Collection, which had been stored in 384-well microplates, were inoculated with a 384-pin (Rotor; Singer Instrument) onto the surfaces of the screening plates. The plates were incubated at 37°C for 18 h. Colony quantification was performed as follows. Cultured plates were scanned in a TIFF format at 300 dpi and 24-bit color in transparency mode. A scanned image was converted to grayscale for image analysis. The colony region was determined by Otsu method thresholding. A colony area is measured by counting the pixel number within the colony region. The colony density is described by colony integrated optical density [20]). Each colony density was normalized by the sum of colony densities in the plate. Edge colonies were further normalized by the ratio of average of edge colony density to average of other colony density (see Table S1 in the supplemental material). A clustering heat map was generated with Cluster 3 (free software) using the complete linkage method. Green represents positive values. For HU-sensitive screening, strongly HU-sensitive clones were selected using a cutoff value of 0 on 7.5 mM HU plates (i.e., the colony density on 7.5 mM HU/the colony density on LB medium). After the primary screening, screened mutants were cultured independently and tested for HU sensitivity by spotting them onto LB medium plus HU at 0 to 10 mM using a series of dilutions (100, 10−2, 10−4, and 10−6). The mutants that did not show sensitivity at a 10−4 dilution on the plate containing 7.5 mM HU were excluded in the present study (see Table S2 in the supplemental material).
Flow cytometry analysis.
Cells harboring a GFP fusion plasmid were grown to exponential phase (optical density at 600 nm of 0.4) in LB-chloramphenicol medium with or without 5 mM HU and then diluted 50-fold with phosphate-buffered saline. All data were collected using a FACScan flow cytometry (Becton Dickinson) with a 488-nm argon laser and a 515- to 545-nm emission filter (FL1) at a low flow rate. The following photomultiplier (PMT) voltage settings were used: E01 (forward scatter [FSC]), 349 (side scatter [SSC]), and 736 (FL1). Flow cytometry analysis measured approximated single-cell portion of cell population defined by the FSC and SSC in order to exclude filamentous cells. The relative fluorescence values were determined by taking the mean fluorescence measurement for each sample, and each value represents the average of the total fluorescent values. To detect hydroxyl radical formation, we used the fluorescent reporter dye 3′-(p-hydroxylphenyl) fluorescein (HPF; Invitrogen [21]).
RESULTS
The wild-type strain is less resistant to HU than the average for the Keio deletion collection.
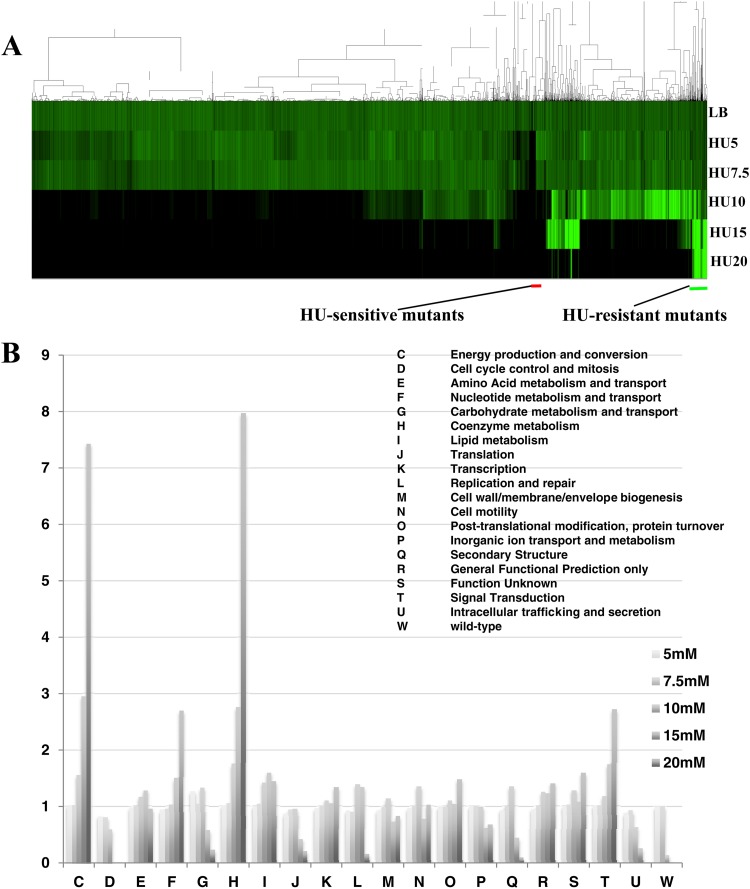
Mutants of the Keio Collection were first screened on solid LB media containing various HU concentrations (0 to 20 mM) at 37°C for 18 h, and plate images were scanned into a computer. Colony densities were quantified and normalized by the average colony size per plate. Edge colonies were further normalized by the ratio of average of edge colonies to average of other colonies. Figure 1A shows the matrix subjected to two-dimensional hierarchical clustering created by Gene Cluster 3.0. Green represents relative colony densities, whereas black represents no growth. Surprisingly, the wild-type strain was less resistant to high concentrations of HU (ca. 10 to 20 mM) than the average for the Keio Collection (Fig. 1B). In order to examine functional enrichment in HU-resistant strains, a clusters of orthologous groups (COG)-based categorization was applied. Values given here are averages of relative colony densities categorized in each COG. As shown in Fig. 1B, gene deletions involved in energy production and coenzyme metabolism (COG-based functions C and H; Fig. 1B) were significantly enriched as the concentration of HU was increased. Strains with deletions of genes encoding pyruvate dehydrogenase, ubiquinone biosynthesis enzymes, and NAD biosynthesis enzymes were particularly resistant to HU challenges, and strains with deletions of the genes for NADH dehydrogenase, succinate dehydrogenase, and cytochrome oxidases showed moderate resistance to HU (see Table S1 in the supplemental material). These results suggested that respiration activity is strongly linked with HU sensitivity.
