Abstract
Characterizing genetic diversity within and between populations has broad applications in studies of human disease and evolution. We propose a new approach, spatial ancestry analysis, for the modeling of genotypes in two- or three-dimensional space. In spatial ancestry analysis (SPA), we explicitly model the spatial distribution of each SNP by assigning an allele frequency as a continuous function in geographic space. We show that the explicit modeling of the allele frequency allows individuals to be localized on the map on the basis of their genetic information alone. We apply our SPA method to a European and a worldwide population genetic variation data set and identify SNPs showing large gradients in allele frequency, and we suggest these as candidate regions under selection. These regions include SNPs in the well-characterized LCT region, as well as at loci including FOXP2, OCA2 and LRP1B.
Understanding how genetic diversity of individuals varies across populations has many important applications in modern population genomics. In particular, measures of population structure are used to correct for population stratification in genome-wide association studies1, to identify associations of genetic variants to disease in the context of admixture mapping2, to detect regions that have undergone recent positive selection3–5 and to illuminate interesting aspects of human population history6,7.
Studies analyzing SNP data sets have been able to extract a considerable amount of information on an individual's ancestral origin. Multiple empirical SNP surveys have shown how an individual's geographic ancestry can be inferred using the first two principal components of the genotype matrix (for example, see refs. 8,9). This relationship between principal components and geographic origin is expected when the underlying genetic variation is spatially structured10,11, that is, when genetic similarity decays with the geographic distance between the origins of the individuals. Spatial structure is widespread in human populations due to histories of spatial expansion and spatially restricted mating. Although principal-component analysis (PCA) can capture the spatial structure of the data, it is not based on an explicit probabilistic model for spatial genetic structure and, as a result, is less amenable to extensions to other applications compared to model-based approaches.
In this paper, we report the development of a probabilistic model for the spatial structure of genetic variation, with which we explicitly model how the allele frequency of each SNP changes as a function of the location of the individual in geographic space (where the allele frequency is a function of the x and y coordinates of an individual on a map). Then, each individual's genotypes are assumed to follow Hardy-Weinberg proportions, with allele frequencies defined by the individual's location. The family of functions we use to model allele frequency over space is deliberately simple but leads to tractable inference algorithms with several applications.
If the geographic origins of the individuals are known, we can use this information to infer their allele frequency functions at each SNP. However, if locations are not known, our model can infer geographic origins for individuals using only their genetic data, in a manner similar in spirit to PCA-based approaches for spatial assignment. This ability provides evidence that our modeling of allele frequencies, albeit simple, is sensitive and captures the information about spatial location that is inherent to most variants. As our approach is model based, the model can predict the geographic origins of an individual, even in the case where the individual is of mixed ancestry. This is not possible in other approaches, such as PCA, which is based on a linear combination of genotypes and, therefore, for example, leads to an individual with an Italian parent and a Swedish parent being assigned to Central Europe. Instead, the approach taken here can identify the disparate parental origins. We also show how our approach can be extended to model spatial structure over a sphere to predict the spatial structure of worldwide populations.
Using this framework, we also can identify loci showing extreme patterns of spatial differentiation, for instance, as a result of recent positive natural selection and/or allele surfing12,13. When we applied our SPA approach to genetic data from human populations, we observed that some of the outlier regions detected by SPA have been found with previous methods designed to detect recent positive selection, such as iHS14, FST (refs. 3,15) and the method presented by Coop et al.16, including, for example, the LCT region and human leukocyte antigen (HLA) regions. In contrast to previous methods, our method is unique in being especially sensitive to strong spatial patterns and works at the level of the individual rather than partitioning individuals into populations. The SPA method is particularly sensitive to SNPs that have steep geographic gradients in allele frequency, whereas FST-based approaches simply highlight loci that have large variation in allele frequency.
