Abstract
Despite increasing evidence that supports the role of different post-translational modifications (PTMs) in modulating α-synuclein (α-syn) aggregation and toxicity, relatively little is known about the functional consequences of each modification and whether or not these modifications are regulated by each other. This lack of knowledge arises primarily from the current lack of tools and methodologies for the site-specific introduction of PTMs in α-syn. More specifically, the kinases that mediate selective and efficient phosphorylation of C-terminal tyrosine residues of α-syn remain to be identified. Unlike phospho-serine and phospho-threonine residues, which in some cases can be mimicked by serine/threonine → glutamate or aspartate substitutions, there are no natural amino acids that can mimic phosphor-tyrosine. To address these challenges, we developed a general and efficient semisynthetic strategy that enables the site-specific introduction of single or multiple PTMs and the preparation of homogeneously C-terminal modified forms of α-syn in milligram quantities. These advances have allowed us to investigate, for the first time, the effects of selective phosphorylation at Y125 on the structure, aggregation, membrane binding and subcellular localization of α-syn. The development of semisynthetic methods for the site-specific introduction of single or PTMs represents an important advance toward determining the roles of such modifications in α-syn structure, aggregation and functions in heath and disease.
INTRODUCTION
α-synuclein (α-syn) aggregation and fibrillogenesis in the brain are the hallmark of Parkinson’s disease (PD) and have been linked to the pathogenesis of several neurodegenerative diseases that are collectively referred to as “synucleinopathies”1. PD is characterized by the formation of intraneuronal inclusions termed Lewy bodies (LB), which are composed primarily of insoluble fibrillar and aggregated forms of α-syn 2, 3. Although three mutations in the α-syn gene (A30P4, A53T5, and E46K6) have been linked to early-onset forms of the disease, they account for only 1% of all PD cases. Therefore, a better understanding of the molecular determinants that govern the structural, aggregation and functional properties of α-syn is crucial to elucidating its role in the pathogenesis of sporadic and inherited forms of PD.
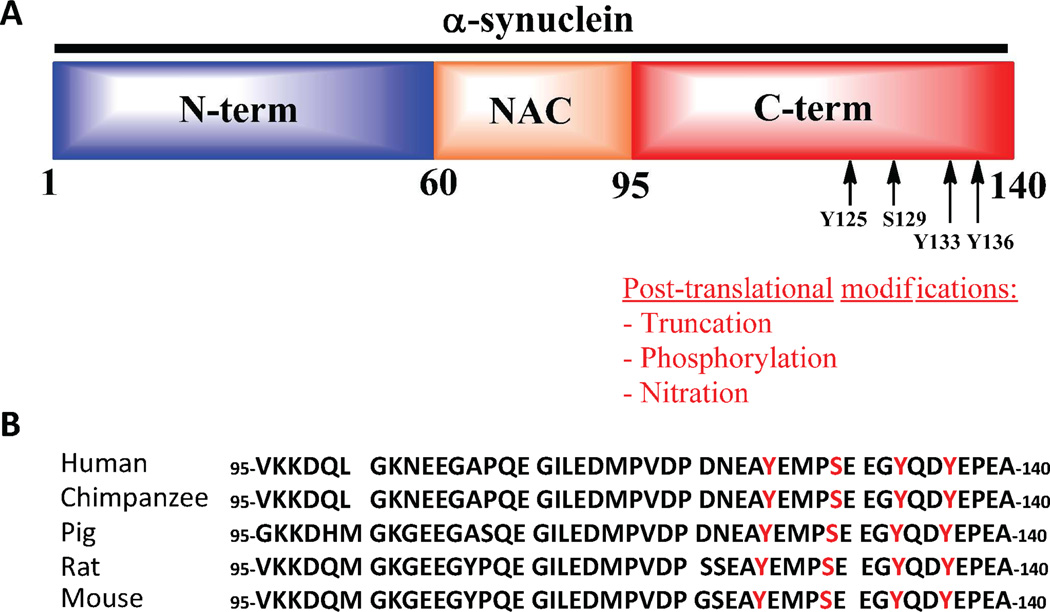
The primary sequence of α-syn can be divided into three domains, which are: 1) The N-terminal domain which contains the sites of three disease-associated mutations and the imperfect 11 repeats of a conserved sequence (KTKEGV) that play an important role in regulating α-syn secretion7, internalization 8, 9 and binding to membranes; 2) The central fragment, also known as the NAC - non amyloid component - (residues 61–95) region, is highly hydrophobic and is essential for α-syn oligomerization and fibrillogenesis 10; 3) The C-terminal region comprising residues 96–140. This region is highly disordered and negatively charged (10 glutamate and 5 aspartate residues). The C-terminal region has been proposed to play a role in regulating α-syn transport to the nucleus and has been shown to mediate α-syn interactions with other proteins 11–14, metal ions 15, 16 and natural ligands (e.g. dopamine and polyamines) 17. The majority of α-syn PTMs (truncation, phosphorylation, methionine oxidation, and nitration) occurs within the C-terminal region. The clustering of PTMs in this functional domain of the protein suggests that these modifications may be involved in the regulation of α-syn function in health and disease. This hypothesis is further supported by the fact that the majority of these post-translational modification sites are highly conserved across different species. Furthermore, the close proximity of these different modifications suggests that they may act in concert, and cross-talk between these modifications may constitute an additional molecular switch for regulating the dynamics of α-syn function and aggregation (Scheme 1).
Scheme 1. The clustering of C-terminal post-translational modifications of α-syn.
A: Schematic depiction of the sequence of α-syn with its 3 domains. The putative and identified sites of C-terminal phosphorylation are indicated with arrows. B: Sequence alignment of the sequences of the C-terminal domain of α-syn from 5 different species. The conserved positions of the phosphorylation sites are highlighted in red.
Despite mounting evidence supporting the role of C-terminal phosphorylation in modulating α-syn aggregation and toxicity, relatively little is known about the functional consequences of tyrosine phosphorylation and whether or not tyrosine and serine phosphorylation in this region are regulated by each other 18. This is partly due to the lack of tools and methodologies for the site-specific introduction of single or multiple PTMs in α-syn. Previously, our group identified the polo-like kinases, PLK2 and PLK3, which phosphorylate α-syn efficiently in vitro and are major contributors to α-syn phosphorylation in vivo 19, 20. However, the kinases that carry out selective and efficient phosphorylation of the other known and putative phosphorylation sites (S87, Y125, Y133, and Y136) remain to be identified.
Protein semisynthesis 21–28, which combines the powers of recombinant protein expression, peptide chemical synthesis and native chemical ligation (NCL) has successfully used to study post-translational modifications on a variety of proteins 27, 29 and offers means of generating site-specifically modified forms of α-syn30, 31 (Scheme 2). To address the aforementioned challenges and to facilitate the investigation of the structural and functional consequences of specific C-terminal PTMs of α-syn, we developed a general and efficient semisynthetic strategy that permits the site-specific introduction of single or multiple PTMs. This strategy also enables the preparation of milligram quantities of homogeneous forms of α-syn modified at the C-terminus that are sufficient for carrying out detailed biochemical, biophysical and cellular studies. We used this strategy to produce α-syn phosphorylated at tyrosine 125 (pY125 α-syn). We choose to investigate Y125 phosphorylation for the following reasons: 1) the levels of pY125 α-syn have been reported to decrease with age 18; 2) studies by Feany and colleagues suggest that phosphorylation at Y125 acts as a counter-regulator of pS129-induced toxicity in the fly model of PD 18, and provide evidence for a potential cross-talk between these two modifications; 3) because there are no effective mutagenesis-based strategies for mimicking tyrosine phosphorylation and thus very little is known about the structural and functional consequences of selective phosphorylation of α-syn at Y125. More specifically, we determined the effect of phosphorylation at Y125 on the structure, fibril formation, and phospholipid membrane association-induced conformational shifts in vitro. In addition, we further probed cross-talk between phosphorylation at S129, Y125, and S87 residues by determining the relative activity of each phosphorylated form of α-syn as a substrate for the kinases that phosphorylate α-syn at other residues within the C-terminus. We also assessed, for the first time, the subcellular localization of pY125 α-syn in mammalian cells and rat hippocampal neurons. Our work demonstrates the power and potential of applying chemical approaches to elucidate the role of C-terminal PTMs in regulating α-syn function in health and disease.
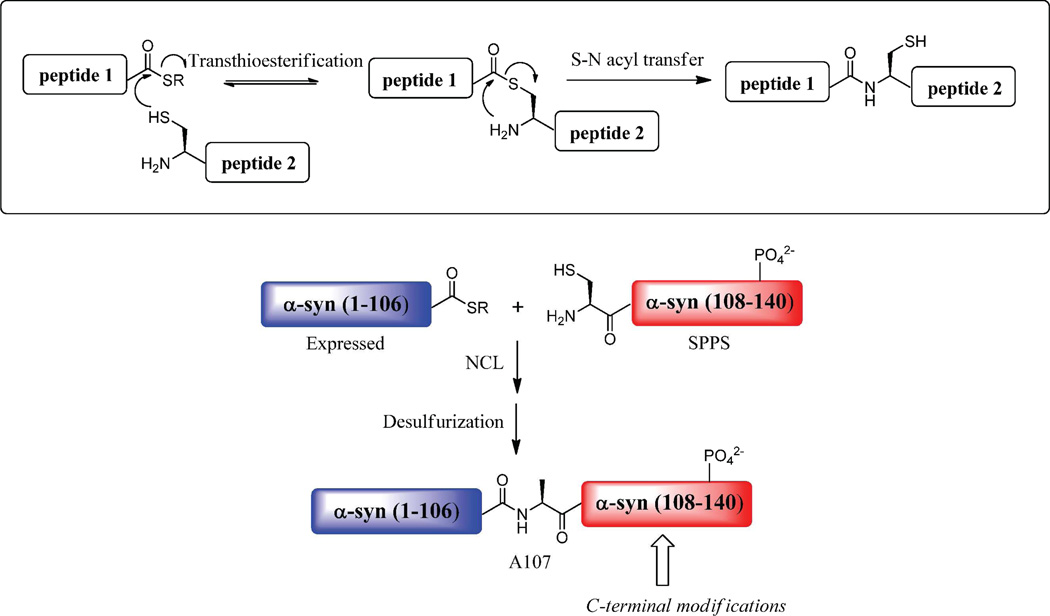
Scheme 2. Mechanism of Native Chemical Ligation.
Top: A peptide thioester and a peptide fragment bearing an N-terminal Cys undergo a thioester exchange followed by an S to N acyl migration to create an amide bond between two unprotected peptides in aqueous solution. Bottom: Semisynthetic strategy to generate derivatives of α-syn site-specifically phosphorylated within the C-terminal region. The protein thioester α-syn(1–106)SR generated by recombinant protein expression is ligated by NCL to the synthetic C-terminal fragment α-syn(A107C-140) bearing the desired phosphorylated side chain. A final desulfurization step restores the native Ala-107.
MATERIAL AND METHODS
Materials
9-Fluorenylmethoxycarbonyl (Fmoc)-amino acids, t-butyloxycarbonyl (Boc)-amino acids, 2 - (1H - benzotriazole - 1 - yl) - 1,1,3,3 - tetramethyluronium hexafluorophosphate (HBTU), were purchased from Novabiochem or Anaspec. The side-chain functionalities of the Fmoc-amino acids were chemically protected as shown in parentheses: Asp(OtBu), Glu(OtBu), Lys(Boc), Thr(tBu), Ser(tBu), Tyr(tBu), Gln(Trt), and Asn(Trt). Phosphorylated Tyr residues were purchased from Novabiochem or Anaspec as protected Fmoc-Tyr[PO(OBzl)]-OH building blocks. All unlisted amino acids were not protected. The Boc-amino acid was Boc-Cys(Trt). N,N-diisopropylethylamine (DIPEA), trifluoroacetic acid (TFA), triisopropylsilane (TIPS), sodium 2-mercaptoethanesulfonate (MESNa), ammonium iodide, 1,2-ethanedithiol (EDT), 2-methyl-2-propanethiol (tert-butyl mercaptan, tBuSH) dimethylsulfide, guanidine hydrochloride (GuHCl) and tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich. Piperidine and diethyl ether were purchased from Acros. 2-2’-Azobis-[2(2-imidazolin-2-yl)propane] dihydrochloride (VA-044) was purchased from Wako. The solvents N-methylpyrrolidone (NMP), dichloromethane, and HPLC-grade acetonitrile were purchased from VWR.