Fig 1.
Profile of the growth of strains from the Keio Collection in response to HU challenges. (A) Clustering analysis of the growth of strains from the Keio Collection. Colony densities were normalized by the average of colony densities per plate. Clustering was performed using Gene Cluster 3.0 with the complete linkage method. Black and green bars represent clustering regions for HU-sensitive and -resistant mutants, respectively. (B) COG-based functional analysis for HU sensitivity. The vertical axis represents the colony density ratio for each HU concentration (HU/LB). The values were calculated as the average of all gene deletions categorized into the COG.
Ribosomal protein deletions are either HU sensitive or HU resistant.
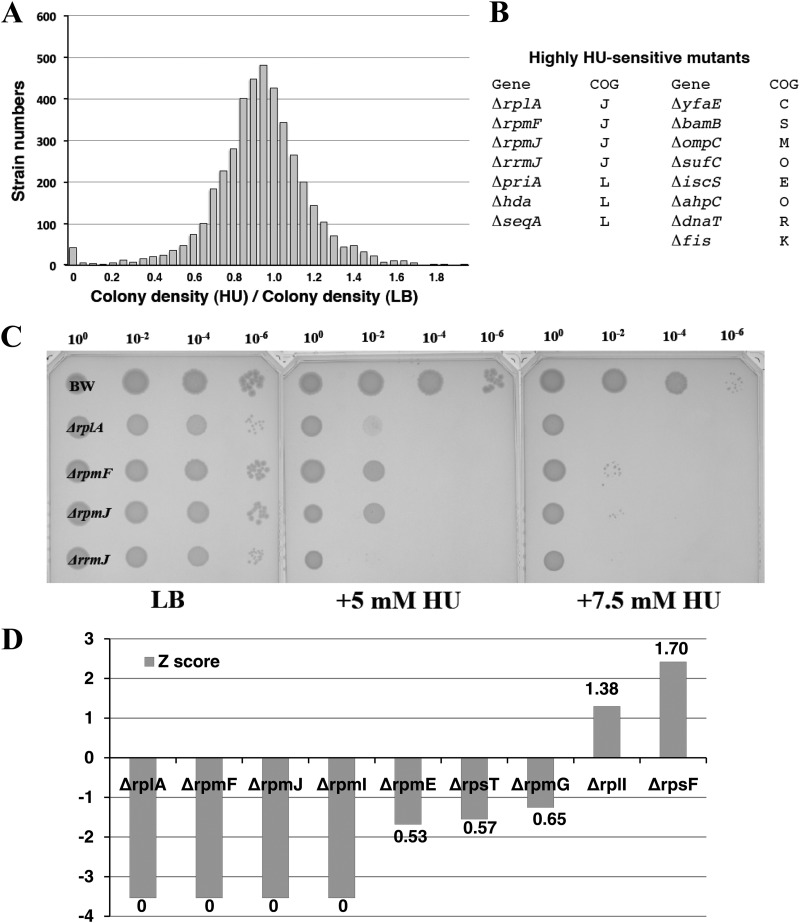
At the concentration of HU where the most mutants survived (5 to 7.5 mM HU), there was no significant COG-functional enrichment (Fig. 1B). Looking at highly HU-sensitive mutants that could not grow on LB plates containing 7.5 mM HU, 38 mutants were unable to form colonies (Fig. 2A; see Fig. S1 in the supplemental material). These 38 mutants were cultured independently and tested for HU sensitivity by spotting them on LB medium plus HU at 0 to 10 mM using a series of dilutions (100, 10−2, 10−4, and 10−6). Mutants that showed clear HU sensitivity were selected (see Table S2 in the supplemental material). In this study, tus mutants (tusABCDE and mnmA strains, classified as group II mutants) were excluded since they were not killed on HU-containing plates and were independently investigated (T. Nakayashiki et al., unpublished data). Finally, 15 mutants were selected as highly HU-sensitive mutants (Fig. 2B). Classified using COG-based functions, four genes (rplA, rpmFJ, and rrmJ: classified as group I) were associated with translation (J) and three mutants (hda, priA, and seqA) were related to DNA replication (L). In particular, rplA (L1), rpmF (L32), and rpmJ (L36) encoded nonessential ribosomal proteins.
Fig 2.
Screening for HU-sensitive mutants. (A) Distribution of colony density ratio (7.5 mM HU/LB) for all of the Keio Collection mutants. Mutants that were unable to grow on the plate (7.5 mM HU) were selected as the first candidates. (B) After individual spotting tests, 15 HU-sensitive mutants were selected. The mutations were classified into COG-based categories. (C) HU sensitivity of mutants that were classified as being in the translation category (COG category J). Portions (5 μl) of overnight cultures were spotted onto agar plates at the indicated dilution rates. The plates were incubated at 37°C for 24 h. (D) Distribution of nonessential ribosomal protein mutants in the Keio Collection. The vertical axis represents the Z score of each mutant. The numbers represent the colony density ratio (HU/LB).
