Abstract
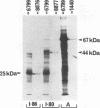
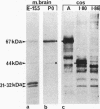
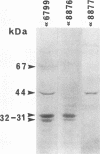
It has been shown that the enzyme glutamic acid decarboxylase (GAD; EC 4.1.1.15), which catalyzes the conversion of L-glutamate to gamma-aminobutyric acid in the central nervous system of vertebrates, can be first detected in rodents at late embryonic stages. In contrast, we have found that the gene coding for the 67-kDa form of GAD is already transcriptionally active at embryonic day E10.5 in the mouse. In addition to the 3.5-kb adult-type mRNA, we have detected two 2-kb embryonic messages that contain alternatively spliced exons of 80 (I-80) and 86 (I-86) bp, respectively. The overlapping stop-start codon TGATG, found in the embryonic exons, converts the monocistronic adult-type transcript into a bicistronic one, coding for a 25-kDa leader peptide and a 44-kDa enzymatically active truncated GAD. A second stop codon at the 3' end of the 86-bp exon abolishes the expression of truncated GAD. The products of the two embryonic mRNAs were identified in a rabbit reticulocyte in vitro translation system, COS cells, and mouse embryos. The two GAD embryonic forms represent distinct functional domains and display characteristic developmental patterns, consistent with a possible role in the formation of the gamma-aminobutyric acid-ergic inhibitory synapses.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony D. D., Merrick W. C. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J Biol Chem. 1991 Jun 5;266(16):10218–10226. [PubMed] [Google Scholar]
- Bell L. R., Horabin J. I., Schedl P., Cline T. W. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991 Apr 19;65(2):229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell. 1988 Dec 23;55(6):1037–1046. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- Boggs R. T., Gregor P., Idriss S., Belote J. M., McKeown M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987 Aug 28;50(5):739–747. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- Bond R. W., Wyborski R. J., Gottlieb D. I. Developmentally regulated expression of an exon containing a stop codon in the gene for glutamic acid decarboxylase. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8771–8775. doi: 10.1073/pnas.87.22.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989 Mar 24;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Choi G. H., Shapira R., Nuss D. L. Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1167–1171. doi: 10.1073/pnas.88.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- Das A., Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 1984 Jun 11;12(11):4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V., Laoide B. M., Masquilier D., de Groot R. P., Foulkes N. S., Sassone-Corsi P. Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4226–4230. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner A. J., Semler B. L., Jackson R. J., Hanecak R., Duprey E., Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984 May;50(2):507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlander M. G., Tobin A. J. The structural and functional heterogeneity of glutamic acid decarboxylase: a review. Neurochem Res. 1991 Mar;16(3):215–226. doi: 10.1007/BF00966084. [DOI] [PubMed] [Google Scholar]
- Frederickson R. M., Mushynski W. E., Sonenberg N. Phosphorylation of translation initiation factor eIF-4E is induced in a ras-dependent manner during nerve growth factor-mediated PC12 cell differentiation. Mol Cell Biol. 1992 Mar;12(3):1239–1247. doi: 10.1128/mcb.12.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Hohn T. Role of an upstream open reading frame in the translation of polycistronic mRNAs in plant cells. Nucleic Acids Res. 1992 Aug 11;20(15):3851–3857. doi: 10.1093/nar/20.15.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Hohn T. Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J. 1991 Dec;10(12):3887–3896. doi: 10.1002/j.1460-2075.1991.tb04958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Scholthof H. B., Wu F. C., Shepherd R. J. Requirement of gene VII in cis for the expression of downstream genes on the major transcript of figwort mosaic virus. Virology. 1991 Dec;185(2):867–871. doi: 10.1016/0042-6822(91)90561-o. [DOI] [PubMed] [Google Scholar]
- Horabin J. I., Schedl P. Regulated splicing of the Drosophila sex-lethal male exon involves a blockage mechanism. Mol Cell Biol. 1993 Mar;13(3):1408–1414. doi: 10.1128/mcb.13.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath C. M., Williams M. A., Lamb R. A. Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 1990 Aug;9(8):2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A., Burgess S. M., Hirsh D. beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katarova Z., Szabo G., Mugnaini E., Greenspan R. J. Molecular Identification of the 62 kd Form of Glutamic Acid Decarboxylase from the Mouse. Eur J Neurosci. 1990;2(3):190–202. doi: 10.1111/j.1460-9568.1990.tb00412.x. [DOI] [PubMed] [Google Scholar]