General Solid Phase Peptide Synthesis and Purification
Solid phase peptide synthesis was performed on an automated CS 336X peptide synthesizer from CS Bio using standard Fmoc protocols including the in situ neutralization protocol described by Alewood and colleagues32. A preloaded Fmoc-Ala Wang resin was used. Syntheses were carried out on a 0.2 mmol scale and coupling steps were performed with 5 eq. Fmoc-amino acid, 5 eq. HBTU/HOBt, and 5 eq. DIPEA in NMP. The N-terminal Cys residues were attached with Boc protecting groups on their N-terminus. Fmoc-group deprotections were carried out with 20% piperidine in DMF. Peptide side chain deprotections, removal of N-terminal Boc group and cleavage from the resin was performed by treatment with 81.5:5:3:5:2.5:2:1.5 of TFA:water:phenol:thioanisole:EDT:Me2S:NH4I for 4 hours on a rotating wheel to generate peptides with a C-terminal acid functionality. The crude product was subsequently precipitated three times with a ten-fold excess volume of cold diethyl ether, dissolved in water and lyophilized. Peptides were then purified by reversed-phase HPLC (Waters 600 system) using a Cosmosil C18 preparative column (20×250 mm, 5µm particles, 300Å pores) with a linear gradient of 20–70% B over 30 minutes (solvent A was water/0.1% TFA and solvent B was acetonitrile/0.1% TFA). Peptide elution was monitored by UV absorbance at 214 nm and 280 nm. Peptide masses were confirmed by MALDI-TOF-MS and ESI-MS analysis. Peptide purity was confirmed by re-injection on an analytical RP-HPLC column (Cosmosil C18, 4.6×250 mm, 5µm particles, 300Å pores).
Expression and Purification of Recombinant α-Syn(1–106)SR
C-terminally truncated human α-synuclein was initially cloned into the pT7-7 vector in fusion with the GyrA Mini-Intein from Mycobacterium Xenopii and a chitin binding domain (CBD) using the following primers: Forward: 5’-GGG CAA GAA TGA AGA AGG ATG CCG AAG AGC AGG AAT TCT G-3’ and reverse: 5’-CAG AAT TCC TGC TCT TCG GCA TCC TTC TTC ATT CTT GCC C-3’. The α-syn fragment (1–106) was isolated by NdeI and SapI digest. The intein was isolated from the pTXB1 vector (New England Biolabs) by SapI and BamHI digest. Both fragments were ligated into pT7-7 digested by NdeI and BamHI to generate the plasmid pT7-7 α-syn-Mxe-CBD. To insert a polyhistidine tag at the C-terminus of the intein instead of CBD, an EcoRI site was introduced using site-directed mutagenesis using the following primers: Forward: 5’ CTG GCC TCA CCG GTC TGG AAT TCG GCC TCA CGA CAA ATC 3’ and reverse 5’ GAT TTG TCG TGA GGC CGA ATT CCA GAC CGG TGA GGC CAG 3’. An oligonucleotide containing six histidines and two stop codons with the digested recognition sequences of EcoRI and BamHI sites was ordered: Forward 5’AA TTC CAC CAC CAC CAC CAC CAC TGA TAA G 3’ and reverse 5’ GA TCC TTA TCA GTG GTG GTG GTG GTG GTG G 3’. The annealed oligonucleotides were ligated into the plasmid pT7-7 α-syn-Mxe-CBD digested with EcoRI and BamHI. Expression was carried out in E.coli BL21(DE3). Protein expression was induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and by growing the cells for 4h at 37°C. The cells were harvested by centrifugation and lysed by ultra-sonication in 40 mM Tris-acetate, 5 mM EDTA pH 8,3. The supernatant was separated from the cell debris by centrifugation (23, 000×g, 30 min, 4°C). 4 equivalents of MgCl2 were added prior to the loading on the Ni-NTA column. The soluble fraction was loaded on a HisPrep FF 16/60 (Amersham) pre-equilibrated with 50 mM HEPES pH 7.5. A washing step of 50 mM HEPES 25 mM imidazole, pH 7.5 for 5 column volumes was included to remove non-specific binding. The protein α-syn-Mxe-His(6) was eluted with a step elution of 50 mM HEPES, 350 mM Imidazole pH 7.5. The splicing reaction was carried out directly in the eluate from the Ni-NTA column for 12 h under agitation at RT in presence of 0.25 M MESNa. The splicing efficiency was around 90% (by SDS-PAGE analysis, data not shown). The sample was subsequently purified by rever-sephase HPLC using a preparative C4 column (VYDAC, 20×250 mm,5µm particles, 300Å pores) with a linear gradient of 20–70% B in 30 min at a flowrate of 10 ml/min (solvent A was water/0.1% TFA and solvent B was acetonitrile/0.1% TFA). The pure protein thioester was obtained in a yield of 10 mg per liter of BL21 (DE3) expression medium.
Expression and purification of 15N labeled proteins
α-syn A107C
The α-syn mutant A107C was generated using single-point mutagenesis with the following primer pairs to generate the plasmid pT7-7 α-syn A107C: forward primer 5’ GCA AGA ATG AAG AAG GAT GCC CAC AGG AAG GAA TTC TG 3’ and reverse primer 5’ CAG AAT TCC TTC CTG TGG GCA TCC TTC TTC ATT CTT GC 3’. BL21 (DE3) cells were transformed with the plasmid and grown in M9 minimal media with 15N labeled ammonium chloride. Protein expression was induced by the addition of 1 mM IPTG for 4 h at 37°C. After harvesting the cells by centrifugation at 6000 × g for 15 min, the cell pellet was resuspended in 40 mM Tris-acetate and 5mM EDTA (pH 8.3) and lysed by ultra-sonication. The insoluble material was removed by centrifugation at 23,000×g for 40 min at 4°C. The supernatant was boiled for 15 min in a water bath at 100°C followed by centrifugation at 23,000×g for 40 min, which precipitates most cellular proteins but not α-syn A107C. The final supernatant was filtered through a PVDF membrane with 0.22 µm pores. Anion-exchange chromatography was performed on a Pharmacia AKTA FPLC system using a 20 mL HiPrep 16/10 Q FF column (Amersham). The protein was eluted with a linear gradient of 0 to 1 M NaCl (the protein elutes at approximately 300 mM NaCl ) and subsequently purified on a Superdex 200 26/60 size-exclusion chromatography column (Amersham) using a mobile phase of buffer containing 50 mM Tris, 150 mM NaCl, pH 7.5. The purified protein was dialyzed extensively against de-ionized water and lyophilized and stored at −20°C until used. From 1 L of expression medium, the average yield of lyophilized material was 30 mg. To remove the glutathatione disulfide adduct, the lyophilized powder was dissolved in a buffer containing 50 mM Tris 150mM NaCl (pH 7.5), with 30eq of TCEP and the solution was incubated for 10 min at RT. The protein was subsequently purified on a C4 preparative column (Vydac) with a linear gradient of 20–70% B in solvent A over 30 min at a flow rate of 10 mL/min (solvent A was water/0.1% TFA and solvent B was acetonitrile/0.1% TFA). The N15 labeled protein was obtained in a final yield of 10 mg per liter of bacterial expression.
α-syn (1–106)SR
The expression of 15N labeled α-syn(1–106) SR was carried out in a similar manner as for the non-labeled protein with an overall yield of pure protein of ~3.5 mg per liter.
Semisynthesis and desulfurization of α-syn Phosphorylated at Tyr 125
The semisynthesis of α-syn site-specifically phosphorylated within the C-terminus (residues 107–140) involved an NCL reaction between recombinant α-syn(1–106)SR and synthetic peptides α-syn (A107C-140) phosphorylated at Tyr-125 or the WT peptide. NCL reactions were carried out in 6 M GuHCl, 200 mM sodium phosphate buffer, pH 7.0 using 1.5 mM of α-syn(1–106)SR and 2 molar excess of C-terminal peptide A107C-140 pY125 or A107C-140 WT peptide in the presence of 30 eq. TCEP in a volume of 1 mL at 37°C with shaking at 1000 rpm. Three ligation reactions were carried out in parallel. To monitor the NCL reactions, aliquots of the reaction mixture (4µl) were removed and quenched with 38 µl of water containing 0.1% TFA and subsequently analyzed by SDS-PAGE (15% polyacrylamide) and MALDI-TOF-MS. RP-HPLC was not used due to the co-elution of α-syn(1–106)SR and the ligated product under all conditions tested in our laboratory. Based on SDS-PAGE analysis, the desired ligated product was produced within 1–2 hrs.
Free radical-based desulfurization of the semi-synthetic α-syn mutant was carried out as previously described by Danishefsky and colleagues33. In our case, the ligation and the desulfurization were carried out in one-pot. 1mL of a solution of 1 M of TCEP (in 6 M GuHCl, 200 mM sodium phosphate pH 7.0) was added to the ligation reaction. 160 µl of 2-methyl-2-propanethiol and 100 µl of a 0.1 M solution of 2-2’-Azobis[2-(2-imidazolin-2-yl)propane] dihydrochloride (VA-044) free radical initiator (in 6 M GuHCl, 200 mM sodium phosphate pH 7.0) were added under argon. The reaction was incubated at 37°C with agitation at 1000 rpm in a ventilated fume hood for 3.5 h. The progress of desulfurization was followed by MALDI-TOF-MS.
Purification of the semisynthetic pY125 α-syn
Prior to purification, the ligated desulfurized products from the 3 ligation/desulfurization reactions (that were carried out in parallel) were desalted through a 5mL HiTrap desalting column (Amersham) with 20 mM sodium dihydrogen citrate pH 4.0 as the mobile phase. The fractions containing the protein were subsequently pooled and manually injected on a 5 mL cation-exchange column HiTrap SP HP (Amersham). We took advantage of the charge difference between the full-length protein, the fragment α-syn(1–106)OH (hydrolyzed thioester) and the C-terminal 107–140 peptide for a single-column purification. At pH 4.0, the peptide is negatively charged (net charge: −2) and will not bind to the column while the full-length protein and the fragment (1–106) are positively charged and hence will bind to the column. At pH 5.0, the charge of the full-length protein is −1 and the fragment (1–106) has a charge of 6 and can be subsequently separated using a gradient of salt. We initially allow the peptide to pass in the flow through by washing the column after injection with 10 column volumes (CV) of 20 mM sodium citrate pH 4 then with a linear gradient of NaCl from 0 to 100% B (where A is a solution of 20 mM sodium citrate pH 4, B the same solution supplemented with 0,5 M NaCl) over 4 CV on a Akta Purifier system. To elute bound proteins, we applied a linear gradient of NaCl from 0 to 70% B (where A is B is a solution of 20 mM sodium citrate pH 5 and B is the same solution supplemented with 0,25 M NaCl) over 30 CV at a flow of 1 mL/min. The full-length protein elutes around 20% B and the fragment (1–106) around 30% B. The fractions containing the protein were extensively dialyzed against de-ionized water and lyophilized. The final yield of pure lyophilized protein is around 19% (9 mg).
Preparation and purification of pS129 and pS87 α-syn
Recombinant WT α-syn (3 mg per reaction, 10 µM) was phosphorylated at S129 in vitro by PLK3 (10 µg per reaction, 4.8 µg/mL) at 30°C in 20 mM HEPES (pH 7.4), 10 mM MgCl22 mM DTT, and 1 mM ATP. The phosphorylated protein was purified by reversed-phase HPLC using a semipreparative C8 column (InertSil 7.6 × 250 mm, 5 µm particles, 300Å pores). The proteins were eluted with a linear gradient from 20% to 40% of solvent B in solvent A over 40 min at a flowrate of 5 mL/min (solvent A was water/0.1% TFA and solvent B was acetonitrile/0.1% TFA). The collected fractions were pooled according to purity assessed by ESI-MS and MALDI-TOF-MS. After purification, pS129 α-syn was lyophilized and stored at −20°C until use. pS87 α-syn was prepared as previously described 34.
Circular dichroism measurements
Protein samples (10 µM) in pH 7.5 buffer containing 50 mM Tris buffer and 150 mM NaCl were analyzed at 25°C using a Jasco J-815 CD spectrometer. An average of 10 spectra was collected in the range of 195–250 nm using a 0.1 mm path length quartz cuvette. Data points were acquired every 0.2 nm in the continuous scanning mode at a speed of 50 nm/min with a digital integration time of 2 s, and a bandwidth of 1 nm. Processed spectra were obtained by subtracting the baseline signal due to the buffer and cell contribution from the protein spectra and a final smoothing was applied (Savitzky-Golay filter; convolution width of 25 data points).
In vitro Phosphorylation assay
pY125 α-syn (6 µM) was incubated in 50 mM Tris pH 7.5, 10 mM DTT, 1 mM ATP, 1 mM MgCl2 in presence of 0,42 µg (1µL) of PLK3 (Invitrogen) in a total volume of 50 µL. For the phosphorylation by CK1, 6 µM of protein were incubated in CK1 buffer 1X supplemented with 1 mM ATP and 1000 units of CK1 (1 µL) in a final volume of 50 µL. The reaction was allowed to proceed for 12 h at 30°C with no agitation. The extent of phosphorylation was monitored by MALDI-TOF-MS and western-blotting using pS87, pS129 and pY125-specific antibodies.
For phosphorylation by tyrosine kinases, WT α-syn, -pS87, and -pS129 were incubated at a concentration of 3 µM in a final volume of 30 µL with the appropriate tyrosine kinase. Kinases were added at a final concentration of 50 nM. The reaction buffers were the following: Syk / Lyn: 50 mM Tris and 5 mM MnCl2 (50µL); Fyn / Src: 20 mM Tris and 5 mM MnCl2 (pH 7.4); Fgr: 50 mM Tris and 5 mM MgCl2 (pH 7.5). The reactions were started by the addition of freshly prepared ATP (1 mM), and incubated for 14 h at 30 °C before analysis.