The Keio Collection includes nine deletions of genes encoding nonessential ribosomal proteins. Seven of the distributions of the ribosomal protein deletions resulted in severe (ΔrplA and ΔrpmFIJ) or moderate (ΔrpmEG and ΔrpsT) HU-sensitive phenotypes, but two of the mutants (the ΔrpsF and ΔrplI mutants) were moderately resistant to HU (Fig. 2C; see Fig. S2 in the supplemental material). The distribution led us to postulate a functional link between nonessential ribosomal proteins and ROS generation. We therefore pursued further analyses to clarify the molecular basis behind the link.
Group I mutants produced increased ROS in the presence of HU.
Davies et al. (15) proposed a model for how HU treatment causes cell death in E. coli. These researchers showed that cell death is directly caused by the generation of hydroxyl radical via membrane stress response and not simply by replication fork arrest. In the model, HU treatment induces mazEF and relBE toxin/antitoxin modules (22), producing incomplete mRNAs and aberrant proteins in the cell. Misfolded proteins incorporated into the membrane trigger the membrane stress response (17). Since translation-related mutations are expected to affect synthesis of proteins, we suspected that these mutant proteins triggered a membrane stress response (15, 17).
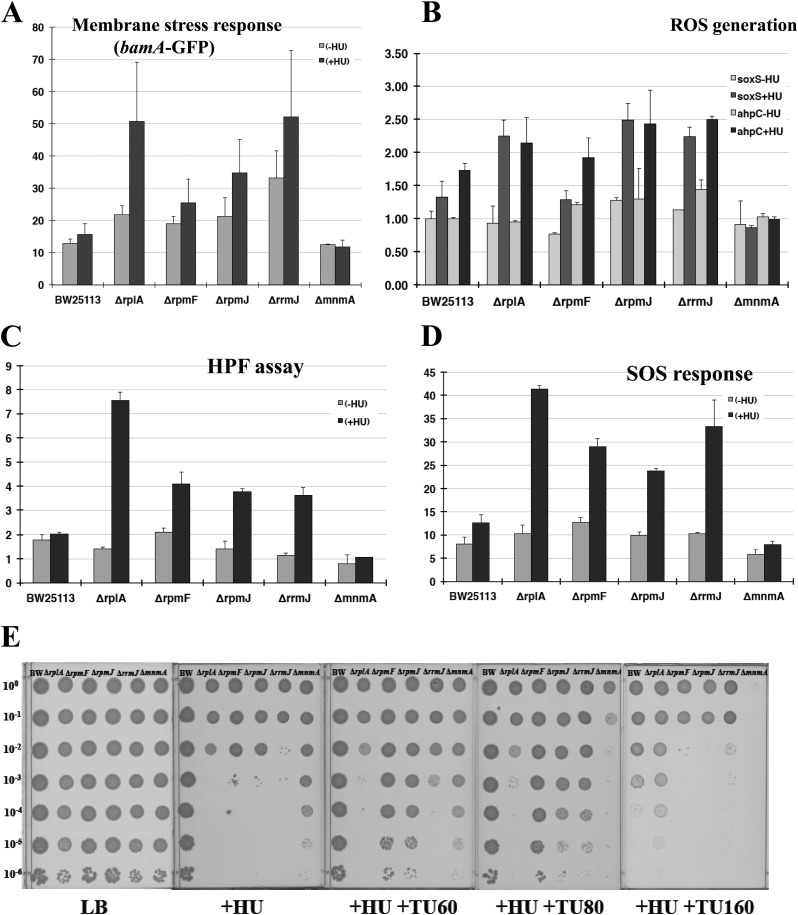
As shown in Fig. 2C, these mutants exhibit clear HU sensitivity. Longer incubation did not result in the appearance of colonies, suggesting that they were killed on HU-containing plates. Considering the model proposed by Davies et al. (15) and the fact that respiration-defective mutants are more resistant to HU, HU-sensitive mutants would be predicted to generate more ROS than the wild-type does. For monitoring membrane stress response, Venus GFP was fused to the bamA promoter region that is under the control of σE. All group I mutants exhibited an increased membrane stress response in response to HU addition (Fig. 3A), suggesting that group I mutations increase the incorporation of misfolded proteins into the membrane. According to the model of Davies et al., membrane stress response produces ROS species (H2O2, O2−, and •OH), which are the direct causes of cell death (15).
Fig 3.
Analysis of group I mutants. (A) Membrane stress response of group I mutants. The membrane stress response was measured by promoter assays of the bamA mutant (pTN246), which is under the control of σE. (B) Promoter assays of soxS and ahpC mutants for monitoring the level of superoxide (green) and hydrogen peroxide (purple). The Venus GFP fusion plasmids used for the assays were pTN247 and pTN249, respectively. The vertical axis shows relative values, with the value for BW25113 (−HU) set as 1.0. (C) HPF assay for the measurement of OH radical formation. The intensity of fluorescence was measured by flow cytometry. (D) SOS response of group I mutants. The Venus GFP fusion plasmid used for the assays was pTN175. (E) Effects of the radical scavenger TU on group I mutants and group II mutants (non-ROS-generating mutants) in HU-containing agar plates. A spotting test was performed as described in Fig. 2C at the indicated dilution rates. HU and thiourea were added at 4 mM and at the indicated concentrations (mM), respectively.