RESULTS
Model implementation
The first assumption of our approach is that the population allele frequency of each SNP can be modeled as a continuous two-dimensional function on a map. In other words, when sampling a chromosome of an individual from a position (x,y) on the map, the probability of observing the minor allele at SNP j on the chromosome can be formulated as fj(x,y), where fj is a continuous function that describes allele frequency behavior as a function of geographic positioning (Online Methods). We then make the simplifying assumption that this function is an instance of a logistic function
where x is a vector of variables indicating geographic locations and a and b are function coefficients. We refer to each of these fj functions as the slope function of SNP j. This function encodes the steepness of the slope by the norm of a, assuming that the offset parameter b is fixed. Moreover, slope directionality is encoded in the value of vector a. In detail, θj = arctan(aj(1) / aj(2)) can be taken as angle degree for SNP j, where aj(1) and aj(2) are the first and second elements in a. Examples of these functions are shown where the parameter a is set to [0.1,–0.1], [1,–1] or [30,–30] and parameter b is set to be zero in all three slopes (Fig. 1).
Figure 1.
Examples of the allele frequency slope model. (a) Flat slope. A SNP with nearly constant allele frequency in all regions of the map. (b) Medium slope. A SNP with gradual allele frequency change. (c) Steep slope. A SNP with a sharp frequency change.
These functions clearly do not capture cases for which SNPs have complicated functions over geographic space, with, for example, multiple modes or peaks in the allele frequency surface; however, these functions should capture general trends in allele frequency where they exist. For spatial assignment applications, as we show, this behavior is not problematic—a substantial amount of information for assignment arises from SNP loci that show gradients across geographic space. Further, when we use our method for detecting extremely differentiated loci, this assumption implies that the method will only detect loci that are extreme in the sense of having steep gradients in allele frequencies.
The advantage of these functions is that they lend themselves to tractable formulations of the likelihood of genotype data, and we were able to implement efficient Newton's- and pseudo-Newton's–based methods for maximizing the likelihood function for various applications (Online Methods). Using other classes of functions is certainly possible in this framework but might lead to very challenging optimization problems.
Mapping using spatial ancestry analysis
As a first application of our approach, we considered a situation similar to that encountered when performing PCA on a set of individuals with unknown spatial coordinates to infer their spatial origins. For the SPA method, a challenge of this type of analysis is that neither the spatial coordinates of the individuals nor the slope function for each SNP is given, and both must be inferred from the genotypes. The ability to jointly estimate both the allele frequency gradients and the spatial positions of individuals only from the genotype data provides evidence that our model captures spatial genetic structure.
We used a maximum likelihood approach to estimate simultaneously the fj functions for every SNP j and the spatial positioning of each of the individuals (Online Methods). We started by placing the individuals in random positions, and we then iteratively used these positions for the estimation of the slope functions, then using the slope functions to update the individual positions.
We applied SPA to the Population Reference Sample (POPRES)17. The individuals of European descent in this data set were previously analyzed9. The data set contains 3,192 individuals who were genotyped at 500,568 SNPs using the Affymetrix 500K SNP chip. For many of the individuals participating in the study, the ancestry of all four of their grandparents is known; we only considered individuals for whom all reported grandparents had the same ancestry.
Both SPA and PCA results on the POPRES samples are shown (Fig. 2). The SPA method was able to converge to the true geographic position of the individuals from a random starting point (Fig. 2a–d). A map of Europe is labeled with the included populations for reference (Fig. 2f). Of note, even though we started the optimization from a set of random positions, after a small number of iterations (~10), the positions of the individuals highly resembled the map of Europe (with only two exceptions, Slovakia and Russia, which did not converge to true geographic position). The results of PCA are shown for comparison (Fig. 2e). The maps (Fig. 2a–e) are rotated counterclockwise by 16 degrees (similarly to the procedure performed in ref. 9) to more closely resemble the map of Europe. The x and y axes are drawn to equal scale; thus, no distortion is involved. The correlation coefficient between the two maps was 0.99, and, thus, the two methods provided similar positioning of the individuals up to an affine transformation. One noticeable difference was that SPA separated individuals from Spain and Portugal more clearly from those from France than the PCA map. Moreover, the five outlier Italians in the PCA map were positioned closer to Italy by SPA.
Figure 2.
Model-based mapping convergence with random initialization. Colors represent the true country of origin of the individual (also represented by country internet code). (a–d) A map generated by SPA. Iteration 1 starts with random positioning of individuals (a). By iteration 4, the northern and southern populations are separated (b). By iteration 7, the positioning of individuals is close to convergence (c). In iteration 10, individuals have reached their final positions (d). (e) A map generated by PCA9. (f) Map of Europe.