NMR spectroscopy
NMR studies were performed as previously described35. Briefly, lyophilized 15N-labeled α-syn was dissolved in sample buffer (100 mM NaCl, 10 mM Na2HPO4, in 90% H2O, 10%, D2O at pH 7.4 to a concentration of 0.1–0.3 mM. For spectra in the presence of SDS, the sample buffer also contained 40 mM SDS. All 1H, 15N HSQC spectra were collected at 10 °C in the absence of SDS and at 40 °C in the presence of SDS on either a Varian INOVA 600 MHz spectrometer (Weill Cornell Medical College) or a Bruker Avance 800 MHz spectrometer (New York Structural Biology Center) equipped with cryogenic probes. NMR data were processed by NMRPipe 36 and analyzed in NMRViewJ 37. Spectra were referenced indirectly to sodium 2,2-dimethyl-2-silapentane-5-sulfonate and ammonia using the known chemical shift of water. Assignments for the spectra of mutant and phosphorylated α-syn at Y125 were estimated by transferring each previously assigned cross-peak in the HSQC spectrum of WT α-syn to the closest unassigned cross-peak in each spectrum. Normalized per residue, the chemical shift change was calculated as Δδ = √(ΔδH1 2 + ΔδN15 2/25) /2). For PRE experiments, α-syn was dissolved in sample buffer without SDS, a 10-fold excess of MTSL spin-label was added and the resulting solution was incubated for 30 minutes. A buffer-exchange column was used to remove unbound spin-label and the sample was divided into diamagnetic and paramagnetic samples. DTT (final concentration 2 mM) was added to the diamagnetic sample to reduce off the spin-label. Matched two-dimensional HSQC spectra were collected on the two samples, and the ratio of the intensity of each cross-peak was determined.
In vitro fibrillization assay
pY125 α-syn and WT α-syn were incubated at 15 µM in a volume of 450 µL under constant agitation at 1200 rpm for 48h or 7 days at 37°C. ThT fluorescence reading was carried out with a ThT concentration of 10 µM and a protein concentration of 1.5 µM in a volume of 70 µL in a pH 8.5 buffer containing 50 mM Glycine. A TECAN spectrometer was used to measure ThT fluorescence at an excitation wavelength of 450 nm and an emission wavelength of 485 nm. Aliquots taken at 0, 24 and 48 h were measured in triplicates. Values were expressed as the ThT fluorescence of three measurements (mean ± S.D.).
Transmission electron microscopy (TEM)
7 µL of proteins were deposited on Formvar-coated 200 mesh copper grids (Electron Microscopy Sciences). Grids were washed with two drops of double-distilled H2O and stained with two drops of freshly prepared 2% (w/v) uranyl acetate (Electron Microscopy Sciences) and then vacuum dried from the edge of the grids. Specimens were viewed on a Philips CIME 12 electron microscope, operated at 80 kV. Digitized photographs were recorded with a CCD camera (Digital Camera Morada by Soft Imaging System, 4008 × 2672 pixels).
Western-blot analysis
Protein samples were separated at 80 V on 15% polyacrylamide 1 mm-thick gels over 1–2 h. The proteins were transferred onto nitrocellulose membranes (Omnilab SA) using a semi-dry transfer system (Bio-Rad) under constant current (~200 mA per membrane) and the membranes were blocked for 1 h at room temperature with Odyssey blocking buffer (Li-COR Biosciences) diluted three-fold in PBS buffer. The membranes were probed with the relevant primary antibodies: mouse anti-α-syn (1:1000 dilution, BD Transduction Laboratories), rabbit anti-α-syn ab51252 (1:1000 dilution, Abcam), rabbit anti-α-syn SA3400 (1:1000 dilution, Biomol) mouse monoclonal anti-phosphorylated α-Syn at Tyr-125 (1:1000 dilution; BD Transduction Laboratories), homemade rabbit anti-pY125 α-syn (1:1000); mouse monoclonal anti-pS129 α-syn (dilution 1:5000; Wako), rabbit anti-pS87 α-syn (1:200 dilution,38) overnight at 4°C. After three washes with PBS buffer containing 0.1% (v/v) Tween 20 (Fluka), the membrane was incubated with secondary goat anti-mouse or anti-rabbit antibodies conjugated to Alexa 680 (dilution 1:5000; Invitrogen). The membranes were then washed three times with PBS buffer containing 0.01% (v/v) Tween 20 and once with PBS buffer and scanned on a LiCOR scanner at 700 nm.
pY125 α-syn dephosphorylation assay
HeLa cells were grown to 90% confluency in a 10cm Petri dish and lysed by occasional vortexing on ice over 30 min in 600 µL of lysis buffer (50 mM tris pH 7.4, 150 mM NaCl, 5% glycerol, 25 mM sucrose, 1% NP-40, and 0.25% sodium deoxycholate with 100 mM PMSF and protease inhibitor cocktail from Sigma). The lysate was clarified by centrifugation at 20800 g for 30 min at 4°C. Incubation of pY125 α-syn (1.2 µg in 100 µL) with HeLa lysate was carried out for 0, 30 min and 1 h at 37°C. The reaction was quenched by the addition of Laemmli sample buffer. For inhibition by sodium orthovanadate, different concentrations of the activated inhibitor (0.1, 1 and 10 mM) were incubated with samples of lysate containing 1.2 µg of pY125 α-syn for 2 h. Lysate from each incubation (300 ng) was loaded onto a SDS-PAGE gel and analyzed by Western-Blot with antibodies against WT α-syn and pY125 α-syn.
Cell culture and transient transfection
Human embryonic kidney cells (HEK 293T) and HeLa cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum and 5% penicillin/streptomycin in a humidified incubator with 5% CO2 at 37 °C. For HEK 293T cells, transient transfection was performed in 6-well plates at a cell confluence of 70–80%, using a standard calcium phosphate (CaPO4) transfection protocol. For HeLa cells, transient transfection using Lipofectamine™ 2000 (Invitrogen) was performed according to the manufacturer’s instructions. The total amount of plasmid used in the transfection was 2 µg/well for 6-well-plates and 0.8 µg/well for 24-well plates.
Detection of pY125 α-syn in HEK 298T and HeLa cells
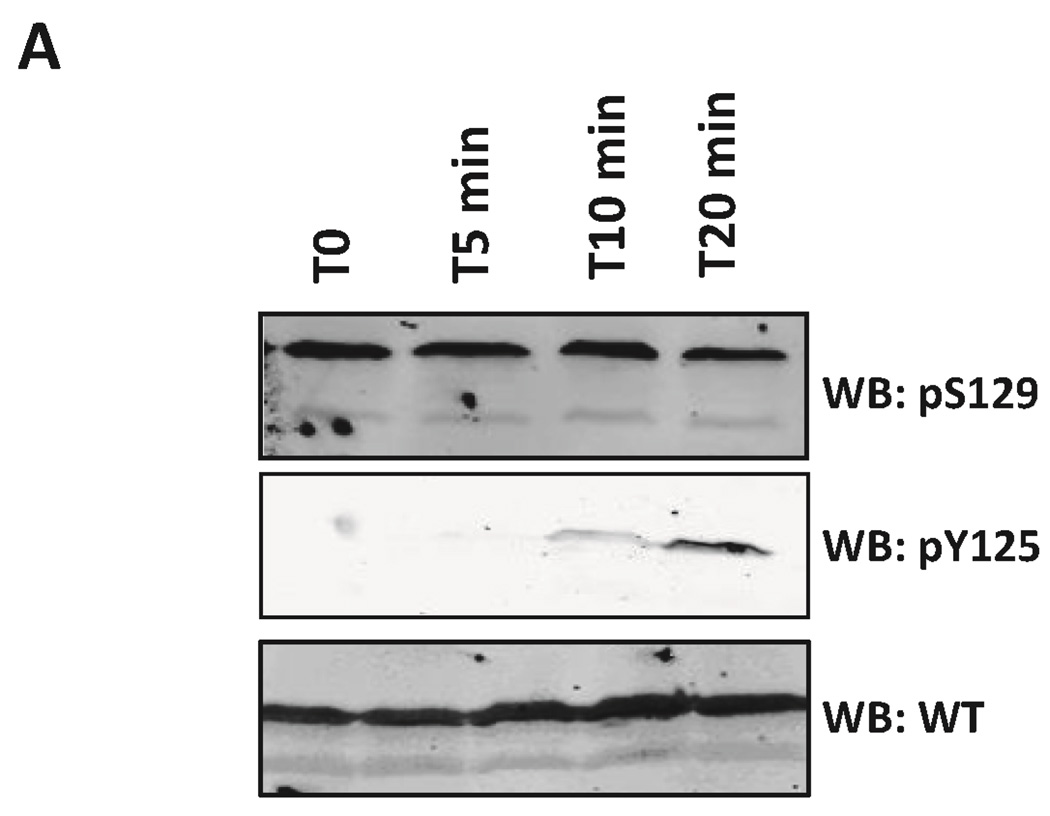
HeLa and HEK 293T cells were grown in 6-well-plates and transfected with the plasmid pAAV α-syn WT and pAAV α-syn Y125F. The cells were treated with pervanadate (100 µM sodium orthovanadate and 4 mM H2O2) 24 h post-transfection for 5, 10 and 20 min, as reported previously 39. The medium containing the inhibitor was removed, and the cells were subsequently harvested by pipetting in PBS or by scraping. The cells were pelleted by centrifugation at 100 g for 3 min, lysed by the addition of 100 µL of Laemmli buffer and boiled at 95°C for 30 min. 30 µL of sample was loaded on SDS-PAGE and analyzed by Western-Blot with antibodies for WT α-syn and pY125 α-syn
Subcellular fractionation
HEK 293 cells were grown in 6-well-plates, transfected with the plasmid pAAV WT α-syn and treated with pervanadate (100 µM sodium orthovanadate and 4 mM H2O2) 24h posttransfection. Subcellular fractionation was carried out using the kit from Calbiochem according to the manufacturer’s instructions and as previously described38. Pervanadate (100 µM sodium orthovanadate and 4 mM H2O2) was also added during the extraction steps.
Primary neuronal cultures
Rat primary neurons were cultured from newborn Sprague Dawley (SD) rat hippocampus, following the regulations of Peking University Animal Care and Use Committee. In brief, rat hippocampal tissues were dissociated with 0.25% trypsin (Invitrogen), which was then inactivated by 10% decomplemented fetal bovine serum (FBS, HyClone). The mixture was triturated through a pipette to produce a homogenous mixture. After filtering the mixture through 0,7 µm sterilized filters, the flow-through was centrifuged. The pellet was then washed once with PBS followed by one wash with Dulbecco's Modified Eagle Medium (DMEM) in Earle’s balanced salt solution containing 0.225% sodium bicarbonate, 1 mM sodium pyruvate, 2 mM L-glutamine, 0.1% dextrose and antibiotic Pen-Strep with 5% FBS (all from Invitrogen). Cells were then plated on poly-L-lysine (Sigma) coated plates or on glass coverslips at a density of 1 × 106 cells/ml. Neurons were incubated at 37°C in DMEM without phenol red with 5% FBS and 5% circulating CO2. Cytarabine was added to culture media 24 hours after plating to a final concentration of 10 µM to inhibit cell growth. The culture medium was changed every 48 hours. Cells were used for experiments at day 7 of culture.
Subcellular localization of pY125 α-syn in HeLa cells
HeLa cells were seeded on coverslips (100 000 cells in a 24-well-plate) pre-coated for 30 min at RT with Poly-L-lysine. The cells were transfected with the plasmid pAAV α-syn WT (0.8 µg ) and subsequently treated with pervanadate prepared as described above for 30 min 24 h post-transfection. The cells were subsequently fixed in 4% paraformaldehyde in PBS for 30 min, washed three times in PBS, blocked for 1 h with 3% BSA, 0.1% Triton X-100 in PBS and incubated overnight at 4°C with a cocktail of primary antibodies (mouse α-syn pY125, BD Pharmingen, 558246, 1:1000; rabbit α-syn, ab51252, Abcam, 1: 1000) in 1% BSA, 0.1% Triton X-100 in PBS. The cells were then washed three times with 1% BSA, 0.1% Triton X-100 in PBS and incubated for 1 h at RT with a cocktail of secondary antibodies (goat antimouse coupled to Alexa Fluor 568, Molecular Probes Inc., 1:1000; goat anti-rabbit coupled to Alexa Fluor 488, Molecular Probes Inc., 1:1000). After three washes in PBS, coverslips were counter-stained with 4,6-diamidino-2-phenylindole (DAPI, 1:5000), washed 3 times with PBS and mounted on glass slides with Vectashield mounting medium (Vector). Cells were imaged using a Zeiss LSM700 confocal microscope.