Next, a series of GFP fusion plasmids was constructed in order to monitor ROS production. For the production of superoxide and peroxide in cells, soxS-GFP and ahpC-GFP plasmids were constructed, regulated by SoxR and OxyR, respectively. In addition, a sulA-GFP plasmid was constructed in order to monitor the SOS response. We also measured •OH production using 3′-(p-hydroxyphenyl) fluorescein (HPF), which is oxidized by hydroxyl radicals with high specificity (21). As shown in Fig. 3B, group I mutants produced more ROS than the wild type in the presence of HU. Moreover, the degree of ROS production was correlated with HU sensitivity. The level of ROS production correlated with •OH production (Fig. 3C) and SOS response (Fig. 3D). Finally, we confirmed that addition of the •OH scavenger thiourea (TU) rescued the HU sensitivity of group I mutants (Fig. 3E). TU is a potent •OH scavenger (23, 24) and was shown to inhibit •OH production in E. coli (15). Hence, we conclude that group I mutants become sensitive to HU because of enhanced membrane stress response, which causes the increased production of •OH in the presence of HU. In group II mutants (ΔmnmA), RNR expression is reduced to one-third (Nakayashiki et al., unpublished). They are sensitive to TU (Fig. 3E), since TU further reduces RNR expression, probably due to a decreased SOS response.
Deficiency of S6 modification causes instability of plasmid maintenance and decreases the basal level of SOS response.
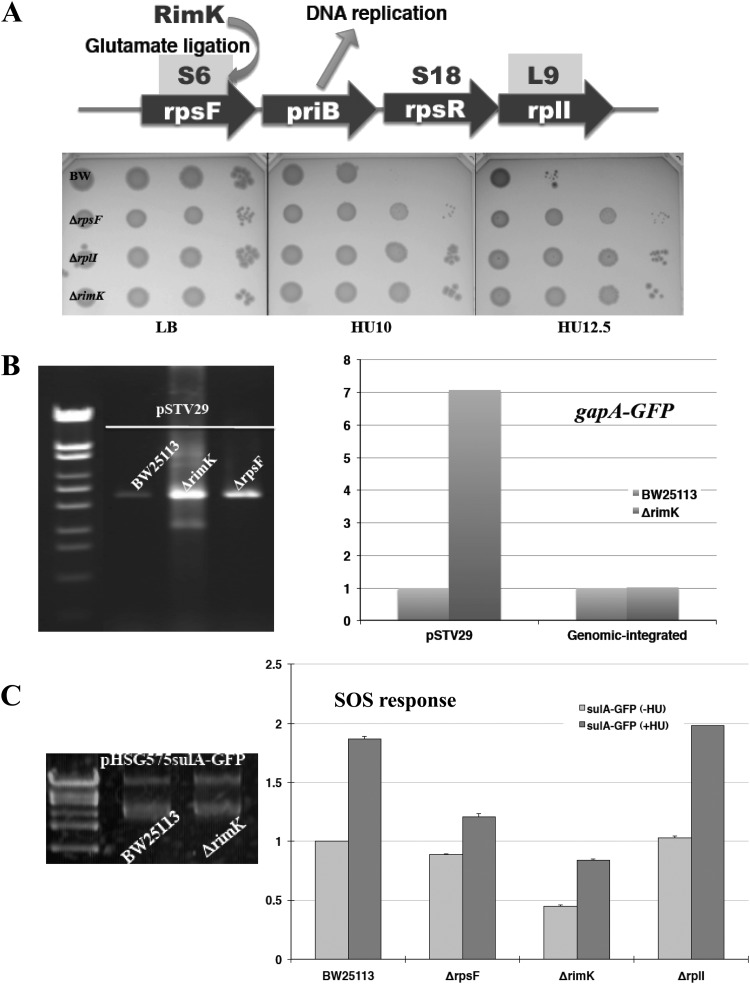
The genes rpsF and rplI are in the same operon, and the gene products S6 and L9 locate near the place called the “platform,” where mRNAs bind to the ribosome. The gene product of rpsR, S18, also locates near the platform, but the ΔrpsR mutant is not included in the Keio Collection because rpsR appears to be an essential gene (1). Interestingly, priB, which encodes a component of the DNA primosome, is located in this operon. The deletion of rpsF, rplI, and rimK, which encodes an S6 modification enzyme, also resulted in a modest HU-resistant phenotype (Fig. 4A). We first attempted to apply the plasmid-based monitoring system to these mutants, but we found that it takes 3 days or more to get ΔrpsF and ΔrimK transformants. In the case of pHSG575-based plasmids (pSC101 replicon; 1 to 5 copy/cell) the copy number was unchanged (Fig. 4C, left), but for pSTV29-based plasmids (p15A replicon; 10 to 15 copy/cell) the copy number was apparently increased in ΔrpsF and ΔrimK mutants (Fig. 4B, left). A comparison of pSTV29-based and chromosomally integrated GFP fluorescence values showed that the plasmid copy number increased ∼7-fold (Fig. 4B, right). Since the SOS monitoring system was a pHSG575-based plasmid, we measured only the SOS response for these mutants. As shown in Fig. 4C, ΔrpsF and ΔrimK mutants exhibited a reduced SOS response in both the absence and the presence of HU. This suggests that deficiency of S6 or S6 modification affects DNA replication, which changes plasmid copy numbers, reduces the basal level of SOS response, and confers HU resistance, probably due to less susceptibility to ROS attack.
Fig 4.
Analysis of HU-resistant gene deletion mutants of ribosomal protein or its modification enzyme. (A) Operon structure of genes encoding ribosomal proteins whose deletion conferred the HU-resistant phenotype. The rimK gene encodes a modification enzyme for S6. The plate images represent the HU resistance of these mutants confirmed by a spotting test. (B) pSTV29 plasmid was extracted from 2 ml of culture, and DNA solutions were applied for agarose electrophoresis (left). The fluorescence values between pSTV29-based and genomic-integrated GFP driven from the gapA promoter are compared. The vertical axis shows relative values, with the value for BW25113 set as 1.0. (C) SOS response of HU-resistant gene-deletion mutants. The Venus GFP fusion plasmid used for the assays was pTN175. The vertical axis shows relative values, with the value for BW25113 (−HU) set as 1.0.