The accuracy of individual placement by SPA was compared to that with PCA, following a described evaluation procedure9 (Table 1). We computed the accuracy on the basis of spatial assignment (Online Methods) that assigns each individual to a country of origin. The results provided support for the notion that the simplified allele frequency functions are capable of extracting the spatial information inherent in the allele frequency data, even when individual spatial coordinates are not provided.
Table 1.
Individual localization result summary
| Geographic origin | Number of individuals | PCA accuracy | SPA accuracy |
|---|---|---|---|
| Italy | 219 | 0.70 ± 0.03 | 0.74 ± 0.03 |
| UK | 200 | 0.44 ± 0.04 | 0.53 ± 0.04 |
| Spain | 136 | 0.71 ± 0.04 | 0.69 ± 0.04 |
| Portugal | 128 | 0.20 ± 0.04 | 0.38 ± 0.04 |
| Switzerland-French | 125 | 0.26 ± 0.04 | 0.33 ± 0.04 |
| France | 89 | 0.70 ± 0.05 | 0.66 ± 0.05 |
| Switzerland-German | 84 | 0.23 ± 0.05 | 0.27 ± 0.05 |
| Germany | 71 | 0.25 ± 0.05 | 0.28 ± 0.05 |
| Ireland | 61 | 0.28 ± 0.06 | 0.28 ± 0.06 |
| Yugoslavia | 44 | 0.25 ± 0.07 | 0.30 ± 0.07 |
| Mean | 115.7 | 0.40 ± 0.05 | 0.45 ± 0.05 |
On the basis of a spatial assignment method, country origin was predicted for each individual (Online Methods). Accuracy ± s.d. is the proportion of individuals from each country of origin correctly assigned to their true country of origin using a leave-one-out procedure.
SPA can also be applied in the case where a subset of the individuals has known spatial origins, and these coordinates can be used to infer the spatial origins of a subset of individuals with unknown origins. In this case, known spatial origins were used for the placement of the individuals, and these placements were then used to estimate the fj functions for every SNP j. We then placed each individual with unknown origins using these functions. We evaluated this approach using POPRES data by performing tenfold cross-validation, where we used the positions of 90% of our individuals to infer the positions of the remaining 10%. The result of SPA placement assuming known positions is shown (Supplementary Fig. 1).
Global genetic spatial structure
Because SPA has explicit geographic coordinates, the approach can be extended to incorporate coordinate systems beyond the two-dimensional plane. As a demonstration of this, we extended SPA to analyze the spatial structure of global populations where a two-dimensional map cannot accurately capture the structure. We mapped each individual to a point on a globe in three-dimensional space. Accordingly, we used a three-dimensional vector x (with the constraint ||x|| equal to a constant) to represent an individual position. We also needed to extend the parameter a to the three-dimensional vector in the logistic function. Examples of these functions with different parameters are shown, where the parameter a is set to [0, 0, 0.1], [0, 0, 3] or [10, 0, 0], and the parameter b is set to be zero in all three spheres (Supplementary Fig. 2). The sphere coordinates are drawn from a unit sphere, where ||x|| = 1.
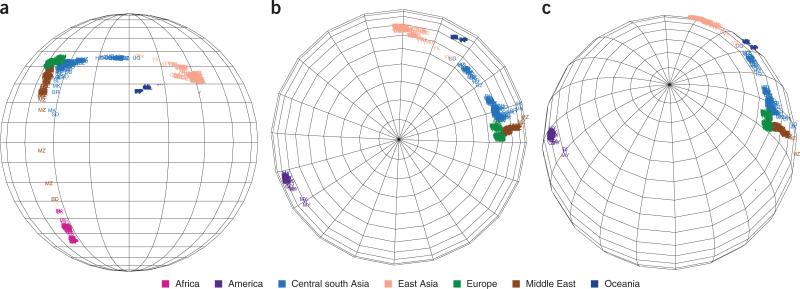
We applied our global genetic spatial structure method to data from the Human Genome Diversity Panel (HGDP)7 in which 940 individuals from 52 populations worldwide were genotyped across the genome using Illumina Infinium HumanHap550 BeadChips (Fig. 3). Notably, even though we started from completely random geographical positioning (Supplementary Fig. 3), we observed that the resulting positioning strongly resembled the world map. In particular, individuals from the same continent were clustered together, and the continents were separated roughly as one would expect.
