Abstract
The synovial joint contains synovial fluid (SF) within a cavity bounded by articular cartilage and synovium. SF is a viscous fluid that has lubrication, metabolic, and regulatory functions within synovial joints. SF contains lubricant molecules, including proteoglycan-4 and hyaluronan. SF is an ultrafiltrate of plasma with secreted contributions from cell populations lining and within the synovial joint space, including chondrocytes and synoviocytes. Maintenance of normal SF lubricant composition and function are important for joint homeostasis. In osteoarthritis, rheumatoid arthritis, and joint injury, changes in lubricant composition and function accompany alterations in the cytokine and growth factor environment and increased fluid and molecular transport through joint tissues. Thus, understanding the synovial joint lubrication system requires a multi-faceted study of the various parts of the synovial joint and their interactions. Systems biology approaches at multiple scales are being used to describe the molecular, cellular, and tissue components and their interactions that comprise the functioning synovial joint. Analyses of the transcriptome and proteome of SF, cartilage, and synovium suggest that particular molecules and pathways play important roles in joint homeostasis and disease. Such information may be integrated with physicochemical tissue descriptions to construct integrative models of the synovial joint that ultimately may explain maintenance of health, recovery from injury, or development and progression of arthritis.
Keywords: Musculo-skeletal system, High throughput screening, Proteomics, Mathematical models, Homeostasis
1. Synovial Joint Lubrication in Health, Injury, and Disease
1.A. General Overview
Synovial joints are the most common joints of the human body and allow load-bearing, low-friction, wear-resistant movement between apposing bone surfaces.1 Synovial joints normally include a cavity filled with synovial fluid (SF). The SF cavity is surrounded by, articular cartilage covering the bone surfaces, and a fibrous capsule including the synovium, the inner lining.
SF has biomechanical, metabolic, and regulatory functions. SF is normally a clear, straw-colored, viscous liquid. In normal human knee joints, the volume of SF is ∼1 ml.2 One main function of SF is to lubricate cartilage in synovial joints, facilitating low-friction and low-wear articulation.1 The molecular and cellular constituents within SF give rise to its unique properties and functions in maintaining joint homeostasis. SF is composed of a blood plasma dialysate and molecules secreted by cells lining and within the synovial joint space, including the lubricant molecules, hyaluronan (HA) and proteoglycan 4 (PRG4, also known as lubricin and superficial zone protein).3-5Additionally, SF serves metabolic functions, facilitating the transport of nutrients, waste products, and other metabolites to and from synovial tissues, as well as enzymes that act upon and within these tissues. Finally, SF contains soluble molecules, such as morphogens, growth factors, and cytokines, which mediate communication between cell populations in the joint.6-9
Joint disease and injury can result in pain and dysfunction of synovial joints. Osteoarthritis (OA) is a degenerative disease that results in destructive changes tojoint structures including cartilage, synovium, and bone. OA has been classified as idiopathic or secondary to other factors, such as joint trauma and congenital diseases. Knee OA has been classified according to clinical, laboratory, and radiographic criteria.10 Rheumatoid arthritis (RA) is a systemic inflammatory disease characterized by joint swelling, joint tenderness, and destruction of synovial joints.11 Synovitisis a key clinical feature of RA, and together with other criteria including the number of joints involved, duration of symptoms, and test results for serology and acute-phase reactants, can be used to classify RA.11, 12 Traumatic joint injury can result in cartilage damage, articular and bone fractures, damage to soft tissues such as ligaments and menisci, and lesions in the joint capsule and synovium. Such damage is associated with a number of mechanical and biological changes in the synovial joint that contribute to the development of post-traumatic OA.13-15
The composition and function of SF are altered in joint injury and disease, both due to changes directly of the SF and changes in tissues of the synovial joint. SF is in direct physical contact with cartilage and synovium, and in some joints, meniscus and ligament. SF interacts with and mediates interactions between synovial joint tissues. These tissues may themselves be altered in injury and disease. Changes in cellular metabolism and structure in these tissues may be reflected by changes in SF composition and function. Such changes in SF may result in a reduced ability to lubricate articulating cartilage and a catabolic environment within the joint, together contributing to joint deterioration. Alterations in joint tissues may be detrimental, propelling the SF to an aberrant state and leading to joint pathology. Thus, the disease-associated changes observed in SF likely are both exacerbated by and contribute to pathology of the synovial joint.
The objectives of this review are to provide an overview of (1) synovial joint lubrication in health, injury, and disease, with a focus on normal SF composition, formation, and rheological and tribological properties, and their alterations with joint injury and disease, and (2) high-throughput transcriptomic and proteomic studies of synovial joint cells and tissues that enable integrative models of synovial joint lubrication and a systems biology approach to studying synovial joint lubrication in health, injury, and disease.
1.B. SF Composition
1.B.i. Plasma Proteins
A major component of SF composition is proteins derived from plasma. Blood plasma and SF share many similarities in their protein composition,16, 17 with a superimposed effect of the sub-synovium and synovium selectively hindering large plasma proteins from entering the synovial joint space from the vasculature. Total protein concentration in normal human SF is 19–28 mg/ml, nearly one-third of that in plasma.2, 18, 19 Quantitatively, the major protein species in SF is albumin, at a concentration of ∼12 mg/ml.19 Other major protein components of normal SF include β1, γ, α1,and α2 globulins, each at concentrations of 1–3 mg/ml in normal SF.19 The size of plasma proteins is a major determinant of their filtration properties through the synovial membrane and entry into SF, and thus their concentrations in SF relative to those in plasma. Large molecular weight plasma proteins, such as fibrinogen (340kDa), are at relatively low concentrations in SF; in contrast, small molecular weight plasma proteins such as albumin (69 kDa) and transferrin (90 kDa), are in relatively high concentrations in SF (the concentration of albumin being ∼37% of that in plasma).19-21
The protein content and concentration in SF is increased with joint inflammation. Total protein concentration in SF from patients with OA, rheumatoid arthritis RA, and traumatic arthritis is higher than normal,19, 21-23 indicating structural and functional changes in the synovium with joint injury and disease. Synovial inflammation, as occurs in many joint diseases, compromises the ability of synovium to selectively retain and filter proteins. For example, SF from RA patients has high levels of globulins and glycoproteins, large molecular weight proteins normally not found in normal SF. The distribution of proteins in SF of RA patients is also altered and more closely resembles that in serum. Large plasma proteins, such as β2 macroglobulin (1,000kDa), fibrinogen (340kDa), β1 lipoprotein (3,200kDa), α2 macroglobulin (820kDa), and α2 glycoprotein (1,000kDa), are present in SF at increased concentrations and amounts in RA patients.20, 21
1.B.ii. Lubricant Molecules
Lubrication of articulating cartilage surfaces by SF is mediated by several lubricant macromolecules synthesized and secreted by synovial cell populations and found in SF. HA24 and PRG425, 26 are the primary lubricant macromolecules in SF and are present in normal SF at mean concentrations of ∼3.2–4.1mg/ml27-29and ∼0.035–0.24 mg/ml,30-32respectively. HA is a non-sulfated glycosaminoglycan composed of repeating disaccharide units of D-glucuronic acid and D-N-acetylglucosamine. HA in normal SF is present as a polydisperse population with a weight average molecular weight of 6–7 MDa, with the majority greater than 4MDa.33 HA contributes to the viscosity of SF and provides outflow buffering.34 Products of the PRG4 gene, which include superficial zone protein (SZP) and lubricin, are mucinous glycoproteins with multiple O-linked β(1-3)Gal-GalNAc oligosaccharides that mediate boundary lubrication of articular cartilage.35 SZP is a ∼345 kDa glycoprotein synthesized and secreted by chondrocytes in the superficial zone of cartilage, and not from other depths of cartilage.3 Lubricin is a ∼220 kDa glycoprotein expressed by synovial fibroblasts and also present in SF.4, 26, 36
OA, RA, and injury are associated with characteristic changes in SF lubricant macromolecules (Table 1A). The mean concentrations of HA in pathological SF are lower than that in normal SF, with SF from OA patients ranging from ∼1.2–2.2 mg/ml27, 37, 38 and RA patients ranging from ∼0.7–2.7 mg/ml.27, 29, 37, 39, 40 The molecular weight distribution of HA in pathological SF is also altered, with a shift to lower molecular weight forms.39 In an ovine meniscectomy model of osteoarthritis, cellular PRG4 immunostaining and mRNA levels were decreased in degenerative cartilage compared to normal cartilage.41 SF analysis following transection of the rabbit anterior cruciate ligament (ACL) and posterior cruciate ligament (PCL) revealed an association between decreased lubricin concentrations and boundary lubricating ability, increased elastase activity, and increased cartilage degradation.42
Table 1.