Microinjection and immunocytochemistry
Thin-walled Borosilicate glass capillaries (outer diameter=1.0 mm, inner diameter=0.5 mm) with a microfilament (MTW100F-4, World Precision Instrument) were pulled with a Flaming/Brown Micropipette Puller (P-97, Sutter) to obtain injection needles with a tip diameter of approximately 0.5 µm. Microinjections into the cytosol of each were performed cell using an Eppendorf Microinjector FemtoJet and Eppendorf Micromanipulator (Eppendorf). Neurons were injected with 25 fl/shot at an injection pressure of 100 hPa, a compensation pressure of 50 hPa, and an injection time of 0.1 s. pY125 α-syn was diluted for a final concentration of 1 µM in phosphate-buffered saline (PBS, 0.14 M NaCl, 0.003 M KCl, 0.01 M Na2HPO4, 0.002 M KH2PO4, pH7.2). The solutions were injected with 100 µg/ml dextran Texas Red (DTR, MW: 3000, Molecular Probes) as a fluorescent marker or with Fast Green as a visible marker to enable the identification of injected cells. The injected cells were treated either with control DMEM medium or with pervanadate (100 µM orthovanadate and 4 mM H2O2) for 30 min. For immunocytochemistry, cells were fixed, permeabilized and blocked with 10% donkey serum at room temperature followed by incubation with anti-α-synuclein antibody (mouse α-syn pY125, BD Pharmingen, 558246, 1:1000; rabbit α-syn, ab51252, Abcam, 1: 1000) at 4°C for 24 h. Cy2 or Cy3-conjugated donkey anti-rabbit antibody was used as the secondary antibody. Nuclei were subsequently stained by Hoechst 33258 (1 µg/ml, Sigma) for 15 minutes in the dark. The coverslips were mounted on glass slides with Immunon™ mounting medium (Shandon) and imaged by fluorescence microscopy (Olympus BH2-RFCA, Olympus) with a digital camera (Olympus DP70 Digital Microscope Camera, Olympus).
RESULTS
The semisynthesis of α-syn site-specifically phosphorylated within the C-terminus (residues 107–140) involved an NCL reaction between the recombinant thioester α-syn(1–106)SR and the synthetic peptide α-syn (A107C-140) phosphorylated at Tyr-125 or the corresponding nonphosphorylated peptide. Because the primary sequence of α-syn does not contain any cysteine residues, we mutated alanine residue 107 to a cysteine to allow the NCL reaction to occur with the juxtaposed residue at position 106 being an unhindered residue (Gly) (Scheme 1)
Generation of semisynthetic pY125 α-syn
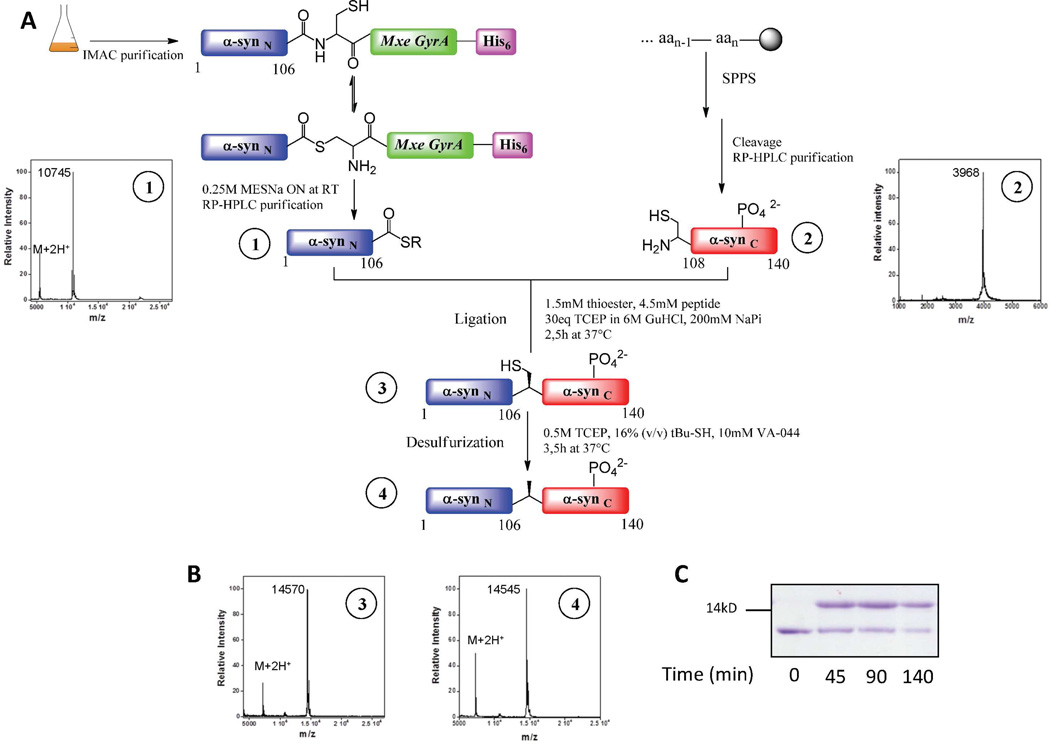
Recombinant α-syn(1–106)SR was expressed in E.coli utilizing the intein approach, which exploits the mechanism of intein self-splicing to generate recombinant proteins with a C-terminal thioester (Scheme 3A) 24. In the case of natural intein splicing, a series of transthioesterification and succinimide formation events that involve an arginine residue at the C-terminus of the intein mediates the splicing of the intein and the linkage of the N-extein and the C-extein by an amide bond 40. Briefly, truncated α-syn(1–106) was expressed in fusion with the GyrA mini-intein (Mxe GyrA) which contained a C-terminal hexahistidine purification tag to facilitate purification from bacterial lysate. Initially, we used the chitin binding domain from the commercially available IMPACT system (New England Biolabs) as a purification tag for the attachment of the fusion protein to the chitin beads followed by splicing on the column with 0.25 M MESNa and subsequent purification of the thioester by reverse-phase HPLC. We then added a C-terminal histidine tag on the intein to facilitate the purification of the protein from larger volumes of E.coli lysates. The purification procedure was therefore modified and cell lysates were passed through a Ni-NTA column to immobilize the fusion protein and remove undesired proteins (Scheme 3A). The α-syn(1–106)SR was released by thiolysis with an excess of MESNa. The protein thioester was then purified by RP-HPLC and characterized by MALDI-TOF mass spectrometry and RP-HPLC (Scheme 3A and Figure S3).
Scheme 3. Semisynthesis and characterization of phosphorylated α-syn derivatives.
A: Expressed Protein Ligation (EPL) approach to generate WT α-syn and pY125 α-syn. The α-syn(1–106)SR fragment was generated using intein-mediated thioester formation. MALDI-TOF analyses of 1) α-syn(1–106)SR (expected mass: 10744 Da [M+H]); The synthetic peptides α-syn (A107C-140) and α-syn(A107C-140)_pY125 were generated by Fmoc-based SPPS. MALDI-TOF analyses of 2) α-syn(A107C-140)_pY125 (expected mass: 3970 Da [M+H]);. Full-length semisynthetic α-synA107C and α-synA107C_pY125 were generated by a NCL step between α-syn(1–106)SR and synthetic peptide α-syn(A107C-140)_pY125 or α-syn(A107C-140). Radical desulfurization of the purified ligation product generated pY125 α-syn and α-syn WT. The final purification step involved a cation-exchange chromatographic step. B: MALDI-TOF analyses of 3) α-synA107C_pY125 (expected mass: 14573 Da [M+H]); and 4) pY125 α-syn (expected mass: 14541 Da ([M+H])). C: Coomassie blue stained SDS-PAGE analysis of the NCL reaction to generate pY125 α-syn. Lane 1, 2, 3, and 4 represent α-syn(1–106)SR; ligation reaction after 45 min, 90 min, and 140 min, respectively.
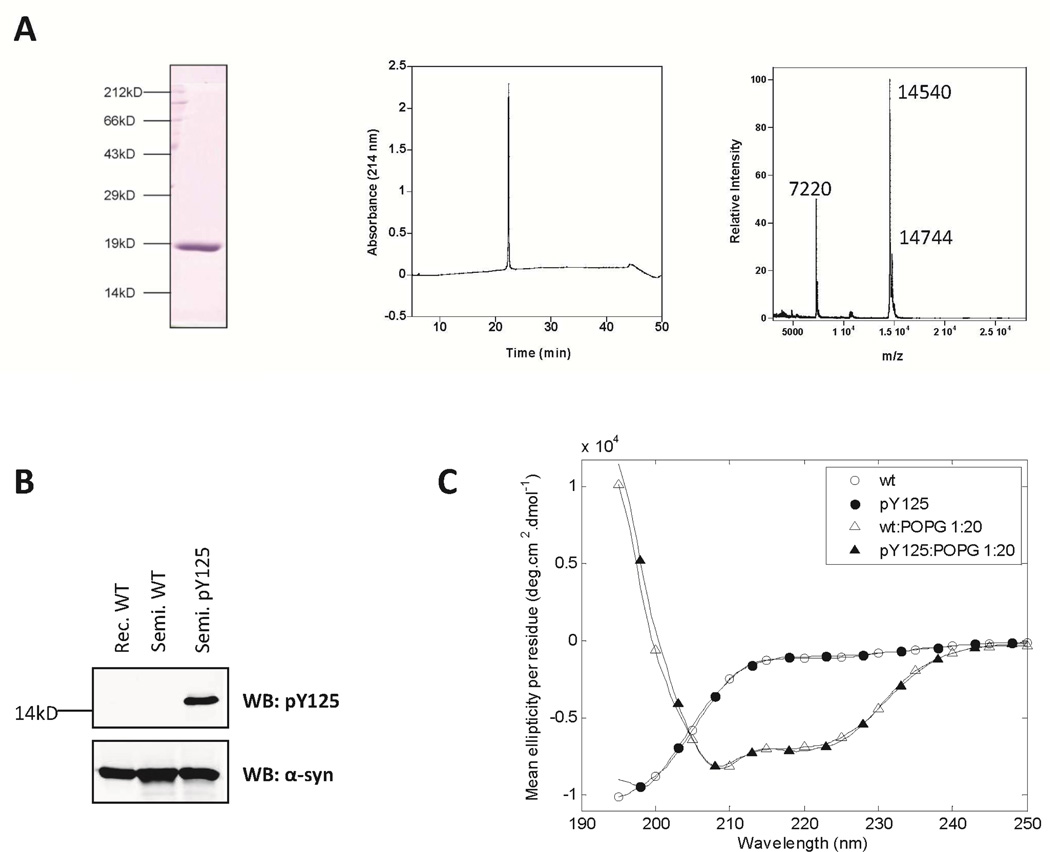
The peptide fragments α-syn (A107C-140) pY125 and α-syn (A107C-140) WT, each with an N-terminal cysteine, were generated by stepwise SPPS (Figures S1 and S2). The expressed fragment α-syn(1–106)SR was then subjected to two separate NCL reactions under denaturing conditions with 2 molar excess synthetic C-terminal peptides α-syn (A107C-140) pY125 and α-syn (A107C-140) (Scheme 3A), generating two variants of full length α-syn A107C selectively phosphorylated at Tyr-125 (pY125 α-syn) and the wild-type (WT) protein, respectively. Hydrolyzed α-syn(1–106)-OH was generated as a minor side product, even when the NCL reaction was carried out at lower pH values. Due to the co-elution of α-syn(1–106)OH and the full-length protein by reverse phase HPLC under all conditions tested in our laboratory, we could not quantitatively assess the yield at each step of the semisynthesis. The final yield of the purified protein is 19%. (Scheme 3C and Figure S5).Radical-initiated desulfurization of Cys-107 in the ligated proteins re-established the native Ala-107 residue in the phosphorylated variant of α-syn and its wild-type counterpart with minimal loss of starting material. A loss of 32 mass units from each protein was observed by MALDI-TOF-MS in each protein, consistent with the loss of a sulfur atom (Scheme 3B and Figures S6 and S8). Separation of the ligated fragment from the hydrolyzed thioester and excess C-terminal peptide was initially achieved by using a combination of different purification methods, including RP-HPLC, anion-exchange, liquid isoelectric focusing and size exclusion chromatography. Only a combination of two or more of these techniques resulted in pure semi-synthetic proteins. However, this approach resulted in very low final yield. Later, the ligation products were purified using cation-exchange chromatography, which permitted single-step chromatographic separation of the full-length fragment, the hydrolyzed thioester, the C-terminal peptide and other contaminants (Figure S9–11). Using optimized NCL, desulfurization and purification conditions, we obtained milligram quantities of the purified ligated proteins. In the case of α-syn phosphorylated at Y125, MALDI-TOF analysis of the purified products demonstrated a major peak with m/z = 14545 corresponding to the desired [M+H+] ions (Figure 1A and Figures S6 and S8). Western-blot analysis using anti-α-syn and anti-pY125 α-syn antibodies further confirmed the identity and purity of the semisynthetic WT α-syn and pY125 α-syn proteins (Figure 1B).
Figure 1. Biochemical and biophysical characterization of semisynthetic WT and pY125 α-syn.