In Bacillus subtilis, the expression of this operon is induced during the SOS response (25). This appears to be true in E. coli, since the expression was elevated in the presence of HU and reduced in the ΔrimK strain (see Fig. S3 in the supplemental material). Autoregulation has also been suggested, because expression is significantly higher in ΔrpsF and ΔrplI strains (see Fig. S3 in the supplemental material).
Analysis of other HU-sensitive mutants.
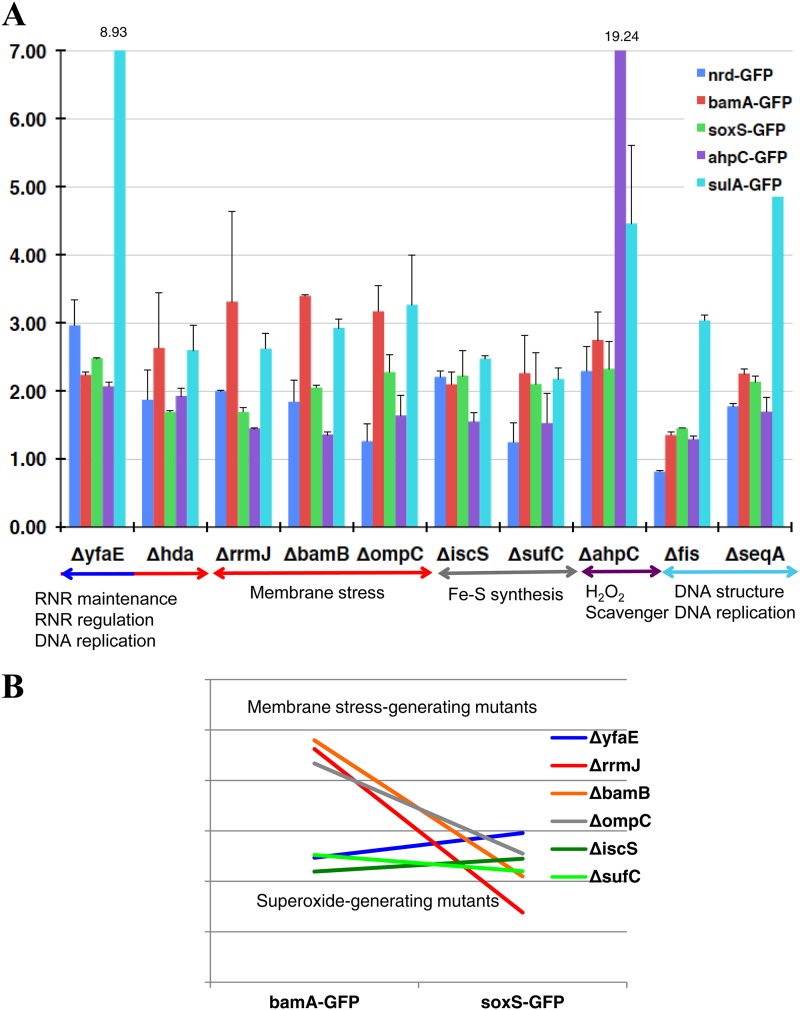
There were other HU-sensitive mutants difficult to classify by COG, but function classification proved fruitful: for instance, both iscS and sufC mutants were involved in the synthesis and repair of iron-sulfur (Fe-S) clusters. The ompC gene encodes outer membrane porin C, and the bamB gene product is involved in outer membrane protein biogenesis (26). As mentioned previously, hda, priA, and seqA are in the category of DNA replication (L) by COG, and DnaT will be put in this group on the basis of its function as a primosome component. YfaE is involved in the maintenance of RNR (27), AhpC is a component of alkyl hydroperoxide reductase, the major player to defend against endogenous levels of oxidative stress (28), and Fis is a small DNA-binding and -bending protein whose main role appears to be the organization and maintenance of nucleoid structure through direct DNA binding. Of 15 HU-sensitive mutants, ΔpriA and ΔdnaT mutants were excluded from the plasmid-based analysis because these mutants are unable to stably maintain plasmids (29). Promoter assays of nrdAB, bamA, soxS, ahpC, and sulA mutants for monitoring the level of RNR expression (blue), membrane-stress response (red), superoxide (green), hydrogen peroxide (purple), and SOS response (light blue) were performed (Fig. 5A). The vertical axis shows relative values, with the value for BW25113 (−HU) being set as 1.0. However, in the case of the ΔiscS mutant, the values were calculated relative to the value for the ΔmnmA mutant (−HU), since the ΔiscS mutant also belongs to group II (the mnm5s2U modification) and the basal expression patterns are different from that of the wild type (Nakayashiki et al., unpublished). Although there were differences in the expression patterns of key enzymes among mutants, it was a common feature in these HU-sensitive mutants that a more-than-double SOS response was provoked in the presence of HU relative to that seen in the wild type. Upon further examination, we found a link between the expression pattern and the function of the deleted gene. For example, YfaE is a [2Fe2S] cluster-containing protein that is involved in the maintenance and possibly in the biosynthesis of the active R2 subunit containing the diferric-tyrosyl radical cofactor within RNR (27). The ΔyfaE mutant showed the highest RNR expression among these mutants, probably due to an autoregulation mechanism (30). OmpC is outer membrane porin C, and BamB is involved in outer membrane protein biogenesis (26). Mutants with deletions of the genes encoding these proteins exhibited high bamA expression, indicating increased membrane stress response. SufC and IscS are involved in synthesis and the maintenance of iron-sulfur clusters, and strains with deletions of genes encoding these proteins showed relatively high soxS expression, suggesting a defect in the iron-sulfur clusters of succinate dehydrogenase or NADH dehydrogenase, which is known to generate superoxide. AhpC is a component of alkyl hydroperoxide reductase, the major player to defend against H2O2 stress (28), and the gene deletion exhibited high self-induction because of activation of the H2O2 sensor OxyR. A fis deletion mutant was also known to be hypersensitive to ROS, likely due to a lack of the change in DNA supercoiling (31), and the SeqA protein is a negative modulator of the initiation of chromosome replication. These deletion strains also showed higher basal levels of SOS expression (see Table S3 in the supplemental material) and high SOS responses upon HU addition, likely due to instability of the DNA structure.