List of concentrations of molecules of interest in healthy, arthritic, and injury synovial fluid. Data are shown as mean±SD, or median. *Concentrations in SF lavages.
| Synovial Fluid Concentrations in Various States | |||||
|---|---|---|---|---|---|
|
| |||||
| Healthy | OA | RA | Injury | ||
|
| |||||
| Lubricant Molecules | HA | 3.2±0.4,27 3.6,28 4.1±1.029 mg/ml | 1.2±4.7,29 1.9±0.5,27 2.237 mg/ml | 0.7±0.3,29 0.8±0.3,39 1.2±0.3,401.2±0.4,27 2.737 mg/ml | 1.1±0.4,29 1.7±1.027 mg/ml |
| PRG4 SZP Lubricin | 0.035±0.028,320.16,24 ∼0.2425mg/ml | 0.15±0.0232 mg/ml | - | variable31,43 | |
|
| |||||
| Signaling Molecules | IL-1α | ∼0*,52 6.6±1.0 51 pg/ml | - | - | variable*52 |
| IL-1β | 0±0,169 ∼5*,5210.0±4.3,51<20 170 pg/ml | 1.0,171 1.4±1.3,17221±13,17027.8±4.5173pg/ml | 9.9±11.3,172 14.4,174 24.0,171 116±240,175 130±22,173193±57170 pg/ml | variable31,43,52,169,176,177 | |
| TGF-β | - | 40.4±8.8178(males),31.8±36.6178(females) pg/ml | - | variable176 | |
| TNF-α | 0±0,169<2*,52<4.4,51 2900±900170 pg/ml | 4.8±3.0,172 50±40,173 80±30170 pg/ml | 19.5±15.1,172 101,174 170±40,170390±40173 pg/ml | variable31,51,169,176,177 | |
| IGF-I | - | 60–80 ng/ml178 | - | - | |
| IFN-γ | 9±7.5 pg/ml,169 38.8±13 U/ml170 | 23.8±14.0 U/ml170 | 0.6±0.2 U/ml170 | variable169 | |
| IL-2 | 0±0169 | 5.2±0.6 U/ml,173 12.5±8.0 pg/ml172 | 4.5±0.6 U/ml,173 85.2±94.4 pg/ml172 | variable169 | |
| IL-4 | 0±0169 | 1.4±1.9 pg/ml172 | 1.4±1.7 pg/ml172 | variable169 | |
| IL-6 | 0±0,169 0.8±0.4 pg/ml51 | 3550±2950,172 212±85178(males), 95±56 pg/ml178(females), 490±324 U/ml179 | 0.99±1.01,175 9.6,174 16.0±9.9172 ng/ml, 5290±1730 U/ml179 | variable31,51,169,176,177 | |
| IL-8 | 0±0,169 21.0±5.151 pg/ml | 1.1±1.8 ng/ml172 | 0.236±0.412,175 21.9±36.8172 ng/ml | variable51,169,176,177 | |
| IL-18 | 36.2±17.4 pg/ml180 | 88.2±34.6 pg/ml180 | 278±65 pg/ml180 | - | |
| GM-CSF | 0.9±0.3,51 4±5169 pg/ml | - | - | variable51,169 | |
| PGE2 | 0.6±0.2 ng/ml180 | 2.8±1.4 ng/ml180 | 5.1±1.8 ng/ml180 | - | |
| NO | 22.6±14.8 μM180 | 57.5±29.4 μM180 | 131±36μM180 | - | |
| PTHrP | - | 1.8±0.2 pM172 | 2.6±0.9 pM172 | - | |
|
| |||||
| Degradative Enzymes | ADAMTS-4 | - | - | - | - |
| MMP-1 | 0.12(pro, active, and bound) ng/ml55 | 0.84(pro, active, and bound),55356(free and pro)54 ng/ml | - | variable55 | |
| MMP-3 | 6.7ng/ml(pro, active, and bound),552(pro, active, and bound),181 2.7(pro, active, and bound)182 nM | 31.2(pro, active, and bound),5511,700(pro, active, free, and bound)54 ng/ml | 88.6±78.5 ng/ml175 | variable43,55,56,181,182 | |
| TIMP-1 (free) | 6.8ng/ml(free),55 4.8(free),181 5.2(free)182 nM | 19.7(free),55 407(free and bound)54 ng/ml | 1040±1410 ng/ml175 | variable55,56,181,182 | |
| TIMP-2 | - | 165(free and bound) ng/ml54 | - | - | |
|
| |||||
| Binding Proteins | Albumin | ∼10 mg/ml69 | - | ∼19 mg/ml21,183 | - |
| TNF-R | - | 8.7±3.7 ng/ml184 | 21.0±9.3 ng/ml184 | - | |
| IL-1RA | ∼67*,52 2520±1470 pg/ml51 | 614,171 486±258178(males), 333±106178(females) pg/ml | 17.0 ng/ml171 | variable51,52,176,177 | |
| IL-2R | - | 606 U/ml184 | 1670 U/ml184 | - | |
| IL-18BP | 179±86 pg/ml180 | 138.4±52.7 pg/ml180 | 56.5±25.8 pg/ml180 | - | |
|
| |||||
| Structural Proteins | CPII | - | - | - | variable158 |
| CTXI | - | - | - | variable158 | |
| CTXII | 1.456ng/ml | 4.856ng/ml | - | variable43,56 | |
| sGAG | ∼0.1,185 ∼0.06186 mg/ml | ∼0.08,185 ∼0.06186 mg/ml | ∼0.04 mg/ml185 | variable31,43,51,186 | |
| ARGS aggrecan | 0.5186nM | 4.6186nM | - | variable43,186 | |
| COMP | 47187μg/ml | ∼90187μg/ml | - | variable43,187 | |
The effects of injury on the concentration of lubricin in SF appear variable. Lubricin concentrations in the SF of patients with ACL injury were reported to be lower compared to SF from the uninjured contralateral knee during the first several months post-injury, and increasing to contralateral values by∼12 months.31 However, SF lubricin concentrations in longitudinal samples from patients with recent severe knee injury decreased from the time of presentation (∼15 days post-injury) to the time of arthroscopic surgery (∼48 days post-injury).43 SF lubricin concentrations may be altered post-injury due to changes in the rates of synthesis, secretion, degradation, and/or loss from the joint, as well as changes in SF volumes. The effects of injury on lubricin synthesis and secretion have been studied with experimental models of cartilage mechanical injury. In a bovine model, lubricin mRNA expression and protein synthesis and secretion increased in explants from superficial regions of cartilage with an intact articular surface after 2 days following injurious compression.44 Further studies are needed to clarify the effects of injury on human SF lubricin concentrations. The absence of PRG4, both in patients with the genetic disease camptodactyly-arthropathy-coxa vara-pericarditis syndrome (CACP),45 an autosomal recessive disease with loss of function mutations in the PRG4 gene, and in PRG4-/- mice,46 results in cartilage degeneration and synovial hyperplasia.
1.B.iii. Cytokines and Growth Factors
Cytokines and growth factors present in SF are important regulatory factors for the cell populations lining and within the synovial joint space, including chondrocytes and synovial cells.6, 9 Regulatory molecules in SF may be derived from plasma or as secreted products of chondrocytes, synovial cells, and other cells within the SF or surrounding tissues. Cytokines may be categorized as pro-inflammatory or anti-inflammatory according to their predominant tissue-specific effects. Pro-inflammatory cytokines in SF include IL-1α, IL-1β, TNF-α, leukemia inhibitory factor (LIF), IL-6, IL-8, IL-17, and IL-18.7, 47, 48 Anti-inflammatory cytokines in SF include IL-4, IL-10, and IL-13. Growth factors found in SF include TGF-β1 and insulin growth factor 1 (IGF-I) and have anabolic effects. Binding proteins are also present in SF and play important roles in cell regulation.