- Kornfeld K., Saint R. B., Beachy P. A., Harte P. J., Peattie D. A., Hogness D. S. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev. 1989 Feb;3(2):243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986 Nov 21;47(4):481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867–19870. [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Latchman D. S. Cell-type-specific splicing factors and the regulation of alternative RNA splicing. New Biol. 1990 Apr;2(4):297–303. [PubMed] [Google Scholar]
- Mattox W., Ryner L., Baker B. S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992 Sep 25;267(27):19023–19026. [PubMed] [Google Scholar]
- Morgan M. J., Earnshaw J. C., Dhoot G. K. Novel developmentally regulated exon identified in the rat fast skeletal muscle troponin T gene. J Cell Sci. 1993 Nov;106(Pt 3):903–908. doi: 10.1242/jcs.106.3.903. [DOI] [PubMed] [Google Scholar]
- Nielsen P. J., Trachsel H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988 Jul;7(7):2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel W. H., Schmechel D. E., Tappaz M. L., Kopin I. J. Production of a specific antiserum to rat brain glutamic acid decarboxylase by injection of an antigen-antibody complex. Neuroscience. 1981;6(12):2689–2700. doi: 10.1016/0306-4522(81)90113-5. [DOI] [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Berg P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol. 1986 Jul;6(7):2695–2703. doi: 10.1128/mcb.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986 Jul;6(7):2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Elledge S. J., Mitchell B. B., Marsh L., Walker G. C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko E. V., Gmyl A. P., Maslova S. V., Svitkin Y. V., Sinyakov A. N., Agol V. I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992 Jan 10;68(1):119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- Prats A. C., Vagner S., Prats H., Amalric F. cis-acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor mRNA. Mol Cell Biol. 1992 Oct;12(10):4796–4805. doi: 10.1128/mcb.12.10.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni M. J., Barthels D., Vopper G., Boned A., Goridis C., Wille W. Differential exon usage involving an unusual splicing mechanism generates at least eight types of NCAM cDNA in mouse brain. EMBO J. 1989 Feb;8(2):385–392. doi: 10.1002/j.1460-2075.1989.tb03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R. A., Miksch J. L., Xie X. L., Cornish J. A., Galewsky S. Expression of the Drosophila gonadal gene: alternative promoters control the germ-line expression of monocistronic and bicistronic gene transcripts. Development. 1990 Apr;108(4):613–622. doi: 10.1242/dev.108.4.613. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Felber B. K., Pavlakis G. N. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol Cell Biol. 1992 Jan;12(1):207–219. doi: 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Solimena M., Aggujaro D., Muntzel C., Dirkx R., Butler M., De Camilli P., Hayday A. Association of GAD-65, but not of GAD-67, with the Golgi complex of transfected Chinese hamster ovary cells mediated by the N-terminal region. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):3073–3077. doi: 10.1073/pnas.90.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Pestova T. V., Maslova S. V., Agol V. I. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology. 1988 Oct;166(2):394–404. doi: 10.1016/0042-6822(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Szabó G., Venetianer P. Translational coupling at the intercistronic boundary of an artificially constructed operon in Escherichia coli. Acta Biochim Biophys Acad Sci Hung. 1985;20(3-4):223–230. [PubMed] [Google Scholar]
- Tahara S. M., Dietlin T. A., Dever T. E., Merrick W. C., Worrilow L. M. Effect of eukaryotic initiation factor 4F on AUG selection in a bicistronic mRNA. J Biol Chem. 1991 Feb 25;266(6):3594–3601. [PubMed] [Google Scholar]
- Thomas K. R., Capecchi M. R. Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature. 1986 Nov 6;324(6092):34–38. doi: 10.1038/324034a0. [DOI] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Van Eden C. G., Mrzljak L., Voorn P., Uylings H. B. Prenatal development of GABA-ergic neurons in the neocortex of the rat. J Comp Neurol. 1989 Nov 8;289(2):213–227. doi: 10.1002/cne.902890204. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981 Dec 21;9(24):6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]