Fig 5.
Analysis of other HU-sensitive mutants. (A) Promoter assays of nrdAB, bamA, soxS, ahpC, and sulA mutants for monitoring the level of RNR expression (blue), membrane stress response (red), superoxide (green), hydrogen peroxide (purple), and SOS response (light blue). The Venus GFP fusion plasmids used for the assays were pTN244, pTN246, pTN247, pTN249, and pTN175, respectively. The vertical axis shows relative values, with the value for BW25113 (−HU) set as 1.0, except for the ΔiscS mutant (for which the value of the ΔmnmA mutant is 1.0). (B) Expression pattern of bamA-GFP (membrane stress) and soxS-GFP (superoxide) in the ΔyfaE mutant, membrane stress-generating mutants (ΔbamB, ΔompC, and ΔrrmJ), and superoxide-generating mutants (ΔiscS and ΔsufC).
The ΔyfaE mutant showed a lower membrane stress response (bamA expression) than did the ΔrrmJ, ΔompC, and ΔbamB mutants but higher soxS expression in the presence of HU. Actually, the ΔyfaE mutant exhibited the highest soxS expression among the mutants. We compared the patterns of bamA and soxS expression for strains with membrane stress-generating mutations and superoxide-generating mutations (ΔiscS and ΔsufC), which were selected in this screening (Fig. 5B). The pattern of the ΔyfaE mutant is similar to that of the superoxide generation mutants. This indicates the possibility that superoxide can be generated not only through the membrane stress response but also directly from the RNR activation step (Fig. 6).
Fig 6.
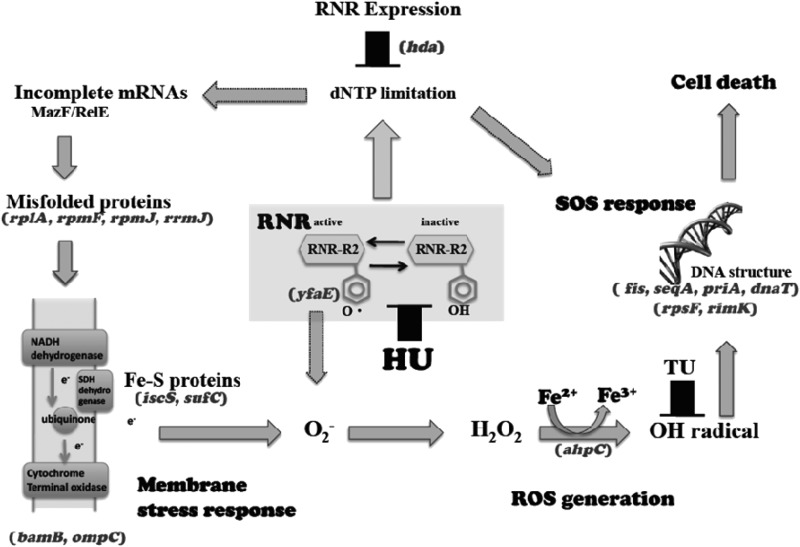
Cell death pathways caused by HU. HU inhibits RNR activity and causes dNTP limitation, which in turn results in induction of RNR expression. HU also accelerates ROS production via membrane stress response. Superoxide and hydrogen peroxide inactivate RNR activity, which also activates pathway II, and the generation of an OH radical provokes an SOS response, which induces RNR expression (pathway I). Superoxide generation directly from RNR was suggested by analysis of the ΔyfaE mutant (Fig. 5). The indicated genes represent mutations that increased HU sensitivity, except for rpsF and rimK, which represent mutations that reduced RNR expression. Gene positions represent the steps where mutations are supposed to affect the circuits of RNR expression from the data, literature, and gene function. Arrows outlined by dotted lines represent the pathways suggested in our study but not confirmed.
DISCUSSION
In this screening, several mutants with deletions of nonessential ribosomal proteins (L1, L32, and L36) were identified as HU-sensitive mutants. We demonstrated that these ribosomal proteins are important not only for translation but also for preventing intracellular ROS generation. However, it remains unclear how a lack of nonessential ribosomal proteins generates an increased membrane stress response. Previous studies reported that mistranslation (17) and incompletely translated products (15) could cause an increased membrane stress response. We measured suppression efficiencies (see Fig. S4 in the supplemental material) and −1 frameshift efficiencies using a programmed −1 ribosomal frameshift sequence within the dnaX region (see Fig. S5 in the supplemental material). Interestingly, all group I mutants showed higher frameshift efficiencies than wild-type samples in both HU+ and HU− conditions. Hence, it is possible that intact translation machinery is required for accurate translation. However, in the cases of ΔrrmJ and ΔrplA mutants, the latter possibility is also likely because it has been suggested that both mutations affect the binding of aminoacylated tRNA to the ribosome (32, 33).