With joint injury and disease, the cytokine profile in SF is altered. Most cytokines and growth factors are at relatively low concentrations in normal SF, and are markedly elevated in joint injury and disease. Cytokines play an important role in disease pathogenesis and acceleration of joint destruction, and have attracted attention as potential therapeutic targets, with some cytokine-directed therapies already in clinical use and others in clinical trials.8, 49 Some pre-clinical studies have demonstrated efficacy in improving synovial joint lubrication. Blocking the effects of TNF-α with etanercept, for example, resulted in increased amounts of lubricin bound to cartilage and decreased sGAG release from cartilage compared with untreated groups in a rat ACL injury model of post-traumatic arthritis.50 Binding proteins such as IL-1 receptor antagonist (IL-1RA) can play chondroprotective roles in the synovial joint and are found at relatively high levels in normal SF, with time-dependent changes in their concentrations after ACL injury.51, 52
1.B.iv. Proteolytic Enzyme Activity
Degradative processes in the synovial joint are mediated primarily by proteolytic enzymes that are carefully regulated, as reviewed by Poole.76 Matrix-degrading enzymes, such as matrix metalloproteinases (MMPs) MMP-1 and MMP-3, are present in normal SF and elevated in joint injury and disease.65, 66 MMPs are secreted primarily from chondrocytes as latent zymogens with propeptide domains that are cleaved during extracellular activation. A distintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) proteinases degrading aggrecan are also secreted as latent zymogens requiring subsequent activation. Other proteinases, such as serine and cysteine proteinases, are involved in activation of proMMPs. For example, plasmin, kallikrein, and cathepsin B activate proMMP-1. Tissue inhibitors of metalloproteinases (TIMPs) and inhibitors of proteinases that activate proMMPs are also present. In RA, polymorphonuclear leukocytes and synovial cells also contribute to proteinase levels in the joint. Changes in the levels and activities of matrix-degrading enzymes, and their associated inhibitors and activators, alter the balance between anabolism and catabolism in joint injury and disease. Changes in proteolytic activity in the synovial joint are indicated by elevated levels of degradation products. Concentrations of fragments of aggrecan and type II collagen are elevated in the SF of patients soon after joint injury and in patients with OA.69, 77
1.B.v. Cells
Normal SF is relatively acellular compared to whole blood, containing <200 leukocytes per mm3 78-80 compared to 3,540–9,060 per mm3 in whole blood.81 Of the nucleated cells in SF, only a minor percentage can be identified as synovial cells.78, 79 Erythrocytes are virtually absent in normal SF, compared to 4.0–5.6 × 106 per mm3 in whole blood.81 OA and RA are usually accompanied by increased synovial effusions. The mean leukocyte count in RA SF is ∼11,000–19,000/mm3,79, 82 compared to <2,000/mm3 in non-inflammatory SF in OA and traumatic arthritis.80 Lymphocytes, macrophages, and shed lining cells are also present in SF. Few neutrophils are found in RA synovium, even though they often comprise the majority of cells in the effusions.83 Erythrocytes may be introduced into SF in cases of trauma.
1.C. SF Formation and Transport through Synovium
1.C.i. SF Formation
SF formation is a process that involves the functioning of synovium as a semi-permeable membrane that is size-selective. SF is an ultrafiltrate of blood plasma with additional molecules secreted by cells lining and within the synovial joint space. SF lubricant molecules are derived from the cell populations lining the synovial cavity, with HA secreted mainly by synoviocytes and PRG4 molecules by chondrocytes in the superficial zone of cartilage,3synoviocytes,4, 36, 84 and meniscal cells.85 Molecular sieving by the synovial membrane matrix is primarily size-dependent.86 High molecular weight species within SF, which include the lubricant molecules HA and PRG4, are selectively retained within the synovial joint, while low molecular weight species, such as most metabolic substrates and byproducts, cytokines, and growth factors, are not. High molecular weight species within plasma, on the other hand, are unable to pass through the synovial membrane easily and are absent or low in normal SF. Joint injury and disease affecting the synovial membrane may impair the SF formation process, resulting in pathologic SF.
1.C.ii. Transport Characteristics of Synovium
Synovium is the main barrier to the transport of molecules in SF and plasma, such as lubricant molecules and plasma proteins, into and out of the synovial joint. Synovium is a vascularized, thin sheet of connective tissue with fibroblast-like (type B) cells and macrophage-like (type A) cells within an extracellular matrix (ECM) composed primarily of HA, collagen, and proteoglycans. The blood-joint barrier has been modeled as a double barrier, in series, consisting of synovial interstitial space that limits diffusion of small molecules, and microvascular endothelium that limits transport of proteins.19, 87 Fluid flow through normal rabbit synovium occurs through interstitial spaces that range from 0.1–11.8 μm and average ∼2 μm,88 which has a hydraulic conductivity of ∼10-14 (m3·s)/(kg).70 Diffusive and convective passive transport of molecules occurs by size-selective molecular sieving through synovial ECM, and can be modeled as transport through pores with a calculated pore radius of ∼25–60 nm.86, 89 High MW lubricant molecules are selectively retained in the joint, as the reflection coefficient of synovium to high MW HA is ∼57–75%.90
In joint injury and disease, alterations in synovium result in pathological SF. In RA, synovium increases dramatically in mass and metabolic activity, with redundant folds, frond-like villi, infiltration with immune cells, and edema. The synovial intimal lining, which directly contacts the synovial fluid compartment, increases greatly in depth up to 10-fold, although there are no tight junctions or basement membranes that interfere with the flux of fluid. Intimal hyperplasia is due to increased numbers of fibroblast-like synoviocytes and macrophage-like synoviocytes. The former express the adhesion protein cadherin-11, which accounts for aggregation of synoviocytes. The intimal lining is the location where most pro-inflammatory cytokines and metalloproteinases are produced in RA. The sublining is infiltrated with T cells, B cells, macrophages, and plasma cells, with CD4+ T lymphocytes as the most prevalent cell type. Lymphocytes can organize into aggregates and lymphoid follicles in ∼20% of patients, with diffuse mononuclear cell infiltration or acellular fibrous infiltration occurring in others. Considerable variation exists throughout the synovium, necessitating careful experimental sampling. An increase in the number of blood vessels is usually prominent, although the capillary network tends to be more disorganized than normal.83
Molecular transport through synovium is markedly altered in diseased synovium. Synovium from joints of RA patients are more permeable to large proteins.19, 87 Increased synovial inflammation in patients with various form of arthritis is associated with proportionately greater increases in permeability to larger plasma proteins.91 Whereas permeability of RA synovium to large proteins is increased, permeability to small molecules (e.g., urea, glucose) is slightly decreased. This may be explained by a combination of increased vascular permeability and synovial hyperplasia and cellular infiltration. Increased Starling pressure due to the reduced oncotic gradient as protein accumulates in SF, increased permeability of capillaries, and an increase in capillary pressure during inflammation contribute to the buildup of fluid and protein in RA SF. HA contributes to outflow buffering in the synovial joint. In arthritis, the size of SF HA is reduced, with an increased rate of HA loss from SF.86 Although synovial membrane inflammation is also recognized as a key factor in OA pathophysiology, the transport characteristics of synovium in OA are less affected than in RA, with apparent permeabilities to proteins over a range of sizes (molecular radii ∼3–9 nm) over 3-fold higher in RA compared to OA.92
1.D. Rheology, Tribology, and Mechanobiology
1.D.i. Rheological and Tribological Properties of SF
The rheological and tribological properties of SF have been characterized through measurements of viscosity, viscoelasticity, and friction. Normal SF is a viscous, non-Newtonian, thixotropic93 fluid. The rheological properties of SF are in large part attributable to the concentration and molecular mass of HA.94, 95Lubricin appears to play an important role in organizing HA and providing SF with the ability to dissipate strain energy.96 SF behaves as a viscous material at low frequencies of oscillation, and as an elastic material at high frequencies.Studies with cartilage-on-cartilage friction tests have identified HA and PRG4 as important boundary lubricants in SF that lower friction in a dose-dependent manner, both alone and in combination.97, 98 Cartilage-on-glass friction testing has revealed that recombinant human lubricin lubricates cartilage in a dose-dependent manner and by two distinct mechanisms, involving lubricin bound to the surface, and in solution.99
Pathological SF exhibits impaired rheological properties. Normal and OA SF display shear-thinning behavior, while RA SF displays Newtonian behavior.95 Viscosity of normal SF is higher than that of OA SF, which is higher than RA SF. In ACL-transected guinea pig knees, increased coefficients of friction and decreased lubricin concentrations were observed.100 In ACL- and PCL-transected rabbit knees, lubricin concentrations and lubricating ability decreased, and collagen degradation increased, at weeks 2 and 3 after injury.42 The lubricating ability of OA SF is not significantly different compared to normal SF, but RA SF lubricates poorly compared to normal SF.101
1.D.ii. Mechanobiology of Lubricant-Secreting Cells
Lubricant-secreting cells of synovial joints are regulated by certain mechanical cues. HA secretion by rabbit synoviocytes increases when primary cultures are subjected to stretch,102 and when rabbit knee joints are exposed to passive cycling.103HAS and PRG4 mRNA expression and HA and PRG4 release into conditioned media increase when scaffolds seeded with bovine chondrocytes are subjected to articulation.104 PRG4 secretion by bovine chondrocytes also increases when cartilage explants are loaded with dynamic shear superimposed upon static compression105 as well as when chondrocytes in three-dimensional culture are subjected to articulation loading;104, 106, 107 in contrast, PRG4 secretion is inhibited by static compression alone.105 The mechanism by which shear loading of cartilage stimulates PRG4 secretion appears to involve signaling pathways mediated by TGF-β.108 Continuous passive motion applied to bovine joints increased PRG4 secretion and the percentage of chondrocytes expressing PRG4 in regions of the femoral condyle that experienced mechanical stimulation from continuous or intermittent sliding against apposing tissues.109
2. Systems Biology of the Synovial Joint
2.A. General Overview
As described in earlier sections, the synovial joint is composed of several cell and tissue types, which are altered in injury and disease. The study of isolated components and their changes can provide detailed information at the molecular, cellular, and tissue scales. However, insights into the behavior and properties of the synovial joint lubricant system require an overall consideration of components and their interactions. A systems biology approach can be employed to study how the relationships between the various components of a synovial joint contribute to produce a load-bearing and friction-lowering joint that, in most people and under normal conditions, provides a lifetime of normal joint function.