A: SDS-PAGE, RP-HPLC and MALDI-TOF analyses of the pure semisynthetic pY125 α-syn. MALDI analysis pY125 α-syn (expected mass: 14541 Da ([M+H]), a sinapinic matrix adduct is also observed as well as the double-charged peak. B: Western blot analyses of WT and pY125 α-syn confirmed that the phosphorylated semisynthetic protein could be detected by the anti-phosphorylated α-syn primary antibody, as well as by a non-phospho-specific antibody C: Circular dichroism spectra of pY125 α-syn in the absence (circles) and presence (triangles) of POPG vesicles. Open and closed symbols indicate WT recombinant α-syn and pY125 α-syn, respectively.
Semisynthetic pY125α-syn adopts an α-helical structure upon biding to membrane
The secondary structure of the semisynthetic pY125 α-syn was investigated using circular dichroism in solution (50 mM Tris buffer and 150 mM NaCl, pH 7.5,) and in the presence of POPG vesicles (with a lipid:protein mass ratio of 20:1). As shown in Figure 1C, pY125 α-syn exhibits a virtually identical CD spectrum to that of the WT recombinant α-syn in solution (random coil) and adopts an α-helical structure upon binding to the lipid vesicles.
Phosphorylation at Y125 does not alter the structure of monomeric α-syn
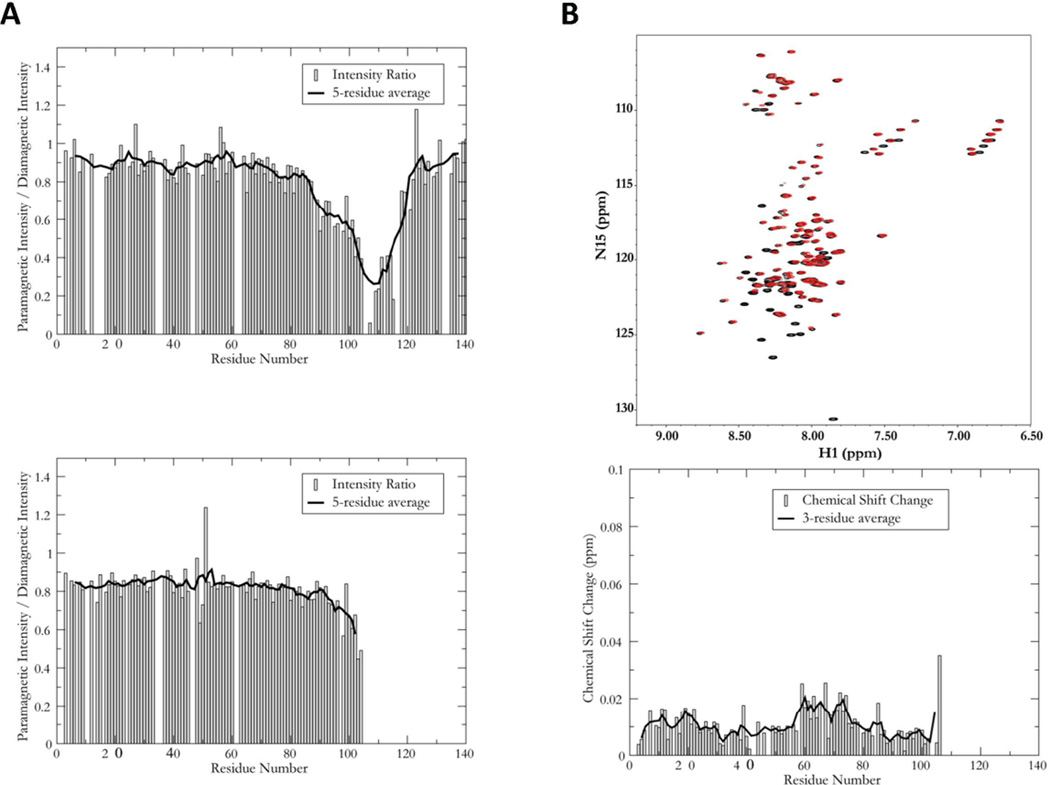
To determine the effect of Y125 phosphorylation on the structural properties of monomeric α-syn, 15N labeled A107C α-syn and A107C/pY125 α-syn were generated for solution NMR studies (Figures S12–13). The NMR spectra of the free proteins were essentially identical and similar to the equivalent spectrum of the WT protein (not shown), indicating that both A107C and A107C/pY125 α-syn remain in the highly disordered ensemble characteristic of this protein41. To assess the potential influence of phosphorylation on the transient long-range structure of the protein, we collected paramagnetic relaxation data for phosphorylated and unphosphorylated α-syn modified at Cys 107 by the spin label MTSL. The paramagnetic spin label increases the relaxation rate and thus broadens the resonance of any residue within ~20 Å. Intramolecular contacts can thus be discerned by measuring the intensity ratio of resonances in a paramagnetically-labeled sample to the equivalent resonances in a diamagnetic control. Plots of the intensity ratio as a function of residue number for A107C and A107C/pY125 α-syn, shown in Figure 2A reveal two interesting results. First, even in the unphosphorylated sample, there is little indication of the previously reported long-range contacts between the C- and N-terminal regions of the protein35. This observation is surprising, but suggests that position 107 may not be highly sensitive to these contacts. However, there is a significant difference in the data between the phosphorylated and unphosphorylated proteins in the regions proximal to the labeling site. Residues close to the labeling site are broadened because they are covalently constrained to close proximity with the spin label, resulting in a well (or minimum) around position 107. It has been argued that the breadth of this well reflects the local compactness of the protein. Despite the fact that only residues prior to the position 107 in A107C/pY125 α-syn are 15N labeled give rise to data, it is evident that the well around position 107 is narrower for the phosphorylated protein. This result indicates that in the phosphorylated protein, the C-terminal region samples a more expanded ensemble of conformations.
Figure 2. NMR characterization of free and micelle-bound WT and pY125 α-syn.
A: Paramagnetic relaxation enhancement profiles for A107C (upper panel) and A107C/pY125 (lower panel) α-syn, derived from the intensity ratios of equivalent resonances between 1H, 15N HSQC spectra collected with and without an attached spin-label. A ratio of 1 indicates no PRE effect, while smaller ratios indicated a PRE effect, resulting from the proximity of the corresponding residue to the site of spin-label attachment. B: Overlaid 1H, 15N HSQC spectra of SDS micelle bound WT (black) and A107C/pY125 (red) α-syn (upper panel) and plots of the normalized chemical shift changes between the two spectra as a function of residue number (lower panel). The solid line in panel B is a three-residue average. Note that each red peak corresponds closely to a black peak, indicating that the structures of the two variants are highly similar in the micelle-bound state. Black peaks without corresponding red peaks arise from residues 108 and beyond, where the semisynthetic A107C/pY125 protein is not 15N-labeled and therefore does not give rise to signals in this spectrum.
Because the PRE data could not be used to monitor changes in long-range contacts due to phosphorylation at position 125, we also measured and compared the hydrodynamic radii of the phosphorylated and unphosphorylated proteins using pulsed field gradient NMR methods. The hydrodynamic radius of A107C/pY125 α-syn was found to be identical, within experimental error, to that of both A107C and WT α-syn, indicating that no significant overall compaction or expansion of the monomer occurs upon Y125 phosphorylation. These observations contrast with observations made in the case of S129 phosphorylation, where a significant degree of expansion occurs35.
We also examined the potential effects of phosphorylation at Y125 on the micelle-bound form of the protein; this form is thought to closely resemble one of several membrane-bound forms of the protein. We observed a high degree of overlap between the 1H,15N-HSQC spectra of wild-type A107C/pY125 α-syn in the presence of SDS micelles (Figure 2B), indicating that no gross structural changes in the protein occur upon phosphorylation at Y125. To more closely assess the effects of Y125 phosphorylation, we plotted the chemical shift changes between the two spectra as a function of residue number (Figure 2B). The results show that while the changes are generally small (on the order of 0.01 ppm), they are larger than those we observed for pS129 35. Furthermore, these changes appear to be slightly greater in regions corresponding to the center of each of the two previously characterized helices of the micelle-bound form of the protein 42.
Phosphorylation at Y125 does not affect the fibrillization of α-syn
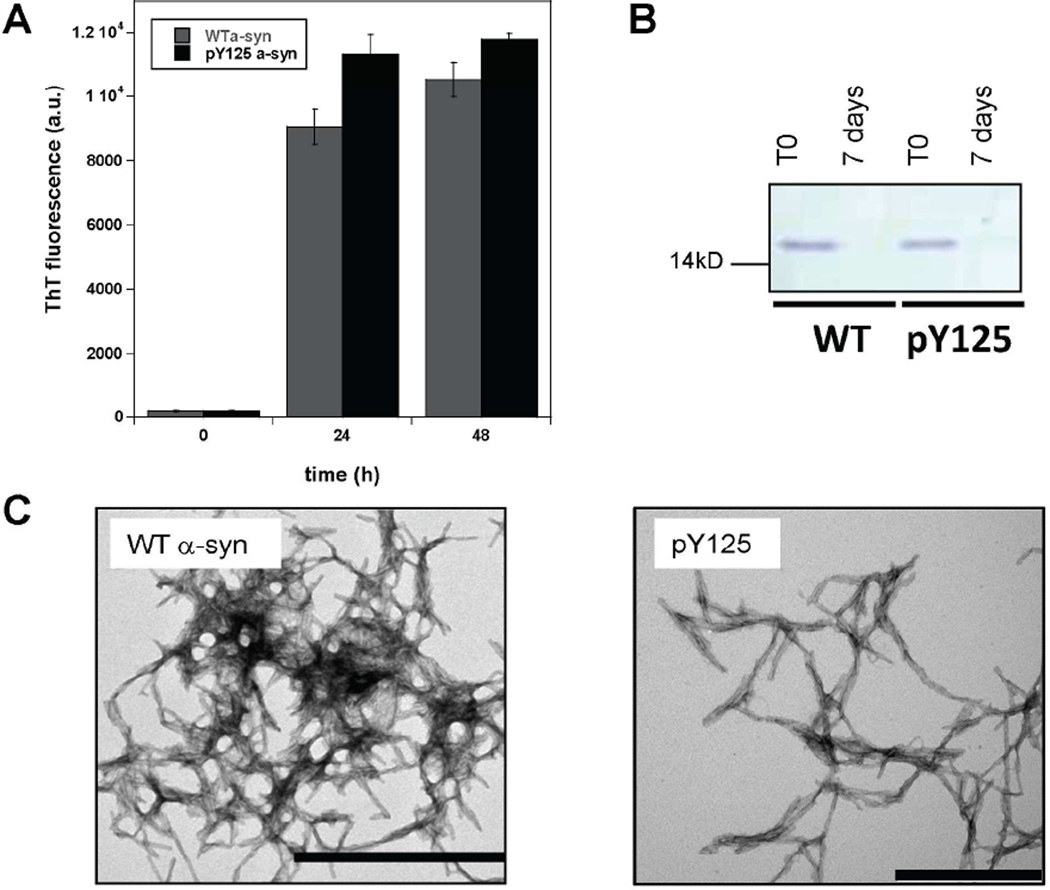
Previously, work from our group and others 35, 43 has shown that phosphorylation at S129 inhibits the fibrillization of α-syn in vitro. To determine whether phosphorylation at Y125 influences the aggregation properties of α-syn, we optimized conditions for α-syn fibril formation at low protein concentrations (15 µM) and compared the extent and kinetics of fibrillization of semisynthetic pY125 α-syn to those of the semisynthetic WT α-syn and recombinant WT proteins. Each protein was incubated with agitation at 37°C and the extent of fibrillization was assessed by ThT fluorescence and TEM after 72 h of agitation. As evidenced by the increase in ThT signal shown in Figure 3, WT and pY125 α-syn forms mature fibrils after 24 h of incubation. Next, we assessed the amount of monomeric protein using a filtration assay and used TEM to characterize the structural properties of the aggregates formed by each protein. On the structural level, both WT and pY125 α-syn formed mature fibrillar structures (Figures 3C and S14). After filtration of the samples through a 100 kDa membrane filter, the amount of monomeric α-syn was assessed by SDS-PAGE analysis. No significant amounts of monomers were detected for WT and pY125 α-syn after 7 days (Figure 3B). Together, these data suggest that phosphorylation at Y125 does not alter the aggregation of α-syn.
Figure 3. Fibrillization of pY125 α-syn.