Of nine mutants with deletions of nonessential ribosomal protein, two showed moderate resistance to HU (Fig. 4A). A deficiency of S6 protein or S6 modification especially affects plasmid copy number, as well as the basal level of SOS response (Fig. 4B and C). There are several reports indicating a link between S6 protein/S6 modification and DNA replication/ROS generation. First of all, the rpsF operon contains priB, which encodes a core component of the primosome, and this operon is induced during the SOS response in Bacillus subtilis (25). The rimK operon is activated by the superoxide sensor SoxS (34) and repressed by the peroxide sensor OxyR (35). A class of mutations in rpsF was reported to suppress the temperature-sensitive growth defect of certain dnaG (DNA primase) mutants (36), suggesting a functional link between S6 protein and primosome activity. It has also been reported that mutations in primosome components affect plasmid maintenance (29) and SOS response (37).
We therefore speculated that the role of S6 protein and its modification is related to DNA primosome activity. Mutations in primosome components result in deficiency of plasmid maintenance, whereas deletion of S6 or S6 modification results in increased copy numbers of pSTV29-based plasmids. The former mutations provoke an SOS response, but the latter mutations reduce the basal level of SOS response. Thus far, we do not know the molecular mechanism behind it. One possibility is that the latter mutations may affect the start codon preference of IF2 (38) and enhance the production of IF2-2. IF2-2 was reported to promote the PriC-dependent restart pathway (39), which preferentially utilizes fork structures with large gaps in the leading strand (40).
In our screening, a total of 15 deletion mutants were selected as HU-sensitive clones. Considering our data and prior knowledge, it is possible to interpret all of the causalities according to the model Davies et al. had proposed (15). As shown in Fig. 6, the ΔrplA, ΔrpmFJ, and ΔrrmJ mutations would increase the incorporation of misfolded proteins into the membrane. The ΔompC and ΔbamB mutations would increase the membrane stress response (26, 41, 42). The ΔsufC and ΔiscS mutations would cause a defect in the iron-sulfur clusters of succinate dehydrogenase or NADH dehydrogenase, which is known to generate ROS species (43, 44). The ΔahpC mutant would accumulate high levels of ROS due to the lack of a major player to defend against oxidative stress. Δfis, ΔseqA, ΔpriA, and ΔdnaT mutants showed high basal levels of SOS expression (37), suggesting that DNA structure is less stable and susceptible to DNA damages. YfaE is involved in diferric-tyrosyl radical cofactor maintenance of RNR (27). As expected, upon exposure to HU, RNR expression was highly increased, suggesting that RNR is more susceptible to HU in the ΔyfaE strain. Since YfaE is directly involved in radical maintenance (45), a lack of YfaE may allow a leakage of radicals to the cytoplasm, directly generating superoxide. Hda stimulates ATP hydrolysis within the DnaA-ATP complex, which inactivates DNA replication and modulates RNR expression (46). Considering the function of Hda, it is possible that DNA structure becomes less stable in the Δhda mutant, but our data suggested that the expression pattern of the Δhda mutant is similar to that of strains with membrane stress-generating mutations.
By genome-wide screening using HU, we found a link between nonessential proteins and intracellular ROS generation. We also were able to map almost all of the identified genes on the model proposed by Davies et al. (15). We believe that the genome-wide screening using the Keio Collection gives us excellent clues to elucidate the intracellular networks that remain to be uncovered.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by a Grant-in-Aid for Scientific Research (A and C) and a Grant-in-Aid for Scientific Research (Kakenhi) on Priority Areas System Genomics from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to the Nara Institute of Science and Technology.
We have no competing financial interests.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02145-12.