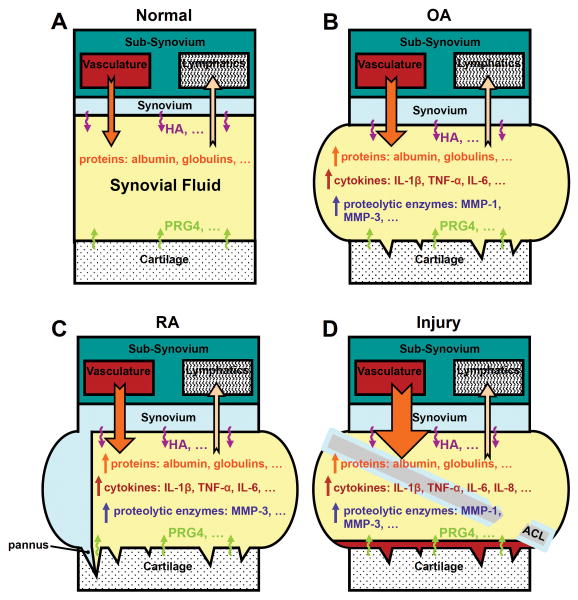
The pathophysiology of the most common rheumatic disorders, OA and RA, and joint injury, involves multiple changes at the molecular, cellular, tissue, and organ levels110 (Figure 1). The normal, healthy, synovial joint involves cartilage, synovium, SF, and other tissues functioning together (Figure 1A). OA is a complex, heterogeneous disease associated with alterations in cartilage degradation and repair, synovial inflammation, and increased production and SF levels of catabolic and pro-inflammatory cytokines (Figure 1B).111 The altered cytokine environment can enhance cartilage breakdown, which in turn may amplify synovial inflammation, leading to a cycle of events resulting in progressive joint destruction. OA develops progressively, but inflammatory flares can occur.111 RA is a systemic inflammatory disease with features of synovitis, pannus formation, and SF volume increase, along with changes in the mass and concentration of plasmaproteins, cytokines, and key proteases such as MMPs and their proenzyme activators and inhibitors (Figure 1C). Acute, traumatic joint injury, such as to the anterior cruciate ligament, result in a number of mechanical and biological changes over time that can contribute to the development of post-traumatic OA (Figure 1D).13-15 Post-traumatic OA is a subset of OA that is associated with an acute traumatic event. Some of the changes that may be involved in post-traumatic OA pathogenesis include increased cell death, increased levels of inflammatory cytokines and ECM-degrading enzymes, and decreased levels of lubricant molecules. Joint injury initiates events that, in many cases, lead to joint destruction, pain, and disability, with pathology and clinical symptoms that are similar to primary OA. In injury and disease states, analysis of whole joint changes can provide an understanding of how joint pathophysiology arises from the interplay between component parts and contributes to the disease phenotype. Following the systems biology paradigm, data on multiple scales and from various sources can be integrated into models that predict emergent properties of the biological system. For the synovial joint lubrication system, this approach may involve integration of transcriptome and proteome data with physicochemical tissue descriptions to construct models that account for the complex interactions between cartilage, synovium, and SF in the synovial joint. At the tissue and whole joint level, physicochemical principles of mass balance and transport, combined with data from transport studies, can be integrated into the mathematical framework of a model. Once constructed, models can be a powerful simulation tool to predict the behavior of the biological system under different conditions, generate new testable hypotheses, and provide insights into the physiology and pathophysiology of the synovial joint.
Figure 1.
Joint injury and disease involve pathologic changes in multiple joint tissues. Schematic view of synovial joint tissue changes in (B) OA, (C) RA, and (D) injury, compared to (A) a normal, healthy joint. Notched, outlined arrows represent fluid flows from vasculature to SF, and from SF to lymphatics. Undulating arrows represent secretion of lubricant molecules HA and PRG4 into SF. The relative sizes of arrows indicate relative magnitudes of flows and secretions. For example, increased flow from the vasculature to SF in (D) injury compared to (A) normal is represented by an orange, notched, outlined arrow that is larger in (D) injury compared to (A) normal. Up-arrows indicate increases in concentrations of the various substances.
McCarty WJ, Nguyen QT, Hui AY, Chen AC, Sah RL. Synovial Joints: Mechanobiology and Tissue Engineering of Articular Cartilage and Synovial Fluid. In: Ducheyne P, Healy KE, Hutmacher DE, Grainger DE, Kirkpatrick CJ, eds. Comprehensive Biomaterials. 1st ed; 2011.
2.B. Data Types and Sources
2.B.i. Conventional Methods
Conventional methods of collecting experimental data provide detailed information about specific entities. ELISA assays, for example, provide valuable quantitative data on specific molecules, such as cytokines and lubricant molecules, in the synovial joint under different conditions. Concentrations of molecules of interest in SF, measured by conventional methods, may be a valuable resource for estimating values of parameters in models of the synovial joint (Table 1A,B). Such methods, however, are typically applied to study a few entities of interest, although multiplex variants of such methods allow study of multiple targets simultaneously. As study of the synovial joint shifts from specific molecules and isolated components to a more comprehensive, systems biology approach, methods that enable the study of many components will be useful.
2.B.ii. Large-Scale, High-Throughput Methods
The application of large-scale, high-throughput technologies to synovial joint tissues is leading to an abundance of data on components of the synovial joint system. Transcriptomic studies aim to characterize gene expression on a large scale, on the order of thousands of genes, in order to capture the global state of the cell. Commonly used microarray technologies include Affymetrix chips and Illumina platforms and enable high-throughput screening by determining the expression of tens of thousands of genes in a massively parallel manner. Much of this data is accessible as datasets in peer reviewed, public repositories such as the Gene Expression Omnibus from the National Center for Biotechnology Information and ArrayExpress from the European Bioinformatics Institute. Several datasets of samples of synovial joint tissues and cells have been published.112-120
Common methods for the large-scale characterization of proteins in synovial tissues include two dimensional polyacrylamide gel electrophoresis (2D-PAGE) or liquid chromatography for protein separation and mass spectrometry (MS) for protein identification. Several software packages are available for spot detection, matching, and quantitation of 2D-PAGE images, and comparisons between two widely used ones, PDQuest and Progenesis, have been made for the analysis of SF samples.121 Advances in protein microarray technologies add another tool for proteomic studies of synovial tissues.
A number of biological issues can complicate the interpretation of data from high-throughput methods, and guidelines for evaluating microarray studies have been suggested.122 Transcriptomic and proteomic studies on complex tissues, such as RA synovium, can be complicated by variation between samples and other issues such as heterogeneity of the cell population in native and diseased tissues. One approach to circumventing this challenge is to focus on individual cell types isolated from tissues. In the case of RA synovium, for example, RA synovial fibroblasts can be studied after being isolated from synovium.