Figure 3.
Mapping spatial structure on a globe using HGDP data. Different colors represent different continents. (a) Africa-Asia-Europe-Oceania view. (b) North Pole view. (c) Atlantic view.
By aligning the map (Fig. 3), we computed the latitude and longitude for each individual and compared these computations with the actual geographic positions of continents. The SPA map distorted the distances between continents but correctly predicted the topology. For example, the longitudinal span of the Eurasia continent was 92 degrees on the SPA globe and approximately 150 degrees on the actual globe. The longitudinal distance between Europe and North America was 167 degrees on the SPA globe and approximately 90 degrees on the actual globe. The summary of these comparisons is given (Supplementary Table 1).
Mapping of individuals of mixed ancestry
Using a PCA-based approach, one can infer the localization of an individual with an average error of a few hundred kilometers9. However, PCA-based methods are not designed for ancestral origin inference, and, particularly if an individual is of mixed ancestry, the PCA map will place the individual at the midpoint between the coordinates of his or her parents.
Because SPA is a model-based approach, it is possible to extend the method to handle individuals of admixed ancestry. As a result, SPA is able to identify which individuals have admixed ancestry and to predict the origin of each of the parents by computing the maximum likelihood estimates of the origin of the father and the mother simultaneously, under the assumption that the slope functions are given (Online Methods). To test this approach, we generated 5,000 admixed individuals by randomly selecting their parents from the POPRES data set. Each of the parents has four grandparents with the same geographic origin, but the four paternal grandparents and four maternal grandparents of the simulated admixed individuals were different. Also, we ignored genders, as we only used autosomal SNPs. The offspring's genotype was simulated using Mendelian segregation considering each locus independently.
We then applied SPA to predict the country of origin of the parents (Table 2), where the slope functions were estimated on the set of non-admixed individuals. We could not compare the performance of PCA on this simulation, as it would only predict one origin for the individual at the midpoint of the true parental origins. Unexpectedly, the accuracy for placing the parents of admixed individuals was comparable to the accuracy in placing non-admixed individuals, as shown (Tables 1 and 2).
Table 2.
Summary of results for the localization of admixed Individuals
| Origin 1 | Origin 2 | Number of individuals | SPA accuracy |
|---|---|---|---|
| Italy | UK | 250 | 0.49 ± 0.03 |
| Italy | Portugal | 147 | 0.49 ± 0.04 |
| Italy | Spain | 142 | 0.68 ± 0.04 |
| Switzerland-French | UK | 138 | 0.21 ± 0.03 |
| Portugal | UK | 137 | 0.41 ± 0.04 |
| Spain | UK | 128 | 0.45 ± 0.04 |
| Portugal | Spain | 104 | 0.78 ± 0.04 |
| France | Italy | 101 | 0.57 ± 0.05 |
| Germany | Italy | 69 | 0.43 ± 0.06 |
| Germany | Portugal | 60 | 0.30 ± 0.06 |
| Mean | 127.6 | 0.48 ± 0.04 |
Using genotypes from the 5,000 simulated admixed individuals, SPA was used to predict the origin of each parent. Origins 1 and 2 represent the countries of origin for each parent. Accuracy ± s.d. is the proportion of parents correctly assigned to their true country of origin.
We also evaluated our method on self-reported admixed individuals from the POPRES data set. We considered individuals who had self-reported maternal origins from one country and paternal origins from a different country. We used PCA to evaluate the accuracy of the self-reported ancestry. PCA should localize an individual of mixed ancestry at the middle point between the parents’ locations. However, out of a total of 190 individuals with mixed ancestry in the data set, only 12 behaved as simple admixtures and were placed by PCA near the midpoint (<200 km away) of the origins of their parents. The remainder were placed at a greater distance from the midpoint between the reported parental origins—perhaps suggesting more complex ancestry. By applying SPA to the individuals who were placed at the midpoint by PCA, we were able to successfully infer the locations of both parents 58.3% of the time, which is comparable to the rate achieved in the simulated results.