A: ThT fluorescence of recombinant WT and pY125 α-syn monitored over 48 h. B: SDS-PAGE analysis of the remaining WT and pY125 α-syn monomers prior to and after aggregation for 72 h and filtration through a 100 kD membrane. C: TEM images of WT and pY125 α-syn after 7 days of aggregation. The scale bar represents 500 nm. The images are representative of 5 independent experiments. The scale bar represents 500 nm.
pY125 α-syn is dephosphorylated within minutes in vitro and in vivo
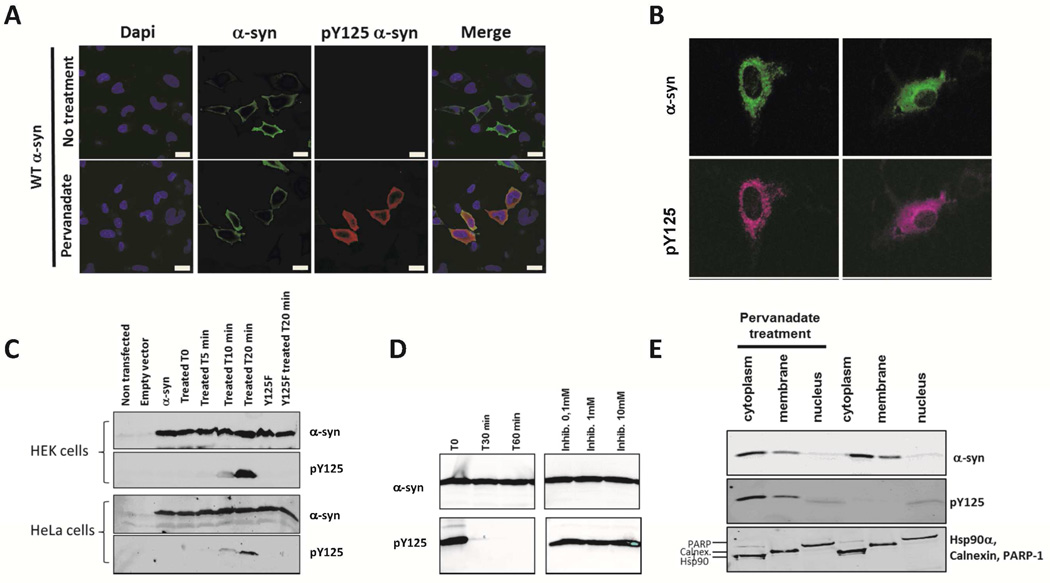
To investigate the subcellular localization in vivo, pY125 α-syn was microinjected into hippocampal primary neurons at a concentration of 1 µM. Because microinjected pY125 α-syn might be rapidly dephosphorylated, we fixed the neurons either immediately or 30 min after microinjection and probed for pY125 α-syn and total α-syn using the appropriate antibodies. Interestingly, in the injected cells, the signal for pY125 α-syn dramatically decreased within 30 min compared to the signal for WT α-syn (data not shown). To confirm these observations and to test for rapid dephosphorylation in vitro, a HeLa cell lysate was spiked with pY125 α-syn and incubated for 0, 30 and 60 min. A loss of signal from the phosphorylated protein but not from the total α-syn protein was observed, indicating that pY125 α-syn is completely dephosphorylated by endogenous phosphatases within 30 min of incubation (Figure 4D). These results indicate tight regulation of the phosphorylation state of α-syn at Y125 by endogenous tyrosine phosphatases. In these experiments, dephosphorylation was effectively inhibited using sodium orthovanadate, a general tyrosine-phosphatase inhibitor.
Figure 4. Dephosphorylation and detection of pY125 α-syn in cells.
A: Confocal images of HeLa cells transfected with WT α-syn plasmid and treated or not with pervanadate for 30min. The scale bar represents 20 µm. Subsequently, the cells were fixed and stained with the indicated antibodies (respectively rabbit anti-α-syn from Abcam ab51252 and mouse anti-pY125 α-syn from BD Pharmingen). B: Detection of pY125 α-syn in primary neurons. Confocal images of rat hippocampal neurons microinjected with 1 µM pY125 α-syn (25 fl/injection) together with the visible marker dye Faster Green (100 µg/ml) to identify the injected cells, the cells were treated with medium containing pervanadate for 30 min. Subsequently, the cells were fixed and stained with the indicated antibodies (respectively rabbit anti-α-syn from Abcam ab51252 and mouse anti-pY125 α-syn from BD Pharmingen). C: Immunoblots of HEK and HeLa cell lysates untreated or treated with pervanadate for 0, 5, 10 and 20min. The membranes were probed with total α-syn and pY125-specific antibodies (respectively rabbit anti-α-syn ab51252 from Abcam and mouse anti-pY125 α-syn from BD Pharmingen) D: Immunoblots of pY125 α-syn dephosphorylated from HeLa cell lysate. Membranes were probed with total α-syn and pY125-specific antibodies. The dephosphorylation was inhibited using different concentrations of sodium orthovanadate. E: Immuboblots of the subcellular fractionation of HEK cells over-expressing WT α-syn and treated with pervanadate for 20 min. Markers for cytoplasmic, membrane and nuclear fraction were used to assess for the purity of each fractions (Hsp90α, calnexin and PARP-1).
pY125 α-syn is localized into the cytoplasm in HeLa cells and primary neurons
Previous attempts to characterize the possible effect of Y125 phosphorylation on the subcellular localization of α-syn were unsuccessful, in part due to lack of pY125-specific antibodies that are suitable for immunocytochemistry 18. In an attempt to overcome this limitation, we generated an antibody specific for pY125 α-syn and used it to immunolocalize of pY125 α-syn in HeLa cells and rat hippocampal neurons. The pattern of pY125 α-syn immunoreactivity produced by this antibody was compared with the pattern obtained using a commercially available antibody from BD Pharmingen. For these experiments, we first identified conditions that prevent rapid dephosphorylation of pY125 α-syn. Previous reports indicated that pY125 α-syn is detectable in HEK293 cells that overexpress WT α-syn upon treatment with pervanadate 39, 44. We validated these findings and found by Western-blot analysis of lysates from HEK293 and HeLa cells overexpressing human WT α-syn, that treatment with pervanadate results in the upregulation of pY125 α-syn in these cells (Figure 4C). In cells over-expressing WT α-syn, but not the mutant Y125F α-syn, we observed a significant increase in the level of pY125 α-syn when the duration of treatment with pervanadate was increased. Non-specific bands of higher molecular weight were also detected with both the commercial (BD Pharmingen) and homemade anti pY125 α-syn antibodies after pervanadate treatment (Figure S15). This precludes the use of these antibodies to accurately assess in more details the subcellular localization of pY125 α-syn in specific cellular organelles. We were able to partially circumvent this problem by studying the subcellular localization of pY125 α-syn in HeLa cells overexpressing α-syn (Figure 4A). Quantification of the average fluorescence intensity of 65 cells (data not shown) treated with pervanadate showed that the increasing levels of cytosolic pY125 α-syn correlated well with increasing α-syn levels, and indicated a predominantly cytosolic localization of pY125 α-syn. We did not observe pY125 α-syn immunoreactivity in the nucleus nor was any cytosolic increase in pY125 α-syn immunoreactivity observed in HeLa cells over-expressing the mutant Y125F α-syn (Figure S16). When semisynthetic pY125 α-syn was microinjected into rat hippocampal neurons at a concentration of 1 µM, it showed a cytoplasmic distribution similar to the distribution of WT α-syn immunoreactivity in HeLa cells (Figure 4B). To more accurately assess the subcellular localization of pY125, we preformed subcellular fractionation studies on HEK 293T cells overexpressing α-synuclein in the presence and absence of pervanadate. Using this approach, we can demonstrate conclusively that pY125 localizes mainly in the cytosolic fraction and to a small extent in the membrane fraction, consistent with the results from immunocytochemistry (Figure 4E).
Phosphorylation at Y125 does not block the phosphorylation at S129 and S87 in vitro and vice-versa
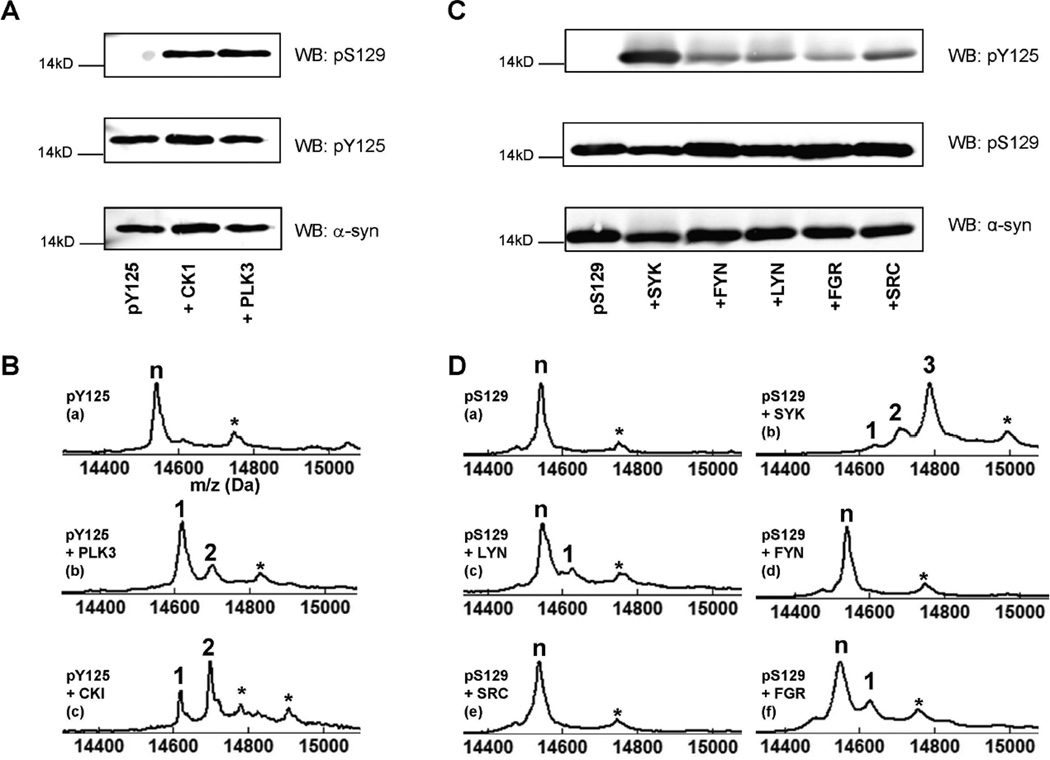
The proximity of the three C-terminal tyrosine residues in α-syn suggests the possibility that the phosphorylation of one of these residues may trigger or modulate the extent of phosphorylation at the other residues. In a recent study, Y125 phosphorylation was suggested to protect against α-syn toxicity 18. Using a transgenic Drosophila model overexpressing the S129D phosphomimetic mutant of human α-syn in retinal and dopaminergic neurons, Chen et al. demonstrated that co-expression of Shark, a Drosophila homolog of the human tyrosine kinase Syk, which was previously reported to efficiently (although non-specifically) phosphorylate α-syn at Y125 45, significantly alleviates neurodegeneration due to S129D α-syn expression. To investigate potential cross-talk between Y125 and S129 phosphorylation sites, pY125 α-syn was incubated with PLK3, CK1 kinases, which are known to phosphorylate α-syn specifically and efficiently at S129 (PLK3) or at S87 and S129 (CK1). Phosphorylation at Y125, S129 and S87 was detected using the appropriate phosphospecific antibodies and confirmed by MALDI-TOF MS (Figure 5A–B). These results indicated that phosphorylation of α-syn at Y125 does not prevent recognition of the protein by PLK3 or CK1, affect the subsequent phosphorylation of pY125 α-syn at S129 and S87, or prevent recognition of the double-phosphorylated species (pY125/pS87 and pY125/pS129) by the appropriate antibodies. To determine whether phosphorylation at Y125 influences S129 phosphorylation by endogenous kinases, we assessed the levels of pS129 in HeLa and HEK293 cells before and after treatment with pervanadate. Despite the significant increase in pY125 α-syn levels observed upon treatment with pervanadate, we did not observe any significant changes in pS129 levels (Figure 6).
Figure 5. Phosphorylation of pY125 and pS129 α-syn.
A: Immunoblots of pY125 α-syn phosphorylated by CK1 and PLK3. Membranes were probed using pS129, pY125, and α-syn antibodies. B: MALDI-TOF-MS analysis of the phosphorylation reaction after 12h. C: Immunoblots of pS129 α-syn phosphorylated by Syk, Lyn, Fyn, c-Src, and c-Fgr. Membranes were probed for pY125, pS129, and α-syn immunoreactivity. D: MALDI-TOF analysis of pS129 α-syn phosphorylated by (a) Syk, (b) Lyn, (c) Fyn, (d) c-Src, (e) and c-Fgr (f) after 14h of reaction. In all MALDI-TOF-MS spectra, the symbol ‘n’ indicates the peak corresponding to the starting material, the numbers above the other peaks correspond to the number of phosphorylation events, which were each detected by a +80 Da mass shift. The symbol ‘*’ indicates a sinapinic acid matrix adduct.
Figure 6. Phosphorylation of pY125 α-syn and pS129 α-syn.
Immunoblots of HeLa cells over-expressing WT α-syn and treated with pervanadate for 0, 5, 10 and 20min. Membranes were probed with pS129 and pY125 α-syn and WT α-syn antibodies.
