Abstract
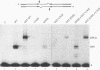
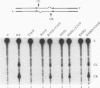
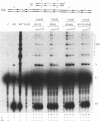
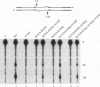
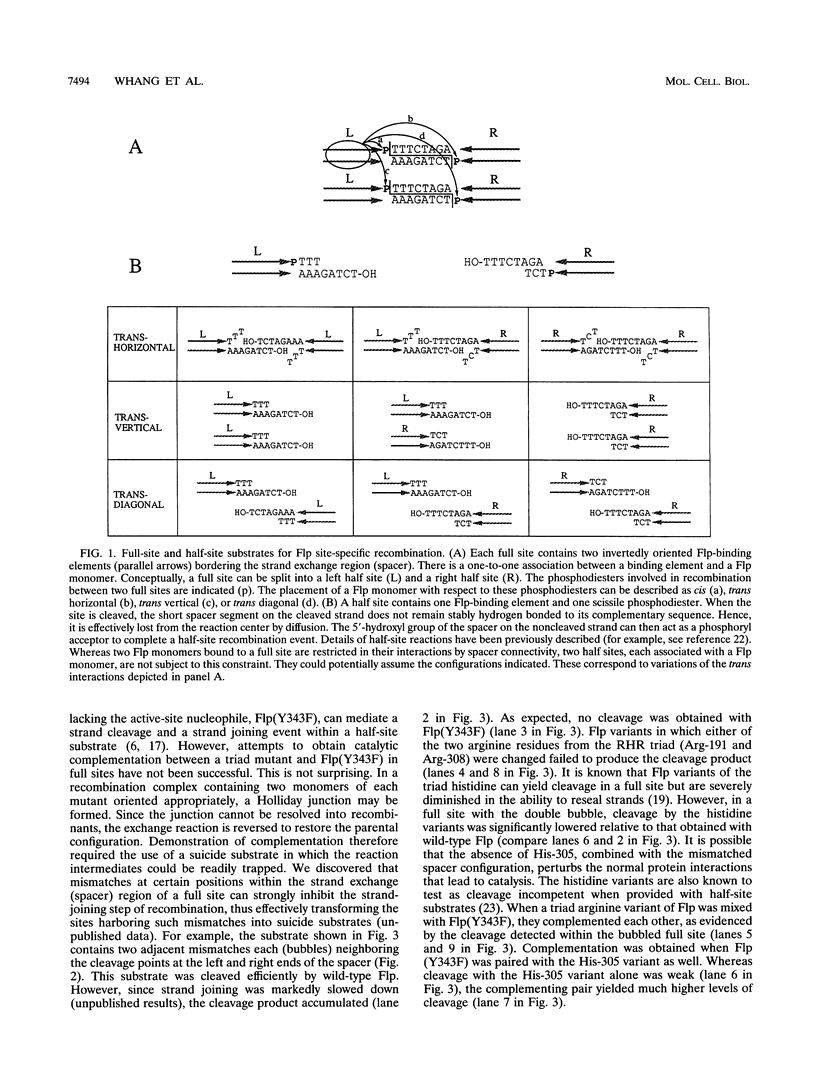
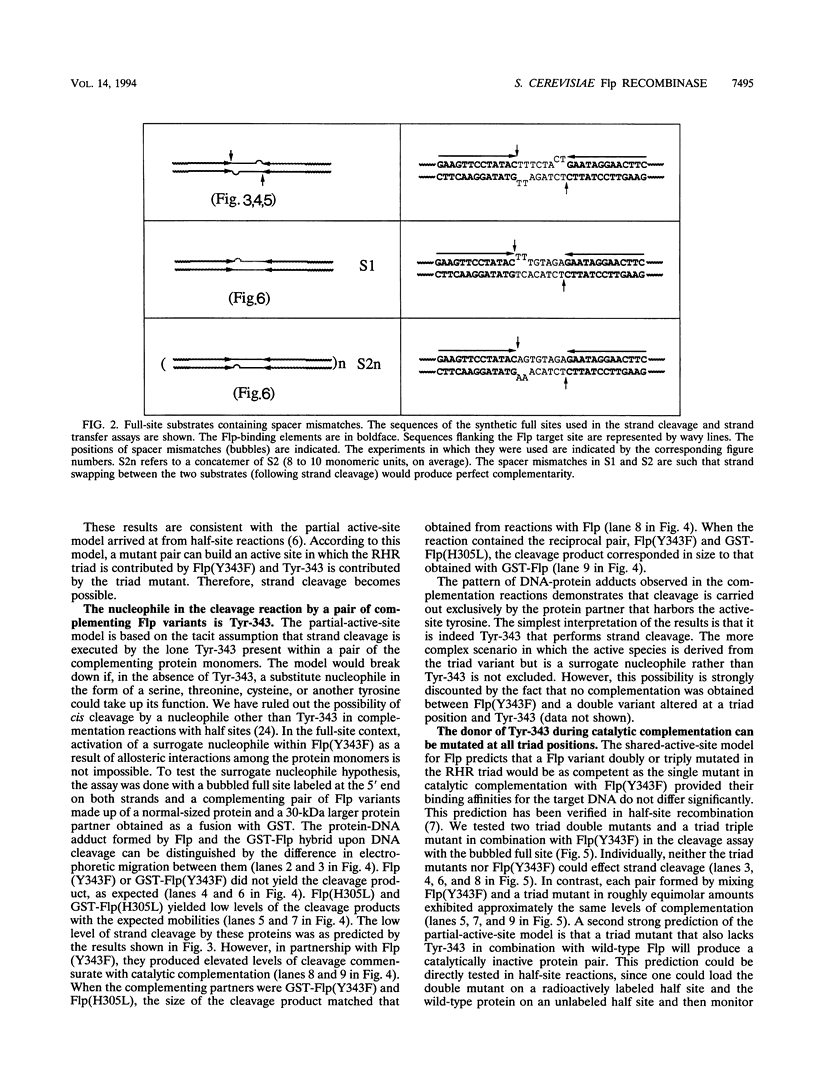
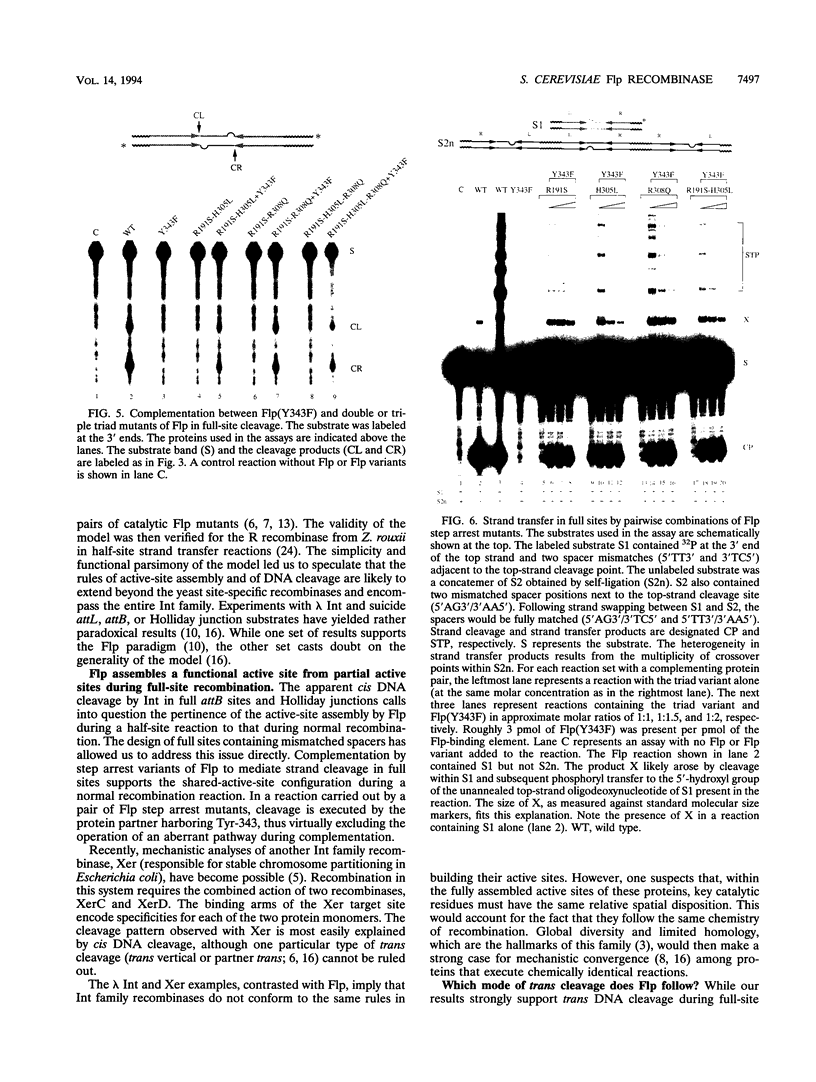
A combination of half-site substrates and step arrest mutants of Flp, a site-specific recombinase of the integrase family, had earlier revealed the following features of the half-site recombination reaction. (i) The Flp active site is assembled by sharing of catalytic residues from at least two monomers of the protein. (ii) A Flp monomer does not cleave the half site to which it is bound (DNA cleavage in cis); rather, it cleaves a half site bound by a second Flp monomer (DNA cleavage in trans). For the lambda integrase (Int protein), the prototype member of the Int family, catalytic complementation between two active-site mutants has been observed in reactions with a suicide attL substrate. By analogy with Flp, this observation is strongly suggestive of a shared active site and of trans DNA cleavage. However, reactions with linear suicide attB substrates and synthetic Holliday junctions are more compatible with cis than with trans DNA cleavage. These Int results either argue against a common mode of active-site assembly within the Int family or challenge the validity of Flp half sites as mimics of the normal full-site substrates. We devised a strategy to assay catalytic complementation between Flp monomers in full sites. We found that the full-site reaction follows the shared active-site paradigm and the trans mode of DNA cleavage. These results suggest that within the Int family, a unitary chemical mechanism of recombination is achieved by more than one mode of physical interaction among the recombinase monomers.