Loci under selection
The detection of genomic regions under natural selection sheds light on the functionality of these regions and provides insights on human history and evolution. A number of methods have been suggested for the detection of selection using genetic variation data, and one particularly common approach leverages the variation in allele frequency between populations through the FST statistic3,15. The FST approach essentially leverages the insight that variation in allele frequencies across populations should follow a background neutral distribution determined by levels of gene flow and divergence and that any regions clearly departing from this distribution are regions that putatively have experienced adaptive differentiation or balancing selection in the recent past.
A disadvantage of FST-based selection detection is that the individual genotypes have to be partitioned into discrete populations. As can be observed (Table 1), the definition of a population, for example, in Europe, is rather subjective. Different groupings of the individuals into populations may result in different results, and, thus, the interpretation of the results is again not straightforward, and particularly important signals of selection may be missed. In addition, FST is not sensitive to whether allele frequency variation is spatially organized into a steep allele frequency gradient or shows a spatially incoherent pattern.
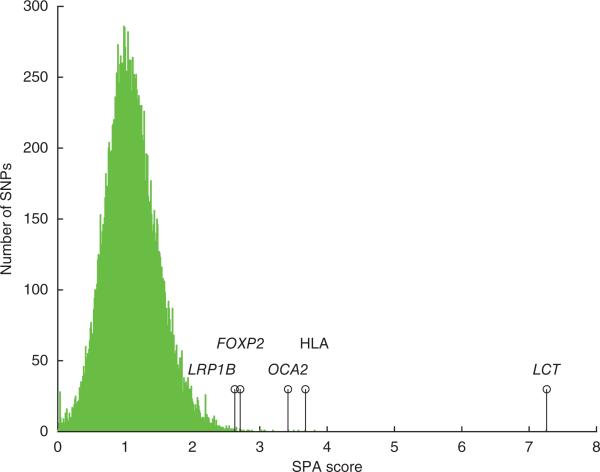
SPA can be used to identify loci showing extreme frequency gradients, which does not require grouping individuals into populations. We used SPA to identify SNPs that show steep slopes of allele frequency change, with the consideration that some of these might show extreme gradients because of the impact of recent positive selection. We developed a new score statistic measuring the slope of each SNP, where large score values correspond to potential regions under selection.
We analyzed the POPRES data set by applying SPA and extracting SNPs with extreme frequency gradients (Online Methods). The distributions of the frequency gradients along with a subset of the SNPs are reported (Fig. 4). We compared the SNPs found by SPA to those identified in the following methods, where (i) we computed FST using two types of population partitions, by country and by geographic regions, as defined9; (ii) we compared SPA scores to the widely used iHS method14, which searches for SNPs with signatures of partial selective sweeps on the basis of haplotype homozygosity, as originally suggested18; and (iii) we compared SPA to Bayenv16, which identifies alleles that correlate strongly with an environmental variable, perhaps due to natural selection. For Bayenv16, we used geographic coordinates as the environmental variable (as if one were searching for latitudinal clines, for example). We obtained outlier signals using latitude, longitude and the individual coordinates corresponding to the first five principal components as the environmental variables.
Figure 4.
The distribution of SPA scores representing allele frequency gradients. The marked positions correspond to genes discussed in the text.
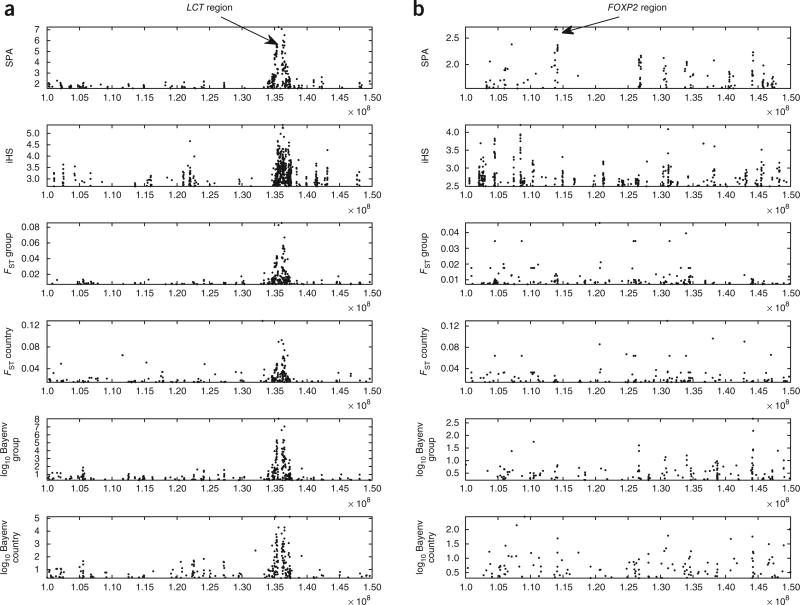
We compared the top results of the four methods applied to chromosomes 2 and 7 (Fig. 5). Note that SPA resulted in a defined cluster of extreme values in 135–138 Mb of chromosome 2, which contains the lactase gene LCT. This region is widely noted as a target of strong selection19, and it was found by all methods. On chromosome 7, SPA detected a strong signal in the FOXP2 region, whereas all other methods did not.