Extraction of biological information from the large volumes of data provided by large-scale and high-throughput methods has been achieved in several ways. Significance Analysis of Microarrays is a statistical technique for determining whether differentially expressed genes in microarray experiments are statistically significant.123 The Significance Analysis of Microarrays method has been applied to identify sets of genes that are differentially expressed in subgroups of RA synovium124, 125 and fibroblast-like synoviocytes,126 providing evidence of the heterogeneous nature of RA. Clustering methods, such as hierarchical and k-means clustering, group together genes that are co-expressed under different conditions, to identify functionally related genes. Principal component analysis reduces the dimensionality of a dataset. Clustering analysis and principal component analysis have been used to identify patterns of gene co-expression by cartilage explants and possible pathways involved under different mechanical loading conditions.127-129 For example, transcription of most matrix proteins is upregulated by dynamic compression and dynamic shear, but downregulated by static compression, while most matrix proteases are upregulated under all three loading conditions.128 Data from high-throughput gene expression studies may be analyzed by gene set enrichment analysis for pathway level analysis, by associating gene expression data with various pathways. Gene set enrichment analysis determines whether sets of genes of pre-defined pathways or processes are enriched or impoverished in a candidate gene list.130
The experimental data from large-scale analyses are useful screens to characterize disease by identifying key molecular constituents that may be mediators of disease on the basis of correlated molecular behavior observed in the datasets. For example, high-throughput methods have suggested the potential pathogenic role of a number of genes and proteins not previously associated with OA, on the basis of differential expression of those genes and proteins (Table 2A,B). Data from high-throughput methods, in particular those concerning key molecules, may be validated with conventional, low-throughput methods such as real-time RT-PCR for quantifying mRNA levels, and ELISA and Western blot for measuring protein. Profiling gene and protein expression patterns of synovial joint tissues has identified key molecular constituents associated with joint injury and disease and provided insights into the molecular basis of synovial joint changes in health, injury, and disease. Important features of the system as suggested by these data sets may then be integrated into model descriptions of joint homeostasis.
Table 2.
Genes not previously associated with OA implicated by high-throughput transcriptomic methods to be involved in OA pathogenesis.
| Functional Classifications | Gene Symbols |
|---|---|
|
| |
| Chemokines | CCL3115 |
| CCL4115 | |
| CKLF1114 | |
| CX3CL1114 | |
| CXCL2115 | |
| CXCL3115 | |
| CXCL14115 | |
| CXCR4114 | |
|
| |
| Cytokines | TNFRSF12A115 |
| TNFRSF21115 | |
|
| |
| Growth Factors | FGF1115 |
| LTBP1115 | |
|
| |
| Matrix Components | COL8A2115 |
| COL13A1115 | |
| COL14A1115 | |
| COL15A1115 | |
| ECM1115 | |
| ECM2115 | |
| EFEMP1115 | |
| FBLN5115 | |
| LAM4115 | |
| SPON2115 | |
|
| |
| Bone related | CDH11115 |
| CHST11115 | |
| CLEC3A115 | |
| CLEC3B115 | |
| GPNMB115 | |
| MSX1115 | |
| MSX2115 | |
|
| |
| Anti-proliferative factor | TOB1116 |
|
| |
| Degradative Enzymes | CTSC114 |
| MMP19115 | |
2.C. Transcriptomic Studies
2.C.i. Synovium and Synovial Cells
In genome-wide gene expression profiling of synovial cells from OA and RA patients, many differentially regulated pathways have been identified, with some validated by conventional techniques such as real-time RT-PCR and Western blots. In RA synovial fibroblasts, upregulation of TGF-β pathway components114 and pathways involved in protection against oxidative stress,136 in particular thioredoxinreductase 1, have been reported.
Gene expression profiling can further be used to examine the effects specific cytokines can have on the synoviocyte transcriptome. Stimulation of synoviocytes with TNF-α, which is important in the pathogenesis of RA, induced elevated Naf1 gene expression in RA compared to OA, suggesting a possible role of Naf1, an inhibitor of NFκB-dependent gene expression, in modulating TNF-α activity in RA.137 In addition, clusters of RA related genes, involved in inflammation, synovial proliferation, and bone and tissue destruction, were upregulated by PGI2-IP signaling in mouse synovial fibroblasts subjected to collagen induced arthritis, suggesting a role for PGI2-IP signaling in RA.138 Gene expression studies on synoviocytes under the influence of TNF-α alone, IL-1β alone, TNF-α and IL-17 together, and TNF-α and IL-1β together, have found that these cytokines induce the same genes that are upregulated in patients who respond poorly to adalimumab therapy, suggesting a possible role of these cytokines in drug resistance.112
Gene expression is different in synovium from normal donors and OA and RA patients. Analysis of inter-individual, intra-group gene expression variances within normal donors and OA and RA patients has identified several potentially disease relevant pathways and complexes by comparing genes with different variances in different patient groups.120 Many of the identified pathways for RA are involved in inflammation, angiogenesis, proliferation, and cell survival. Synovial membranes from OA patients showed higher gene expression variances in genes involved in pathways such as oxidative phosphorylation, MAPK signaling pathway, folate biosynthesis, and starch and sucrose metabolism, representing a desynchronization of metabolic processes. Synovial membranes from RA patients, on the other hand, were found to have increased gene expression variances in pathways involved in melanogenesis, B-cell receptor signaling, and VEGF signaling. Microarrays have shown differential gene expression profiles in synovium from RA patients, identifying two molecular subtypes of RA synovium, with one subgroup upregulating clusters of genes involved in immune activation pathways and the other upregulating genes, such as collagens and SOX9, suggesting fibroblast dedifferentiation.124 Synoviocytes with molecularly distinct expression patterns in synovium have been reported in RA, with an association between α-SMA expressing myofibroblast-like synoviocytes and highly inflamed synovial tissue.125, 126 Distinct expression profiles have also been observed in early compared to late RA, suggesting different biological processes and pathways may be involved with RA disease progression.119
2.C.ii. Cartilage and Chondrocytes
Gene expression profiling has been used to study chondrocyte transcriptome in health, injury, and disease. Chondrocytes from OA patients compared to those from patients undergoing autologous chondrocyte transplantation for focal cartilage lesions are less differentiated in monolayer culture, as assessed by gene expression profiling, but the number of differentially expressed genes decreases following chondrogenic differentiation in 3D culture scaffolds, with TIMP3 the most notable persistently differentially expressed gene.113
Determining expression profiles in response to specific manipulations in vitro is one approach to elucidate important molecular mediators in signaling pathways relevant to joint disease. In OA and RA, TNF-α levels in SF are elevated. Microarray technology has been used to study the effects of TNF-α stimulated MEK/ERK signaling on global chondrocyte gene expression, resulting in the discovery that among the TNF-α modulated genes, proinflammatory genes are MEK/ERK independent, while genes encoding proteins with proteinase activity and HA binding activity, contributing to matrix catabolism, are MEK/ERK dependent.115 Further studies showed that TNF-α regulation of the transcription factors SOX9 and NFκB are MEK/ERK independent, whereas regulation of EGR-1 is MEK/ERK dependent. Another cytokine implicated in OA and RA, IL-1β, has been studied for its effects on chondrocyte gene expression.116 Chondrocytes stimulated by IL-1β increased expression of GCSF, SELE, LIF, IL-1β, IL-6, IL-8, and a number of chemokines, and decreased expression of type II collagen and ADAMTS-5. OA cartilage had gene expression patterns similar to normal cartilage stimulated with low dose IL-1β. IL-1β induced altered gene expression in chondrocytes were partially reversed by TGF-β1, but not by BMP-2.116
A number of transcriptomic studies have been conducted on cartilage tissue. In experimental models of OA in rats, 1619 significantly differentially expressed probe sets were found compared to control, including genes expressed in hypertrophic growth plate chondrocytes genes for cytokine and chemokine signaling.132 Newly identified genes with potential roles in OA pathogenesis were identified, including chemokine signaling factors CXCR4, CKLF1, CX3CL1, and CTSC. Comparing cartilage from OA and normal donors, new genes not previously associated with OA, including some involved in bone formation and collagen synthesis,131 were found upregulated in OA samples (Table 2A). TOB1,133 which is an anti-proliferative factor not previously associated with OA, SOX6, and SOX9139 were repressed in OA. The role of different degradation pathways in late vs. early OA have been suggested by microarray analysis showing downregulation of MMP3 and upregulation of MMP2 and MMP13 in late-stage disease.140 Comparisons of intact and damaged regions of OA cartilage have revealed upregulation of genes involved in wound healing, cell proliferation, and collagen synthesis in damaged regions.141 Cartilage samples with induced mechanical injury compared to normals showed differentially expressed genes, including upregulation of WNT16 upon injury.142 Approximately 30% of the differentially regulated genes in OA vs. normal were also altered in injury vs. normal.