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K. E., Hoess R. H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992 Jan;5(1):87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- Amin A., Roca H., Luetke K., Sadowski P. D. Synapsis, strand scission, and strand exchange induced by the FLP recombinase: analysis with half-FRT sites. Mol Cell Biol. 1991 Sep;11(9):4497–4508. doi: 10.1128/mcb.11.9.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Chen J. W., Lee J., Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992 May 15;69(4):647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- Chen J. W., Yang S. H., Jayaram M. Tests for the fractional active-site model in Flp site-specific recombination. Assembly of a functional recombination complex in half-site and full-site strand transfer. J Biol Chem. 1993 Jul 5;268(19):14417–14425. [PubMed] [Google Scholar]
- Doolittle R. F. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994 Jan;19(1):15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Evans B. R., Chen J. W., Parsons R. L., Bauer T. K., Teplow D. B., Jayaram M. Identification of the active site tyrosine of Flp recombinase. Possible relevance of its location to the mechanism of recombination. J Biol Chem. 1990 Oct 25;265(30):18504–18510. [PubMed] [Google Scholar]
- Han Y. W., Gumport R. I., Gardner J. F. Complementation of bacteriophage lambda integrase mutants: evidence for an intersubunit active site. EMBO J. 1993 Dec;12(12):4577–4584. doi: 10.1002/j.1460-2075.1993.tb06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee J., Jayaram M. Mechanism of site-specific recombination. Logic of assembling recombinase catalytic site from fractional active sites. J Biol Chem. 1993 Aug 15;268(23):17564–17570. [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Pan G., Luetke K., Sadowski P. D. Mechanism of cleavage and ligation by FLP recombinase: classification of mutations in FLP protein by in vitro complementation analysis. Mol Cell Biol. 1993 Jun;13(6):3167–3175. doi: 10.1128/mcb.13.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Parsons R. L., Prasad P. V., Harshey R. M., Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol Cell Biol. 1988 Aug;8(8):3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre M. C., Evans B. R., Araki H., Oshima Y., Jayaram M. Half-site recombinations mediated by yeast site-specific recombinases Flp and R. J Mol Biol. 1992 Jun 5;225(3):621–642. doi: 10.1016/0022-2836(92)90390-6. [DOI] [PubMed] [Google Scholar]
- Serre M. C., Jayaram M. Half-site strand transfer by step-arrest mutants of yeast site-specific recombinase Flp. J Mol Biol. 1992 Jun 5;225(3):643–649. doi: 10.1016/0022-2836(92)90391-v. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Jayaram M. Generality of the shared active site among yeast family site-specific recombinases. The R site-specific recombinase follows the Flp paradigm [corrected]. J Biol Chem. 1994 Apr 29;269(17):12789–12796. [PubMed] [Google Scholar]