Figure 5.
Selection results of six methods in two chromosomes. The SPA, FST and Bayenv methods were run across the POPRES data set, and the iHS14 approach used HapMap Europe data. The plot is for 2% of POPRES SNPs and 1% of HapMap Europe SNPs. (a,b) Results are shown for chromosome 2 (a) and chromosome 7 (b).
Overall, the different scores provided by these four methods were moderately correlated (r2 < 0.4; Supplementary Fig. 4 and Supplementary Table 2), even though they each measure unique aspects of genetic variation. Most signals found by SPA analysis were also found by the FST method and by Bayenv. However, some of the strong signals that were found by SPA analysis were found by iHS and were not found by FST or by Bayenv, suggesting that our SPA method captures loci in some regions with iHS signals, which are outliers with respect to allele frequency gradient (SPA) but not with respect to overall allele frequency variation (as would be detected by FST or Bayenv16). In addition, there were, as expected, signals that were found using SPA but not using iHS (Supplementary Table 3). We note that the SNPs found by FST and not by other methods were mostly rare SNPs with one or two occurrences of the minor allele in populations with small sample size.
Notably, we observed that the FST and Bayenv scores were sensitive to the manner in which individuals were partitioned into populations. In particular, defining populations on the basis of country of origin led to a different set of genes compared to defining populations on the basis of general geographic regions. In contrast, the analysis performed using SPA was oblivious to the partitioning of individuals into populations, as the approach treats ancestry as a continuous variable and not as a categorical variable.
We provide a list of genes that were detected by our SPA method but were not detected by iHS, FST or Bayenv methods (Supplementary Table 3) and a full list of loci with extreme frequency gradients that were identified in the SPA analysis (Supplementary Table 4). Among the loci with the most extreme gradients were the HLA, LCT and OCA2 regions, which are widely known to have undergone recent positive selection and show differentiation among populations. Of note, SPA analysis also indicated an extreme gradient for SNPs in the FOXP2 gene; FOXP2 is associated with speech, and the FOXP2 protein was suggested to have had important aminoacid changes in early human evolution20. In addition, LRP1B21, a gene associated with lipid function that is relevant to cancer, was found to have an extreme allele frequency gradient. The above loci are a few examples out of a longer list of genes that the SPA method highlighted as having strong gradients in allele frequency across space (Supplementary Fig. 5 and Supplementary Table 4).
DISCUSSION
In this paper, we present spatial ancestry analysis (SPA), a new method for modeling the spatial structure of genetic variation. Unlike previous methods that use PCA to model spatial structure, our approach explicitly models allele frequency in space and uses this model to place individuals on a two-dimensional map or three-dimensional sphere. We show that our method for localization of samples in space is slightly more accurate than PCA and, notably, can be used to localize individuals of mixed ancestry in space, which is not the case for PCA.
Accurate spatial localization of individuals on the basis of genetic data is important in many applications in genetics, including population stratification in genome-wide association studies, admixture mapping and personalized genomics. We show that a model-based approach has additional applications, as it characterizes the spatial behavior of each of the SNPs separately. In particular, we show that the modeling can be used to identify SNPs with rapidly changing allele frequencies across geographic space.