2.D. Proteomic Studies
2.D.i. Synovial Fluid
The characterization of protein and peptide components in SF in a comprehensive and high-throughput manner has provided investigators with another tool to study the underlying pathophysiology of joint injury and disease. Endogenous peptides are of interest because they are major regulators of biological functions in health and disease. Cytokines and growth factors, for example, are critical in cell signaling and metabolism, and alterations in their abundance and activity levels in the synovial joint and their role in injury and disease have been described earlier. Additionally, peptides and peptide fragments may serve as surrogate markers of enzymatic and proteolytic activity, with potential applications as biomarkers to diagnose, prognosticate, or monitor disease progression and treatment. One approach to discovering sets of proteins associated with joint injury or disease is comparative proteomics, which aims to detect and characterize the differences in abundance and activities of expressed proteins and peptides in samples of SF from donors in normal and injured or diseased states. These systems-wide analyses have confirmed the involvement of previously known participants and opened new lines of investigation by implicating novel and previously unassociated molecular pathways and mediators that contribute to joint disease.
In recent years, several proteomic studies of SF have contributed insights into the molecular mediators of joint disease. Several peptide cleavage products from proteins previously associated with OA were elevated in SF from OA patients compared to SF from cadavers without joint disease, including type II collagen, PRG4, serum amyloid A (SAA), tubulin, vimentin, and matrix gla protein.143 In studies of SF and plasma from patients with RA, OA, or reactive arthritis, calgranulin B was exclusively identified in RA SF, and SAA was identified in RA SF, but not OA SF.144 Comparing the SF proteome in health and OA, cystatin A, aggrecan, and dermicidin levels were lower in OA SF compared to healthy SF.145 No duration-dependent changes or disease-stage specific differences were found in OA SF, but two distinct patterns of protein expression were observed. Proteomic studies have identified biomarkers of disease severity, with C-reactive protein, S100A8, S100A9, and S100A12 elevated in the SF and serum of patients with erosive RA compared to nonerosive RA or normal.146, 147
2.D.ii. Cartilage and Chondrocytes
The proteome of normal human chondrocytes has been characterized by 2D-PAGE and MS under basal and stimulated conditions. A reference map showing 136 spots and 93 identified proteins is available.148 The proteome of normal chondrocytes in response to cytokine stimulation has been investigated.149 Stimulation by IL-1β differentially modulated 22 proteins involved in cytoskeletal rearrangement and cellular metabolism. TNF-α modulated 20 proteins involved in transcription, synthesis, and protein turnover, cytoskeletal rearrangement, and stress responses. The combination of both cytokines modulated 18 proteins related to metabolism and energy and transcription, protein synthesis, and turnover. Nine novel proteins not previously known to be synthesized by human chondrocytes were discovered in cytokine-stimulated chondrocytes.
Proteomic analyses have also been applied to the study of conditioned media from cultured explants of normal and injured or diseased cartilage. In one study, conditioned media was analyzed in order to characterize the proteins that are secreted and released by osteoarthritic cartilage.150 Proteins previously known to be secretion products of cartilage, such as YKL-39 and osteoprotegerin, were identified, along with a number of novel proteins not previously known to be secreted by cartilage, such as serum amyloid P-component, pigment epithelium derived factor, fibrinogen, lyl-1, thrombopoietin, fibrinogen, angiogenin, gelsolin, and osteoglycin. Medium from explants subjected to injurious compression, IL-1β, or TNF-α contained ECM proteins such as aggrecan, collagens, perlecan, fibronectin, and link protein. Novel cartilage proteins were also identified, including CD109, platelet-derived growth factor receptor-like protein, angiopoietin-like 7, and peptidoglycan-recognition protein long. CHI3L1, CHI3L2, complement factor B, MMP-3, ECM-1, haptoglobin, serum amyloid A3, and clusterin were increased in the medium of explants treated with IL-1β and TNF-α. Type VI collagen subunits, cartilage oligomeric matrix protein, and fibronectin were increased in explants subjected to injurious compression. Proteins involved in inflammation and stress response were increased in the medium of IL-1β and TNF-α treated explants. Release of intracellular proteins into the medium was increased in compression-injured explants, suggesting cell death and loss of cell membrane integrity.151
Proteomic approaches may be applied to the study of the proteomes of different cellular components of chondrocytes. The proteome of articular cartilage vesicles was characterized in one study.152 170 proteins were identified in articular cartilage vesicles from cartilage of 10 OA and 10 normal patients. A number of proteins were discovered with significantly different levels in OA and normal patients. Six proteins were found exclusively in vesicles from normal cartilage, and 9 proteins, including inflammation related proteins such as fibrinogen, complement, immunoglobulins, and apolipoproteins, were found exclusively in vesicles from OA cartilage. In another study, the mitochondrial proteome from chondrocytes isolated from healthy donors was obtained by 2D-PAGE and MS and a reference map was constructed.153
Recently, a method for the selective extraction of proteins from cartilage has been described, allowing direct proteome characterization of human articular cartilage by 2D-PAGE and MS.154 To study differences in protein synthesis in cartilage from OA patients and normal donors, cartilage explants were incubated in medium containing [35S] methionine/cysteine to label newly synthesized proteins and analyzed.134 Collagen II and activin A were strongly upregulated in OA cartilage compared to controls. Analysis of normal cartilage from 7 donors and OA cartilage from 7 OA patients revealed 59 proteins that were differentially expressed, including HtrA serine protease 11 which was found at levels 8 fold higher in OA compared to normal cartilage. A number of proteins not previously associated with OA were also identified (Table 2B), including fibulin 3, osteonectin, secreted modular calcium-binding protein 2, vitrin, and tenomodulin.135
2.D.iii. Synovium and Synovial Cells
Proteomic approaches have been used to characterize protein expression in synovial cells isolated from healthy donors and patients suffering from OA and RA. Analysis by 2D-PAGE and MS of synovial cells from RA patients confirmed the presence of a number of proteins previously known to be important in synovial cell function. From this study, a protein product of chromosome 19 ORF10 was identified, whose biological activity and role in synovial cell function is unclear.155 In synovial fibroblasts obtained from 6 OA, 5 RA, and 7 control patients, 2D-PAGE revealed 15 and 34 proteins that were significantly elevated in OA and RA synovial fibroblasts, respectively, compared to controls.156 Validation by Western blot showed that levels of enolase-α, S100A4, S100A10, annexin I, cathepsin D, mitochondrial superoxide dismutase, and peroxiredoxin 2 were significantly higher in OA and RA synovial fibroblasts compared to controls, while mitochondrial manganese superoxide dismutase and cathepsin D were significantly higher in OA compared to RA synovial fibroblasts.
A few studies have investigated the proteome of synovium from OA and RA patients. 2D-PAGE of synovium from 6 OA and 6 RA patients showed that 5 gel spots were significantly differentially expressed.157 Hierarchical cluster analysis showed RA patients to cluster separately from OA patients, and OA patients to be divided into two subclusters. Levels of calgranulin A were significantly higher in synovial tissues from RA patients compared to OA patients, and validated by real-time RT-PCR in a separate cohort of 10 OA and 9 RA patients. Using a panel of 791 mouse antibodies, the proteins expressed in synovium samples from one OA and one RA patient were compared, with 260 antibodies detecting their target proteins in the samples.158 Levels of 71 proteins differed significantly between the OA and RA patients. Validation with Western blots of 8 OA and 8 RA patients showed that protein levels of Stat1, p47phox, and MnSOD were significantly increased, and cathepsin D decreased, in RA synovial tissue compared to OA.
2.E. Interactions between Joint Tissues
The production and function of SF involves interactions between different joint tissues, cytokines, and the physical transport processes that connect all joint components. As new mechanistic details of SF, joint tissues, and cytokine effects are discovered, a comprehensive model of synovial fluid in health, injury, and disease will be needed to account for how the interactions between components give rise to joint function and behavior.
Interactions and regulatory influences between various synovial joint tissues play an important role in joint health and disease. The involvement of synoviocytes in the breakdown of cartilage in inflammatory arthritis has long been recognized,159 with the finding that synovial tissue culture medium contains a catabolic protein capable of inducing chondrocytes to degrade cartilage ECM.160 More recently, the effects of RA synoviocyte metabolic products on chondrocyte gene expression have been studied in vitro by culturing chondrocytes in alginate beads suspended in conditioned medium from RA synovial fibroblasts, normal synovial fibroblasts, and anti-rheumatic drug treated synovial fibroblasts.117, 118 Chondrocytes cultured in RA synovial fibroblast conditioned medium, compared to normal synovial fibroblast conditioned medium, differentially expressed 110 genes, upregulating genes associated with immunological and catabolic processes and downregulating genes associated with cell proliferation and differentiation.