A recent study by Chen and colleagues suggested that phosphorylation at S129 does not affect the tyrosine phosphorylation of α-syn18. However, these results were based on expression of a phosphomimetic mutant (S129D) that does not reproduce all aspects of phosphorylation at this site 35. Therefore, we sought to determine whether prior phosphorylation at S129 effects further phosphorylation at Y125 by the non-receptor tyrosine kinases Syk and members of the Src family (Lyn, Fyn, c-Fgr and c-Src), which have been previously shown to phosphorylate tyrosine residues in the C-terminus of α-syn. The protein was first quantitatively phosphorylated using PLK3, purified by reversed-phase HPLC and incubated at 3 µM for 14 h at 30°C in the presence of 100 nM of each individual tyrosine kinase. Western blot and MALDI-TOF-MS analyses (Figure 5C–D), showed the same efficiency and site-specificity of phosphorylation by these kinases as reported in previous in vitro studies. Of these kinases, Syk is by far the most efficient at phosphorylating α-syn with a reported Kcat/Km of around 30, it also phosphorylated multiple tyrosines within the C-terminal region of α-syn 39, 45. We consistently observed phosphorylation at Y125 and at two other sites by MALDI-TOF MS (Figure 5D (b)). The other kinases were more specific but lacked efficiency. For example, the Kcat/Km for Lyn is around 0.5. Most importantly, we observed that S129 phosphorylation did not block the activity of any of the tyrosine kinases. As a control, WT α-syn was incubated under the same conditions, a similar phosphorylation pattern was obtained (data not shown). Western-blot-based detection of Y125 phosphorylation appeared to be more sensitive than MALDI-TOF-MS because pY125 α-syn could be detected in all of the enzymatic reactions (Figure 5C). Together, these data suggest that S129 phosphorylation of α-syn does not interfere with further modifications of the protein by tyrosine kinases. Moreover, because the consensus recognition sequences of tyrosine kinases such as Syk include acidic residues 46, pS129 might act to stabilize the electrostatic interactions between the kinases and α-syn.
We performed a similar experiment aimed at determining whether phosphorylation at S87, another disease-related phosphorylation site within α-syn that was recently identified by our group 34, influence the recognition and processing of α-syn by the soluble tyrosine kinases mentioned above. We observed that phosphorylation of α-syn at S87 does not significantly influence the protein’s activity as a substrate for the non-receptor tyrosine kinases Syk, Fyn, Ly, c-Fgr, and c-Src or the site-specificity of its phosphorylation by these enzymes (Figure S17).
DISCUSSION
Recent studies indicate that α-syn phosphorylation at serine residues S87 and S129 can dramatically influence α-syn structure 34, 35, turn-over 47, membrane binding 34, 35, aggregation 34, 35 and protein-protein interactions 48. Notably, phosphorylation at S87 and S129 have emerged as a defining hallmark of PD and other synucleinopathies 49, 50. The use of phosphomimetic mutations (S→E/D) combined with the identification of kinases that specifically phosphorylate α-syn in vitro and in vivo has facilitated detailed studies aimed at investigating the structural and functional consequences of phosphorylation of the protein at these residues in vitro and in vivo 18, 20, 38, 51–55. In addition to serine residues, α-syn has also been shown to undergo phosphorylation at its C-terminal tyrosine residues Y125, Y133, Y136 in vitro, with a preference for Y125 as the major phosphorylation site 39, 44, 45, 56. However, only phosphorylation at Y125 has been detected in vivo 18.
Several kinases have been reported to phosphorylate tyrosine residues in the C-terminus of α-syn. However, all known kinases are either inefficient or lack specificity to phosphorylate α-syn at specific tyrosine residues in its C-terminus. The tyrosine kinase Syk phosphorylates α-syn at multiple tyrosine residues (Y125, Y133 and Y136) 45. In contrast, Fyn, Lyn, c-Fgr and c-Src phosphorylate α-syn specifically at Y125, but these kinases are less efficient45. The lack of specificity and efficiency of tyrosine phosphorylation of α-syn has hampered efforts to obtain homogenous preparations of phosphorylated α-syn and to elucidate the structural and functional consequences of phosphorylation at Y125. Unlike phospho-serine and phosphor-threonine, which in some cases can be mimicked by serine → glutamate or aspartate substitutions, there are no natural amino acids that can mimic phospho-tyrosine. Inspired by the pioneering work of Muir and Cole on the use of expressed protein ligation to study the effect of site-specific phosphorylation and other post-translational modifications24, we developed, for the first time, an efficient semisynthetic approach based on a one-pot native chemical ligation and desulfurization followed by a single chromatographic purification step for the production of pY125 α-syn. This strategy enabled us to obtain preparations of α-syn protein site-specifically modified at single or multiple residues within the C-terminal region comprising residues 107–140, the region that contains the majority of the reported PTMs of the protein. Using this approach, we were able to obtain milligram quantities of semisynthetic WT and pY125 α-syn and to carry out detailed studies aimed at elucidating the effect of this modification on the structure, membrane binding, aggregation and subcellular localization of α-syn.
The results presented here demonstrate that phosphorylation at Y125 does not significantly alter the structure of monomeric α-syn in solution (Figure 1B and 2). Although the hydrodynamic radius of pY125 α-syn is virtually identical to that of the wild-type protein, the paramagnetic relaxation data presented here indicate that the C-terminal region of pY125 α-syn samples a more expanded ensemble of conformations than in the wild-type protein. Interestingly, these observations contrast with the case of S129 phosphorylation, where a significant degree of expansion occurs 35. Phosphorylation at Y125 did not interfere with the ability of α-syn to form an α-helical structure upon binding to synthetic phosphatidylglycerol (POPG) or SDS micelles as assessed by CD spectroscopy. These results are consistent with previous observations regarding pS129 and with the fact that both S129 and Y125 lie outside the region involved in α-syn binding to membranes. However, the 1H,15N-HSQC spectra of the protein reveal small structural changes that are larger than those observed for the case of pS12935. Furthermore, the structural changes in the protein that result from Y125 phosphorylation appear to be slightly greater in regions corresponding to the centers of each of the two previously characterized helices of the micelle-bound form of the protein 57. Thus, while phosphorylation at Y125 does not result in large-scale structural changes in the micelle-bound form of the protein, it may cause slight rearrangements of the two micelle-bound helices. Such small changes may be consistent with results that indicate that modifications occuring in the C-terminal tail of the protein can influence the N-terminal lipid-binding domain 58.
Previous studies by Negro et al. showed that co-incubation of α-syn with Syk kinase inhibits α-syn aggregation in vitro 45. Co-expression of Syk and α-syn was also reported to inhibit α-syn oligomerization and inclusion formation in cell culture 45 and in Drosophila expressing Shark, the fly homolog of Syk 18. However, it is difficult to discern the relative contribution of phosphorylation at Y125 in these studies, because the Syk kinase phosphorylates all three C-terminal tyrosine residues (Y125, Y133 and Y136) to different extents. In subsequent studies by Negro and colleagues, it was reported that the addition of other tyrosine kinases (Lyn and c-Fgr) did not influence the aggregation of α-syn in vitro 45. However, the significantly reduced efficiency of these kinases for the phosphorylation of α-syn makes it difficult to draw any conclusions regarding the effect of phosphorylation at Y125 on α-syn aggregation and fibrillization. In this work, the availability of semisynthetic pY125 α-syn allowed us to address this question directly for the first time. WT α-syn and semisynthetic pY125 α-syn exhibited similar aggregation properties as discerned by TEM and ThT fluorescence, demonstrating that pY125 α-syn behaves similarly to the recombinant protein produced in E. Coli, unlike phosphorylation at S129, which inhibits α-syn fibrillization. Interestingly, pY125 α-syn exhibited similar aggregation properties as WT α-syn, consistent with the NMR results demonstrating that phosphorylation at Y125 induces only subtle structural changes on monomeric α-syn.
The availability of semisynthetic pY125 α-syn provided unique opportunities to assess the subcellular localization of pY125 α-syn and to explore the potential cross-talk between phosphorylation at S129 and Y125. Recent studies by Chen et al. suggested that Y125 phosphorylation may protect against α-syn toxicity by decreasing α-syn phosphorylation at S12918. To investigate possible cross-talk between phosphorylation at S129 and Y125, we combined semisynthetic approaches and enzymatic phosphorylation assays. Employing optimized procedures for the in vitro phosphorylation of α-syn, we used semisynthetic pY125 α-syn to examine the effect of phosphorylation at Y125 on S129 phosphorylation by subjecting pY125 α-syn to phosphorylation by kinases that phosphorylate α-syn efficiently at S129 (PLK3) and/or S87 and S129 (CK1) 35, 38. To determine whether phosphorylation at S129 influence Y125 phosphorylation, we prepared pS129 α-syn by in vitro phosphorylation with PLK338 and examined the extent of Y125 phosphorylation (pS129/pY125) by the tyrosine kinases Fyn, Lyn, Src, c-Fgr and Syk39, 44, 45, 56. Our results demonstrate that phosphorylation at Y125 does not attenuate α-syn phosphorylation at S129 or S87 in vitro and vice-versa. These findings are consistent with the results of studies from Feany and colleagues in Drosophila suggesting that S129 phosphorylation of α-syn does not influence Y125 phosphorylation. The levels of pY125 α-syn were similar in flies overexpressing WT, S129A or S129D, suggesting that S129 phosphorylation does not influence phosphorylation at Y125. This study confirms these findings in mammalian cells (HeLa and HEK 293T), where no changes in pS129 levels were observed after treatment with phosphatase inhibitors that significantly enhance pY125 α-syn levels.
The lack of significant interest in phosphorylation of α-syn at Y125, compared to phosphorylation at S87 and S129 can be attributed to difficulties in detecting significant levels of pY125 α-syn in human brain tissues and lack of evidence for a direct correlation between pY125 α-syn levels and disease progression. Proteomic and mass spectrometry analyses of α-syn from Lewy bodies failed to detect phosphorylation at Y12549. The rapid dephosphorylation of α-syn in post-mortem tissues may explain the limited success of previous studies in detecting pY125 α-syn in human brains and by biochemical analysis of brain-derived materials using mass spectrometry. Due to the dynamic regulation of tyrosine phosphorylation in vivo, detection of α-syn phosphorylation at Y125 appears to be dependent on the use of tyrosine phosphatase inhibitors or on the time elapsed prior to postmortem analyses. Recently, Y125 phosphorylation was shown to occur both in human and Drosophila brains and pY125 α-syn levels was reported to decrease with aging and in PD brains, suggesting a possible protective role for this modification 18. Interestingly, in the presence of phosphatase inhibitors, 30% of α-syn in Drosophila appears to be phosphorylated at Y125 supporting the role of Y125 phosphorylation in modulating α-syn properties. In this study, we showed that upon addition of pY125 α-syn to cell lysates or microinjection into primary neurons, pY125 α-syn is almost completely dephosphorylated within 5 min. This effect is blocked in the presence of pervanadate. These results are consistent with a very rapid dephosphorylation of α-syn both in vitro and in vivo. Phosphorylation at Y125 appears to be tightly regulated such that the levels of pY125 α-syn are maintained at very low levels but increases significantly in cell culture, primary neurons, and brain tissues, upon treatment with the phosphatase inhibitor pervanadate 18, 39, 44. Together, these studies demonstrate that α-syn undergoes phosphorylation at Y125 in vivo and suggest that experiments aimed at evaluating and correlating specific PTMs with disease progression must take into account the stability and dynamics of these modifications, and that the protocols used for the analysis and preparations of samples must be modified accordingly.
Nakamura et al reported that the levels of pY125 α-syn increases by approximately 15 fold within the first few minutes in response to hyperosmotic stress, but then declines rapidly after 10 min. These observations indicate the existence of efficient dephosphorylation mechanisms that regulate pY125 α-syn levels 59. However, the identities of the phosphatases responsible for regulating pY125 α-syn phosphorylation remain unknown. The availability of semisynthetic pY125 α-syn as a substrate should facilitate the development of assays to screen focused libraries of tyrosine phosphatase inhibitors and identify natural phosphatases that are involved in regulating Y125 phosphorylation in vivo. To prevent the rapid dephosphorylation of pY125 α-syn, we will replace phosphotyrosine by nonhydrolyzable phosphonate analogues 60, 61 that, once incorporated into α-syn, will render it resistant to phosphatase activity62. Cole and colleagues have successfully introduced non-hydrolyzable tyrosine analogues into proteins using expressed protein ligation strategies and demonstrated their utility to investigate the role tyrosine phosphorylation in regulating the activity of different tyrosine phosphatases and their interaction with other proteins 29, 62–66.
Whether phosphorylation at Y125 plays an important role in regulating α-syn aggregation and toxicity in vivo remains unknown. This question can only be answered by experimental conditions that permit site-specific regulation of phosphorylation at Y125 in vivo. To determine the functional consequences of Y125 phosphorylation, Feany and colleagues overexpressed WT α-syn, and mutant (α-syn YF) in which all C-terminal tyrosines were mutated to phenylalanine (Y125F/Y133F/Y136F) and compared the effects of the mutations on α-syn aggregation, toxicity, and locomotor deficit in flies. The mutant (α-syn YF) exhibited enhanced toxicity and caused a significant reduction in climbing ability compared to flies overexpressing the wild-type protein. Additionally, overexpression of the Shark kinase, the Drosophila homolog of Syk, increased the levels of pY125, decreased oligomer formation and rescued the neurotoxicity of WT α-syn and the more toxic S129 phosphomimetic S129D. It is important to note that in both approaches, the three tyrosines were either mutated or phosphorylated to different extent, making it difficult to decipher the effects of selective modification at Y125. Identification and expression of the kinases that allow selective modification of pY125 and/or coexpression of such kinases with Y133F/Y136F is crucial for gaining insight into the role of Y125 phosphorylation in regulating α-syn aggregation and toxicity in vivo. The availability of semisynthetic pY125 α-syn and our improved understanding of the dynamics of Y125 phosphorylation should aid in the development of assays to identify the natural kinases and phosphatases involved in the regulation of Y125 phosphorylation and will facilitate a quantitative evaluation of the extent of pY125 α-syn during disease progression.