REFERENCES
- 1. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol. Syst. Biol. 2:0008. doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu A, Tran L, Becket E, Lee K, Chinn L, Park E, Tran K, Miller JH. 2010. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 54:1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tamae C, Liu A, Kim K, Sitz D, Hong J, Becket E, Bui A, Solaimani P, Tran KP, Yang H, Miller JH. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190:5981–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Inoue T, Shingaki R, Hirose S, Waki K, Mori H, Fukui K. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 189:950–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niba ET, Naka Y, Nagase M, Mori H, Kitakawa M. 2007. A genome-wide approach to identify the genes involved in biofilm formation in Escherichia coli. DNA Res. 14:237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. 2008. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 4:e37 doi:10.1371/journal.ppat.0040037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pérez-Mendoza D, de la Cruz F. 2009. Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any? BMC Genomics 10:71 doi:10.1186/1471-2164-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiriyathanawudhiwong N, Ohtsu I, Li ZD, Mori H, Takagi H. 2009. The outer membrane TolC is involved in cysteine tolerance and overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 81:903–913 [DOI] [PubMed] [Google Scholar]
- 9. Sharma O, Datsenko KA, Ess SC, Zhalnina MV, Wanner BL, Cramer WA. 2009. Genome-wide screens: novel mechanisms in colicin import and cytotoxicity. Mol. Microbiol. 73:571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Y, Minami T, Honda K, Omasa T, Ohtake H. 2010. Systematic screening of Escherichia coli single-gene knockout mutants for improving recombinant whole-cell biocatalysts. Appl. Microbiol. Biotechnol. 87:647–655 [DOI] [PubMed] [Google Scholar]
- 11. Eydallin G, Montero M, Almagro G, Sesma MT, Viale AM, Muñoz FJ, Rahimpour M, Baroja-Fernández E, Pozueta-Romero J. 2010. Genome-wide screening of genes whose enhanced expression affects glycogen accumulation in Escherichia coli. DNA Res. 17:61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Odsbu I, Morigen Skarstad K. 2009. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One 4:e7617 doi:10.1371/journal.pone.0007617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eklund H, Uhlin U, Färnegårdh M, Logan DT, Nordlund P. 2001. Structure and function of the radical enzyme ribonucleotide reductase. Prog. Biophys. Mol. Biol. 77:177–268 [DOI] [PubMed] [Google Scholar]
- 14. Rosenkranz HS, Winshell EB, Mednis A, Carr HS, Ellner CJ. 1967. Studies with hydroxyurea. VII. Hydroxyurea and the synthesis of functional proteins. J. Bacteriol. 94:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, Walker GC. 2009. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell 36:845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alba BM, Gross CA. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613–619 [DOI] [PubMed] [Google Scholar]
- 17. Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. 1986. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63–74 [DOI] [PubMed] [Google Scholar]
- 20. Lawless C, Wilkinson DJ, Young A, Addinall SG, Lydall DA, Lu M, Campbell JL. 2010. Colonyzer: automated quantification of micro-organism growth characteristics on solid agar. BMC Bioinformatics 11:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC. 2006. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 25:868–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Novogrodsky A, Ravid A, Rubin AL, Stenzel KH. 1982. Hydroxyl radical scavengers inhibit lymphocyte mitogenesis. Proc. Natl. Acad. Sci. U. S. A. 79:1171–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Touati D, Jacques M, Tardat B, Bouchard L, Despied S. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindner C, Nijland R, van Hartskamp M, Bron S, Hamoen LW, Kuipers OP. 2004. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J. Bacteriol. 186:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charlson ES, Werner JN, Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 188:7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu CH, Jiang W, Krebs C, Stubbe J. 2007. YfaE, a ferredoxin involved in diferric-tyrosyl radical maintenance in Escherichia coli ribonucleotide reductase. Biochemistry 46:11577–11588 [DOI] [PubMed] [Google Scholar]
- 28. Seaver LC, Imlay JA. 2001, Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee EH, Kornberg A. 1991. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc. Natl. Acad. Sci. U. S. A. 88:3029–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torrents E, Grinberg I, Gorovitz-Harris B, Lundström H, Borovok I, Aharonowitz Y, Sjöberg BM, Cohen G. 2007. NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 189:5012–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weinstein-Fischer D, Elgrably-Weiss M, Altuvia S. 2000. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 35:1413–1420 [DOI] [PubMed] [Google Scholar]
- 32. Sander G. 1983. Ribosomal protein L1 from Escherichia coli: its role in the binding of tRNA to the ribosome and in elongation factor G-dependent GTP hydrolysis. J. Biol. Chem. 258:10098–10103 [PubMed] [Google Scholar]
- 33. Widerak M, Kern R, Malki A, Richarme G. 2005. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene 347:109–114 [DOI] [PubMed] [Google Scholar]
- 34. Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 87:6181–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Britton RA, Lupski JR. 1997. Isolation and characterization of suppressors of two Escherichia coli dnaG mutations, dnaG2903 and parB. Genetics 145:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53:1343–1357 [DOI] [PubMed] [Google Scholar]
- 38. Sacerdot C, Vachon G, Laalami S, Morel-Deville F, Cenatiempo Y, Grunberg-Manago M. 1992. Both forms of translational initiation factor IF2 (α and β) are required for maximal growth of Escherichia coli: evidence for two translational initiation codons for IF2β. J. Mol. Biol. 225:67–80 [DOI] [PubMed] [Google Scholar]
- 39. North SH, Kirtland SE, Nakai H. 2007. Translation factor IF2 at the interface of transposition and replication by the PriA-PriC pathway. Mol. Microbiol. 66:1566–1578 [DOI] [PubMed] [Google Scholar]
- 40. Heller RC, Marians KJ. 2005. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17:733–743 [DOI] [PubMed] [Google Scholar]
- 41. Egler M, Grosse C, Grass G, Nies DH. 2005. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187:2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onufryk C, Crouch ML, Fang FC, Gross CA. 2005. Characterization of six lipoproteins in the sigmaE regulon. J. Bacteriol. 187:4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Esterházy D, King MS, Yakovlev G, Hirst J. 2008. Production of reactive oxygen species by complex I (NADH:ubiquinone oxidoreductase) from Escherichia coli and comparison to the enzyme from mitochondria. Biochemistry 47:3964–3971 [DOI] [PubMed] [Google Scholar]
- 44. Yankovskaya V, Horsefield R, Törnroth S, Luna-Chavez C, Miyoshi H, Léger C, Byrne B, Cecchini G, Iwata S. 2003. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299:700–704 [DOI] [PubMed] [Google Scholar]
- 45. Stubbe J. 2003. Di-iron-tyrosyl radical ribonucleotide reductases. Curr. Opin. Chem. Biol. 7:183–188 [DOI] [PubMed] [Google Scholar]
- 46. Olliver A, Saggioro C, Herrick J, Sclavi B. 2010. DnaA-ATP acts as a molecular switch to control levels of ribonucleotide reductase expression in Escherichia coli. Mol. Microbiol. 76:1555–1571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.