Responses to cytokines are specific to tissues, cell types, and culture conditions, and may be influenced by interactions between multiple cytokines. TGF-β1 generally upregulates HA secretion by normal synoviocytes.6, 161 In chondrocytes and synoviocytes, TGF-β1 and IL-1β increased HA secretion, individually and synergistically.162 In rheumatoid fibroblastic synovial lining cells, IL-1β stimulates and TGF-β1 inhibits HA synthesis.163 Three isoforms of hyaluronan synthase (HAS), the enzyme responsible for HA synthesis, have been identified, and HAS1 mRNA predominates in synovial cells, whereas HAS2 mRNA predominates in chondrocytes.162 TGF-β1 has a differential effect on HAS in human fibroblast-like synoviocytes, upregulating HAS1 and downregulating HAS3 in a dose dependent manner.164 Cytokines in SF modulate PRG4 as well, as TGF-β1 upregulates and IL-1βdownregulates PRG4 gene expression and protein secretion in superficial zone cartilage, synovium, meniscus, and the anterior and posterior cruciate ligaments.165 The effects of cytokines on cartilage matrix and chondrocyte metabolism have also been studied, with IL-1α, IL-1β, and TNF-α inhibiting proteoglycan synthesis and increasing proteoglycan degradation, while IGF-1 and TGF-β1 had the opposite effects.166
2.F. From Data to Models
The systems biology approach culminates in the construction of a model, a simplified representation of the basis for a biological phenomenon. Data of multiple types from multiple sources are organized within a mathematical framework. The most useful models reproduce experimentally obtained data, provide quantitative predictions of the behavior of the system under new conditions, and serve as a platform for the generation of new hypotheses that may be experimentally validated, all of which may lead to further refinement of the model. Synovial joint phenomena that may benefit from a systems biology approach of study include processes such as the formation of synovial fluid and signaling networks, which involve the interaction of multiple component parts. Ultimately, models aim to provide insight into mechanisms underlying specific biological phenomena. Several models of the component parts of the synovial joint in isolation have been constructed, including models with a focus on the rheological properties of SF,167-171 lubrication mechanisms,172-174 and mass transport in cartilage175-182 and synovium.183-186 More recently, models of the whole joint with interacting components have been constructed to enable understanding of synovial joint function and behavior at the whole joint level.
2.F.i. Model Construction
Models aim to recapitulate some aspect of the properties of a biological system or process that is of interest to the modeler. In constructing models, the advantages of simplicity and complexity must be weighed with the primary modeling goal in mind. Simple, idealized models may provide clearer insight into the system and reveal the main features and relationships of the system. Complex, detailed models may provide increased predictive power of the behavior of the system, but often at the cost of understanding how the system functions.
Data and insights gained from the detailed study of isolated components are useful in the construction of mechanistic models of a process of interest. For example, studies from synovial tissues in isolation have provided information on the transport properties of synovium, which can be used to construct models of fluid and mass transport processes in synovium. Data from high-throughput screens can be used to refine and validate the model. Knowledge of key molecular constituents, processes, and pathways in the synovial joint that are altered in injury and disease may suggest relevant details and parameters to incorporate into models.
2.F.ii. Models of the Synovial Joint System
A few integrative models of the synovial joint have been developed recently and illustrate a systems biology approach to studying the synovial joint. As data from large-scale, high-throughput analyses and conventional analyses become integrated with current models, new models of the synovial joint are anticipated.
A quantitative, compartmental model of the synovial joint to predict steady-state SF lubricant concentrations in the human knee joint has been constructed.187 The model consists of four compartments: (i) SF surrounded by (ii) articular cartilage and (iii) semi-permeable synovium, which provides the outflow resistance and filtration of SF to the (iv) subsynovium. Lubricants are secreted into the SF compartment by chondrocytes in the articular cartilage compartment and synoviocytes in the synovium compartment. The governing equations of the model were formed by mass balances of the lubricant molecules HA, PRG4, and SAPL, and depend on rates of secretion, degradation, and flux across compartments. Values for the model parameters are found from a thorough search of the available literature. The system of linear, first order differential equations with three state variables is solved numerically with Matlab. Predictions of steady state concentrations of lubricant molecules under basal, TGF-β, and IL-1 conditions, and transient concentrations with simulated joint lavage and therapeutic HA injection were found to be consistent with experimental data.
A mathematical model of RA has been constructed with the EntelosPhysioLab® platform, with representations for inflammatory cells, endothelium, synovial fibroblasts, and chondrocytes within a synovial joint compartment.188 Cytokines and growth factors, including TNF-α, IL-1β, IL-6, IFN-γ,TGF-β, GM-CSF, VEGF, chemokines, MMP proteases and TIMP inhibitors, and cell surface molecules are represented in the model. Program simulations can predict synovial hyperplasia, cartilage degradation rate, bone erosion index, and soluble factor concentrations. The model can be used to determine the critical pathways that produce the predicted disease outcomes, suggest potential new targets for pharmaceutical research, and evaluate the expected clinical efficacy of candidate pharmacological targets.
Conclusion
The synovial joint is a complex biological system composed of several highly specialized cell and tissue types. Normal joint loading and articulation depends on the precise interplay between the various components of a synovial joint. Pathologic alterations in joint injury and disease involve changes in multiple components across a range of scales. Developments in large-scale, high-throughput technologies provide an extensive enumeration of the changes that occur in joint injury and disease. Diverse data types from multiple sources can be integrated in defining the components and parameters of a mathematical model of the synovial joint system. A systems biology approach will be an invaluable tool to the study of joint behavior and function in health, injury, and disease.
Table 3.
Proteins not previously associated with OA implicated by high-throughput proteomic methods to be involved in OA pathogenesis.
Contributor Information
Alexander Y. Hui, Email: alexhui@ucsd.edu, Department of Bioengineering, University of California-San Diego
William J. McCarty, Email: wmccarty@ucsd.edu, Department of Bioengineering, University of California-San Diego
Koichi Masuda, Email: komasuda@ucsd.edu, Department of Orthopaedic Surgery, University of California-San Diego.