We note that our proposed model for slope functions is only one choice for such a model, and there may be other natural choices. The fact that our algorithm converges to a map that is highly similar to the map of Europe suggests that our choice is sensible but not necessarily optimal. In particular, some loci under selection might in principle have a spatial structure, where the maximum allele frequency occurs in the middle of the region under study and decays in all directions. Such patterns of spatial structure would not be detected by SPA. Further exploration of other choices of slope functions may potentially provide better characterization of each SNP's spatial behavior, yielding a better localization of samples to space and enhanced ability to identify SNPs with unique and interesting spatial distributions.
ONLINE METHODS
Data sets
We applied our methods to a data set collected from European populations, which was assembled and genotyped as part of the larger POPRES project17. A total of 3,192 European individuals were genotyped at 500,568 loci using the Affymetrix 500K SNP chip. After removing SNPs with low-quality scores, the same stringency criteria as in a previous study9 were applied to avoid sampling individuals from outside of Europe, to create more even sample sizes across Europe and to remove individuals whose grandparents had different geographic origins. When available, we used identical geographic origins of the grandparents as the geographic origin for each individual. Otherwise, we used self-reported country of birth. As a result, we focused our analysis on genotype data from 447,245 autosomal loci in 1,385 individuals from 36 populations.
For three-dimensional globe mapping, we used HGDP data7 consisting of 56 populations from Europe, Africa, the Middle East, central Asia, east Asia, Oceania and America. American samples are from both native North American and South American populations. In our experiments, we used genotypes at 572,139 autosomal SNPs in 940 individuals.
Genetic spatial structure model
We assume we are given genotypes at L SNPs from N unrelated individuals drawn from different populations distributed across the geographic regio uder consideration. We assume that the allele frequency of a SNP j is a function
| (1) |
where aj and bj depend on the SNP j and x is the K-dimensional vector of coordinates describing the spatial positioning of an individual. Typically, K = 2 for geographic position. This function has a range [0,1] that can be interpreted as a probability, and, thus, the likelihood of the data can be easily expressed as a function of the values of a, b and x.
We let gij represent the observed number of minor alleles at SNP j of individual i and let fij be a shorthand for fj(xi), where xi is the position of individual i. As the individuals are independently sampled from the population, the log likelihood of the entire observed sample can be calculated from the log likelihood for each genotype.
| (2) |
The parameter matrices X = {xik}, A = {ajk} and B = {bj} are N × K, L × K and L × 1 matrices, respectively. Specifically, each row of X contains the geographic location for each individual. Each row of A and B contains the coefficient for each allele frequency function.
Maximum likelihood estimation
Given the above likelihood model and a set of genotypes, we are interested in the matrices X, A and B that maximize the log likelihood. The above likelihood function is not concave, and it is therefore hard to optimize. We note, however, that when X is fixed or when A and B are fixed, the objective function (2) is concave. We therefore use alternative maximization in conjunction with Newton's method. Furthermore, with fixed A and B, the objective function in X can be decomposed into a series of unrelated parts, each of which corresponds to one row in X, and, therefore, the optimization problem in variable X can then be decomposed into a series of much smaller problems, further simplifying optimization.
After simplification of the above alternative maximization and variable separations of the function (2), we arrive at the following two unconstrained convex programming problems in only K variables and K + 1 variables, respectively.
| (3) |
| (4) |
The smooth and continuous property of this problem allows us flexibility in the choice of optimization method. We apply Newton's method, which is widely known for fast convergence, as it utilizes the first and second order derivatives. Details of the algorithm are given in the Supplementary Note.
An extended model for an admixed individual
Instead of identifying one origin for an admixed individual, our method can infer two geographic origins for the parents. First, we let x and y denote the locations of the two parents of a given admixed individual, and two shorthands pj = fj(x) and mj = fj(y) denote the allele frequency of those two parents at marker j, where the function fj(.) is defined in equation (1).
Therefore, again under the assumption of independent NPs, the genotype of the admixed individual is drawn from the following distribution.
This distribution assumes that the two alleles of admixture individuals are drawn from the parents independently. Finally, we can infer the location of the parents by maximizing the log-likelihood function.
| (5) |
This likelihood function is not concave. Thus, instead of directly using Newton's method that would cause numerical problems, we use Pseudo-Newton's method to optimize this function in x and y. Details of the algorithm are given in the Supplementary Note.