CONCLUSIONS
We recently reported a semisynthetic approach that enabled site-specific modification of the N-terminus of α-syn and used this approach to prepare and characterize an α-syn form that is mono-ubiquitinated at Lysine 6 30. In this study, we describe a general and efficient semisynthetic strategy that enables the site-specific introduction of single or multiple PTMs of α-syn and the preparation of homogeneously modified forms of α-syn at its C-terminus in milligram quantities. These two approaches make it possible to prepare all the known disease-associated PTMs of α-syn, except pS87, and to study potential cross-talk between these modifications. These advances have allowed us to investigate for the first time the effect of selective phosphorylation at Y125 α-syn on the structure, aggregation, membrane binding and subcellular localization of the protein. In addition, these advances provide unique opportunities to investigate PTM-dependent protein-protein interactions involving α-syn and to study the cross-talk between different PTMs. Furthermore, the availability of these proteins is crucial for advancing current research programs with the following aims: 1) characterization of PTMs in humans and in animal models of PD; 2) development of biomarkers and target identification; and 3) development of sensitive assays for the detection and quantitative assessment of specific PTMs in vivo and in biological fluids. Therefore, the development of these semisynthetic strategies represents an important advance toward unraveling the roles of PTMs in determining α-syn structure, aggregation and functions in health and disease. We hope that this work will encourage other research groups to pursue similar approaches to elucidate the role of PTMs in regulating the function of other proteins, particularly proteins linked to neurodegenerative diseases.
Supplementary Material
Acknowledgments
The authors wish to acknowledge Nathalie Jordan for preparation of hippocampal primary neurons, Drs. Abid Oueslati and Marc Moniatte and Prof. Ashraf Brik for stimulating discussions, and Drs. Clay Bracken and Shibani Bhattacharya for assistance with NMR experiments. This work was supported by NIH grants AG019391 and AG025440 (D.E.), and an ERC starting grant (H.L, M.H, 243182).
Footnotes
Supporting Information Available: Complete Ref. 5, 20, 34, 44 & 49. This material is available free of charge via the Internet at http://pubs.acs.org/."
REFERENCES
- 1.Trojanowski JQ, Lee VM. Ann N Y Acad Sci. 2003;991:107. doi: 10.1111/j.1749-6632.2003.tb07468.x. [DOI] [PubMed] [Google Scholar]
- 2.Dawson TM. Cell. 2000;101:115. doi: 10.1016/S0092-8674(00)80629-7. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Nature. 1997;388:839. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 4.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen JT, Schols L, Riess O. Nat Genet. 1998;18:106. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 5.Polymeropoulos MH, et al. Science. 1997;276:2045. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, Llorens V, Gomez Tortosa E, del Ser T, Munoz DG, de Yebenes JG. Ann Neurol. 2004;55:164. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 7.Desplats P, Lee HJ, Bae EJ, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee SJ. Proc Natl Acad Sci U S A. 2009;106:13010. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. J Neurosci. 2010;30:6838. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang A, Lee HJ, Suk JE, Jung JW, Kim KP, Lee SJ. J Neurochem. 2010;113:1263. doi: 10.1111/j.1471-4159.2010.06695.x. [DOI] [PubMed] [Google Scholar]
- 10.Del Mar C, Greenbaum EA, Mayne L, Englander SW, Woods VL., Jr Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15477. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goers J, M-B A, McCormack AL, Millett IS, Doniach S, Di Monte DA, Uversky VN, Fink AL. Biochem J. 2003;42:8465. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 12.Cherny D, Hoyer W, Subramaniam V, Jovin TM. J Mol Biol. 2004;344:929. doi: 10.1016/j.jmb.2004.09.096. [DOI] [PubMed] [Google Scholar]
- 13.Jensen PH, Hager H, Nielsen MS, Hojrup P, Gliemann J, Jakes R. J Biol Chem. 1999;274:25481. doi: 10.1074/jbc.274.36.25481. [DOI] [PubMed] [Google Scholar]
- 14.Giasson BI, F M, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM. Science. 2003;300:636. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 15.Paik SR, Shin HJ, Lee JH, Chang CS, Kim J. Biochem J. 1999;340(Pt 3):821. [PMC free article] [PubMed] [Google Scholar]
- 16.Brown DR. FEBS J. 2007;274:3766. doi: 10.1111/j.1742-4658.2007.05917.x. [DOI] [PubMed] [Google Scholar]
- 17.Goers J, Uversky VN, Fink AL. Protein Sci. 2003;12:702. doi: 10.1110/ps.0230903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Periquet M, Wang X, Negro A, McLean PJ, Hyman BT, Feany MB. J Clin Invest. 2009;119:3257. doi: 10.1172/JCI39088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbefo MK, Paleologou KE, Boucharaba A, Oueslati A, Schell H, Fournier M, Olschewski D, Yin G, Zweckstetter M, Masliah E, Kahle PJ, Hirling H, Lashuel HA. J Biol Chem. 285:2807. doi: 10.1074/jbc.M109.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inglis KJ, et al. J Biol Chem. 2009;284:2598. doi: 10.1074/jbc.C800206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackenberger CP, Schwarzer D. Angew Chem Int Ed Engl. 2008;47:10030. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 22.Komarov AG, Linn KM, Devereaux JJ, Valiyaveetil FI. ACS Chem Biol. 2009;4:1029. doi: 10.1021/cb900210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berrade L, Camarero JA. Cell Mol Life Sci. 2009 doi: 10.1007/s00018-009-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci U S A. 1998;95:6705. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang KP, Jensen MS, McGinty RK, Muir TW. Chembiochem. 2009;10:2182. doi: 10.1002/cbic.200900238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broncel M, Krause E, Schwarzer D, Hackenberger CP. Chemistry. 2012 doi: 10.1002/chem.201103032. [DOI] [PubMed] [Google Scholar]
- 27.Muir TW. Annu Rev Biochem. 2003;72:249. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 28.Spasser L, Brik A. Angew Chem Int Ed Engl. 2012 doi: 10.1002/anie.201200020. [DOI] [PubMed] [Google Scholar]
- 29.Szewczuk LM, Tarrant MK, Cole PA. Methods in enzymology. 2009;462:1. doi: 10.1016/S0076-6879(09)62001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hejjaoui M, Haj-Yahya M, Kumar KS, Brik A, Lashuel HA. Angew Chem Int Ed Engl. 2011;50:405. doi: 10.1002/anie.201005546. [DOI] [PubMed] [Google Scholar]
- 31.Butterfield S, Hejjaoui M, Fauvet B, Awad L, Lashuel HA. the Journal of molecular biology. 2012 doi: 10.1016/j.jmb.2012.01.051. [DOI] [PubMed] [Google Scholar]
- 32.Alewood P, Alewood D, Miranda L, Love S, Meutermans W, Wilson D. Methods in enzymology. 1997;289:14. doi: 10.1016/s0076-6879(97)89041-6. [DOI] [PubMed] [Google Scholar]
- 33.Wan Q, Danishefsky SJ. Angew Chem Int Ed Engl. 2007;46:9248. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 34.Paleologou KE, et al. J Neurosci. 2010;30:3184. doi: 10.1523/JNEUROSCI.5922-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, Lansbury PT, Jr, Fernandez CO, Eliezer D, Zweckstetter M, Lashuel HA. J Biol Chem. 2008;283:16895. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA. Methods Mol Biol. 2004;278:313. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 38.Mbefo MK, Paleologou KE, Boucharaba A, Oueslati A, Schell H, Fournier M, Olschewski D, Yin G, Zweckstetter M, Masliah E, Kahle PJ, Hirling H, Lashuel HA. J Biol Chem. 2010;285:2807. doi: 10.1074/jbc.M109.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis CE, Schwartzberg PL, Grider TL, Fink DW, Nussbaum RL. J Biol Chem. 2001;276:3879. doi: 10.1074/jbc.M010316200. [DOI] [PubMed] [Google Scholar]
- 40.Evans TC, Jr, Xu MQ. Biopolymers. 1999;51:333. doi: 10.1002/(SICI)1097-0282(1999)51:5<333::AID-BIP3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 41.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. J Mol Biol. 2001;307:1061. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 42.Bisaglia M, Tessari I, Pinato L, Bellanda M, Giraudo S, Fasano M, Bergantino E, Bubacco L, Mammi S. Biochemistry. 2005;44:329. doi: 10.1021/bi048448q. [DOI] [PubMed] [Google Scholar]
- 43.Levitan K, Chereau D, Cohen SI, Knowles TP, Dobson CM, Fink AL, Anderson JP, Goldstein JM, Millhauser GL. J Mol Biol. 2011;411:329. doi: 10.1016/j.jmb.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn BH, et al. J Biol Chem. 2002;277:12334. doi: 10.1074/jbc.M110414200. [DOI] [PubMed] [Google Scholar]
- 45.Negro A, Brunati AM, Donella-Deana A, Massimino ML, Pinna LA. FASEB J. 2002;16:210. doi: 10.1096/fj.01-0517fje. [DOI] [PubMed] [Google Scholar]
- 46.Brunati AM, Donella-Deana A, Ruzzene M, Marin O, Pinna LA. FEBS Lett. 1995;367:149. doi: 10.1016/0014-5793(95)00555-n. [DOI] [PubMed] [Google Scholar]
- 47.Chau KY, Ching HL, Schapira AH, Cooper JM. J Neurochem. 2009;110:1005. doi: 10.1111/j.1471-4159.2009.06191.x. [DOI] [PubMed] [Google Scholar]
- 48.McFarland MA, Ellis CE, Markey SP, Nussbaum RL. Mol Cell Proteomics. 2008;7:2123. doi: 10.1074/mcp.M800116-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JP, et al. J Biol Chem. 2006;281:29739. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 50.Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, Shen J, Takio K, Iwatsubo T. Nat Cell Biol. 2002;4:160. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 51.Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, Meijer L, Kahle PJ, Haass C. J Biol Chem. 2000;275:390. doi: 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- 52.Pronin AN, Morris AJ, Surguchov A, Benovic JL. J Biol Chem. 2000;275:26515. doi: 10.1074/jbc.M003542200. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Feany MB. Nat Neurosci. 2005;8:657. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 54.Gorbatyuk OS, Li S, Sullivan LF, Chen W, Kondrikova G, Manfredsson FP, Mandel RJ, Muzyczka N. Proc Natl Acad Sci U S A. 2008;105:763. doi: 10.1073/pnas.0711053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azeredo da Silveira S, Schneider BL, Cifuentes-Diaz C, Sage D, Abbas-Terki T, Iwatsubo T, Unser M, Aebischer P. Hum Mol Genet. 2009;18:872. doi: 10.1093/hmg/ddn417. [DOI] [PubMed] [Google Scholar]
- 56.Nakamura T, Yamashita H, Takahashi T, Nakamura S. Biochem Biophys Res Commun. 2001;280:1085. doi: 10.1006/bbrc.2000.4253. [DOI] [PubMed] [Google Scholar]
- 57.Bussell R, Jr, Eliezer D. J Mol Biol. 2003;329:763. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 58.Sevcsik E, Trexler AJ, Dunn JM, Rhoades E. J Am Chem Soc. 2011;133:7152. doi: 10.1021/ja2009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura T, Yamashita H, Nagano Y, Takahashi T, Avraham S, Avraham H, Matsumoto M, Nakamura S. FEBS Lett. 2002;521:190. doi: 10.1016/s0014-5793(02)02861-2. [DOI] [PubMed] [Google Scholar]
- 60.Berkowitz DB, Eggen M, Shen Q, Shoemaker RK. J Org Chem. 1996;61:4666. doi: 10.1021/jo9604752. [DOI] [PubMed] [Google Scholar]
- 61.Desmarais S, Friesen RW, Zamboni R, Ramachandran C. The Biochemical journal. 1999;337(Pt 2):219. [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng W, Schwarzer D, Lebeau A, Weller JL, Klein DC, Cole PA. The Journal of biological chemistry. 2005;280:10462. doi: 10.1074/jbc.M412283200. [DOI] [PubMed] [Google Scholar]
- 63.Schwarzer D, Zhang Z, Zheng W, Cole PA. Journal of the American Chemical Society. 2006;128:4192. doi: 10.1021/ja0585174. [DOI] [PubMed] [Google Scholar]
- 64.Shen K, Hines AC, Schwarzer D, Pickin KA, Cole PA. Biochim Biophys Acta. 2005;1754:65. doi: 10.1016/j.bbapap.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Zheng W, Zhang Z, Ganguly S, Weller JL, Klein DC, Cole PA. Nat Struct Biol. 2003;10:1054. doi: 10.1038/nsb1005. [DOI] [PubMed] [Google Scholar]
- 66.Lu W, Shen K, Cole PA. Biochemistry. 2003;42:5461. doi: 10.1021/bi0340144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.