Globe mapping
For globe mapping, we have to extend the two-dimensional vector x to three dimensions. Then, by similar derivation as for two-dimensional mapping, we can obtain the log-likelihood function in the same form as in equation (2) but in a different number of dimensions. To guarantee the placement of individuals in a sphere, we need to enforce the constraint ||x||2 = 1 while maximizing the log likelihood. However, this additional constraint and its non-convexity do not allow us to use Newton's method. Instead, we turn to another widely known optimization technique called gradient projection22, which can handle simple constraints in the optimization problem. Basically, this technique modifies the line search step in the gradient descent method to ensure that the current solution is in the feasible region. One key step is the projection from any point to the feasible region. The projection to a sphere can be very efficiently computed by P(x) = x / ||x||2.
Evaluation of individual mapping
SPA can be applied in the case that the geographic origins of the individuals are known, as well as in the case where the geographic origins are unknown. If the geographic origins are known, the slope functions parameterized by aj and bj are estimated using these known locations and will be concordant with actual geography. In this case, the output of individual mapping is immediately latitude and longitude.
If the geographic origins are unknown to SPA, the mapping coordinates might be different from real geography in latitude and longitude up to an affine transformation. In order to perform spatial assignment, we follow the approach taken in ref. 9 and assume the following model between mapping coordinates and geographic locations
where u and v are latitude and longitude, respectively. x = (x1,x2) are the coordinates from our model. The parameters α and β can be estimated from a few individuals with known mapping coordinates and geographic locations. The same model was used in ref. 9 in order to estimate the accuracy of geographic assignments with PCA.
The accuracy evaluations (Tables 1 and 2) are computed on the basis of spatial assignment. We follow a similar leave-one-out strategy to the one used in ref. 9. First, we estimate the coefficients α and β by performing a least-square regression from the mapping coordinates to the true geographic location in latitude and longitude with a leave-one-out training set of individuals. Then, for a test individual, we make a prediction of geographic location using the obtained regression coefficients α and β. We also predict population origin by assigning this individual to the nearest country center. The assignment accuracy for a given population is calculated as the number of correct predictions divided by the total number of individuals in that population.
Characterization of extreme allele frequency gradients
The outputs of the SPA model would be individual mapping coordinates X and coefficients A and B for allele frequency slope functions. On the basis of these two outputs, all individuals in the model will have allele frequencies fj = {fj(x1),fj(x2), ..., fj(xN)} organized in a slope corresponding to each SNP j.
A straightforward statistic to quantify the steepness of allele frequency slope is as follows
where stands for the allele frequency for individual i at locus j. This score is exactly proportional to the s.d. of fj by a constant .
Supplementary Material
ACKNOWLEDGMENTS
W.-Y.Y. and E.E. are supported by grants from the US National Science Foundation (0513612, 0731455, 0729049, 0916676 and 1065276) and the US National Institutes of Health (K25 HL080079, U01 DA024417, P01 HL30568 and PO1 HL28481). J.N. is supported by National Science Foundation grant (0933731) and by the Searle Scholars Program. E.H. is a faculty fellow of the Edmond J. Safra Program at Tel Aviv University and was supported in part by the Israeli Science Foundation (grant 04514831) and by IBM open collaborative research award program.
Footnotes
AUTHOR CONTRIBUTIONS
W.-Y.Y., J.N., E.E. and E.H. designed the methods and experiments. W.-Y.Y. implemented the methods. W.-Y.Y., J.N., E.E. and E.H. jointly performed the analysis. All authors discussed the results and contributed to the writing of the manuscript.
URLs. SPA software, http://genetics.cs.ucla.edu/spa.
Note: Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 2.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat. Rev. Genet. 2011;12:523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coop G, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 7.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 8.Lao O, et al. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 9.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat. Rev. Genet. 2009;10:745–755. doi: 10.1038/nrg2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Excoffier L, Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008;23:347–351. doi: 10.1016/j.tree.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson MR, et al. The population reference sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 2008;83:347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 19.Bersaglieri T, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 21.Liu CX, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic organization of a new candidate tumor suppressor gene, LRP2B. Genomics. 2000;69:271–274. doi: 10.1006/geno.2000.6331. [DOI] [PubMed] [Google Scholar]
- 22.Nocedal J, Wright SJ. Numerical Optimization. Springer; New York: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.