Gary S. Firestein, Email: gfirestein@ucsd.edu, Division of Rheumatology, Allergy and Immunology, University of California-San Diego
Robert L. Sah, Email: rsah@ucsd.edu, Department of Bioengineering and Institute of Engineering in Medicine, University of California-San Diego
References
- 1.Ateshian GA, Mow VC. Friction, lubrication, and wear of articular cartilage and diarthrodial joints. In: Mow VC, Huiskes R, editors. Basic Orthopaedic Biomechanics and Mechano-Biology. 3. Philadelphia: Lippincott Williams & Wilkins; 2005. pp. 447–494. [Google Scholar]
- 2.Ropes MW, Rossmeisl EC, Bauer W. The origin and nature of normal human synovial fluid. J Clin Invest. 1940;19:795–799. doi: 10.1172/JCI101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacher BL, Block JA, Schmid TM, Aydelotte MB, Kuettner KE. A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch Biochem Biophys. 1994;311:144–152. doi: 10.1006/abbi.1994.1219. [DOI] [PubMed] [Google Scholar]
- 4.Jay GD, Britt DE, Cha CJ. Lubricin is a product of megakaryocyte stimulating factor gene expression by human synovial fibroblasts. J Rheumatol. 2000;27:594–600. [PubMed] [Google Scholar]
- 5.Meyer K, Smyth EM, Dawson MH. The isolation of a mucopolysaccharide from synovial fluid. J Biol Chem. 1939;128:319–327. [Google Scholar]
- 6.Hyc A, Osiecka-Iwan A, Niderla-Bielinska J, Jankowska-Steifer E, Moskalewski S. Pro- and anti-inflammatory cytokines increase hyaluronan production by rat synovial membrane in vitro. Int J Mol Med. 2009;24:579–585. doi: 10.3892/ijmm_00000268. [DOI] [PubMed] [Google Scholar]
- 7.Goldring MB. Osteoarthritis and cartilage: the role of cytokines. Curr Rheumatol Rep. 2000;2:459–465. doi: 10.1007/s11926-000-0021-y. [DOI] [PubMed] [Google Scholar]
- 8.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7:429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 9.Blewis ME, Lao BJ, Schumacher BL, Bugbee WD, Sah RL, Firestein GS. Interactive cytokine regulation of synoviocyte lubricant secretion. Tissue Eng Part A. 2010;16:1329–1337. doi: 10.1089/ten.tea.2009.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, et al. Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum. 1986;29:1039–1049. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 11.Neogi T, Aletaha D, Silman AJ, Naden RL, Felson DT, Aggarwal R, Bingham CO, 3rd, Birnbaum NS, Burmester GR, Bykerk VP, et al. The 2010 American College of Rheumatology/European League Against Rheumatism classification criteria for rheumatoid arthritis: Phase 2 methodological report. Arthritis Rheum. 2010;62:2582–2591. doi: 10.1002/art.27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe F, Cush JJ, O'Dell JR, Kavanaugh A, Kremer JM, Lane NE, Moreland LW, Paulus HE, Pincus T, Russell AS, et al. Consensus recommendations for the assessment and treatment of rheumatoid arthritis. J Rheumatol. 2001;28:1423–1430. [PubMed] [Google Scholar]
- 13.Lotz MK, Kraus VB. New developments in osteoarthritis Posttraumatic osteoarthritis: pathogenesis and pharmacological treatment options. Arthritis Res Ther. 2010;12:211. doi: 10.1186/ar3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furman BD, Olson SA, Guilak F. The development of posttraumatic arthritis after articular fracture. J Orthop Trauma. 2006;20:719–725. doi: 10.1097/01.bot.0000211160.05864.14. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DD, Chubinskaya S, Guilak F, Martin JA, Oegema TR, Olson SA, Buckwalter JA. Post-traumatic osteoarthritis: Improved understanding and opportunities for early intervention. J Orthop Res. 2011;29:802–809. doi: 10.1002/jor.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid K, Macnair MB. Characterization of the proteins of human synovial fluid in certain disease states. J Clin Invest. 1956;35:814–824. doi: 10.1172/JCI103334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmid K, Macnair MB. Characterization of the proteins of certain postmortem human synovial fluids. J Clin Invest. 1958;37:708–718. doi: 10.1172/JCI103657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holley HL, Patton FM, Pigman W, Platt D. An electrophoretic study of normal and post-mortem human and bovine synovial fluids. Arch Biochem Biophys. 1956;64:152–163. doi: 10.1016/0003-9861(56)90251-x. [DOI] [PubMed] [Google Scholar]
- 19.Levick JR. Permeability of rheumatoid and normal human synovium to specific plasma proteins. Arthritis Rheum. 1981;24:1550–1560. doi: 10.1002/art.1780241215. [DOI] [PubMed] [Google Scholar]
- 20.Schur PH, Sandson J. Immunologic studies of the proteins of human synovial fluid. Arthritis Rheum. 1963;6:115–129. doi: 10.1002/art.1780060204. [DOI] [PubMed] [Google Scholar]
- 21.Decker B, McKenzie BF, McGuckin WF, Slocumb CH. Comparative distribution of proteins and glycoproteins of serum and synovial fluid. Arthritis Rheum. 1959;2:162–177. doi: 10.1002/1529-0131(195904)2:2<162::aid-art1780020208>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Sitton NG, Dixon JS, Bird HA, Wright V. Serum and synovial fluid histidine: a comparison in rheumatoid arthritis and osteoarthritis. Rheumatol Int. 1986;6:251–254. doi: 10.1007/BF00541315. [DOI] [PubMed] [Google Scholar]
- 23.Shtacher G, Maayan R, Feinstein G. Proteinase inhibitors in human synovial fluid. Biochim Biophys Acta. 1973;303:138–147. doi: 10.1016/0005-2795(73)90155-4. [DOI] [PubMed] [Google Scholar]
- 24.Ogston AG, Stanier JE. The physiological function of hyaluronic acid in synovial fluid: viscous, elastic and lubricant properties. J Phys. 1953;119:244–252. doi: 10.1113/jphysiol.1953.sp004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swann DA, Sotman S, Dixon M, Brooks C. The isolation and partial characterization of the major glycoprotein (LGP-I) from the articular lubricating fraction from bovine synovial fluid. Biochem J. 1977;161:473–485. doi: 10.1042/bj1610473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swann DA, Slayter HS, Silver FH. The molecular structure of lubricating glycoprotein-I, the boundary lubricant for articular cartilage. J Biol Chem. 1981;256:5921–5925. [PubMed] [Google Scholar]
- 27.Decker B, McGuckin WF, McKenzie BF, Slocumb CH. Concentration of hyaluronic acid in synovial fluid. Clin Chem. 1959;5:465–469. [PubMed] [Google Scholar]
- 28.Hamerman D, Schuster H. Hyaluronate in normal human synovial fluid. J Clin Invest. 1958;37:57–64. doi: 10.1172/JCI103585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stafford CT, Niedermeier W, Holley HL, Pigman W. Studies on the Concentration and Intrinsic Viscosity of Hyaluronic Acid in Synovial Fluids of Patients with Rheumatic Diseases. Ann Rheum Dis. 1964;23:152–157. doi: 10.1136/ard.23.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid T, Lindley K, Su J, Soloveychik V, Block J, Kuettner K, Schumacher B. Superficial zone protein (SZP) is an abundant glycoprotein in human synovial fluid and serum. Trans Orthop Res Soc. 2001 [Google Scholar]
- 31.Elsaid KA, Fleming BC, Oksendahl HL, Machan JT, Fadale PD, Hulstyn MJ, Shalvoy R, Jay GD. Decreased lubricin concentrations and markers of joint inflammation in the synovial fluid of patients with anterior cruciate ligament injury. Arthritis Rheum. 2008;58:1707–1715. doi: 10.1002/art.23495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu CP, Reddi AH, Komvopoulos K, Schmid TM, Di Cesare PE. Increased friction coefficient and superficial zone protein expression in patients with advanced osteoarthritis. Arthritis Rheum. 2010;62:2680–2687. doi: 10.1002/art.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HG, Cowman MK. An agarose gel electrophoretic method for analysis of hyaluronan molecular weight distribution. Anal Biochem. 1994;219:278–287. doi: 10.1006/abio.1994.1267. [DOI] [PubMed] [Google Scholar]
- 34.McDonald JN, Levick JR. Effect of intra-articular hyaluronan on pressure-flow relation across synovium in anaesthetized rabbits. J Physiol. 1995;485(Pt 1):179–193. doi: 10.1113/jphysiol.1995.sp020722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jay GD, Harris DA, Cha CJ. Boundary lubrication by lubricin is mediated by O-linked beta(1-3)Gal-GalNAc oligosaccharides. Glycoconj J. 2001;18:807–815. doi: 10.1023/a:1021159619373. [DOI] [PubMed] [Google Scholar]
- 36.Jay GD, Tantravahi U, Britt DE, Barrach HJ, Cha CJ. Homology of lubricin and superficial zone protein (SZP): products of megakaryocyte stimulating factor (MSF) gene expression by human synovial fibroblasts and articular chondrocytes localized to chromosome 1q25. J Orthop Res. 2001;19:677–687. doi: 10.1016/S0736-0266(00)00040-1. [DOI] [PubMed] [Google Scholar]
- 37.Gomez JE, Thurston GB. Comparisons of the oscillatory shear viscoelasticity and composition of pathological synovial fluids. Biorheology. 1993;30:409–427. doi: 10.3233/bir-1993-305-612. [DOI] [PubMed] [Google Scholar]
- 38.Bollet AJ. The intrinsic viscosity of synovial fluid hyaluronic acid. J Lab Clin Med. 1956;48:721–728. [PubMed] [Google Scholar]
- 39.Dahl LB, Dahl IM, Engstrom-Laurent A, Granath K. Concentration and molecular weight of sodium hyaluronate in synovial fluid from patients with rheumatoid arthritis and other arthropathies. Ann Rheum Dis. 1985;44:817–822. doi: 10.1136/ard.44.12.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swann DA, Radin EL, Nazimiec M, Weisser PA, Curran N, Lewinnek G. Role of hyaluronic acid in joint lubrication. Ann Rheum Dis. 1974;33:318–326. doi: 10.1136/ard.33.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young AA, McLennan S, Smith MM, Smith SM, Cake MA, Read RA, Melrose J, Sonnabend DH, Flannery CR, Little CB. Proteoglycan 4 downregulation in a sheep meniscectomy model of early osteoarthritis. Arthritis Res Ther. 2006;8:R41. doi: 10.1186/ar1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elsaid KA, Jay GD, Warman ML, Rhee DK, Chichester CO. Association of articular cartilage degradation and loss of boundary-lubricating ability of synovial fluid following injury and inflammatory arthritis. Arthritis Rheum. 2005;52:1746–1755. doi: 10.1002/art.21038. [DOI] [PubMed] [Google Scholar]
- 43.Catterall JB, Stabler TV, Flannery CR, Kraus VB. Changes in serum and synovial fluid biomarkers after acute injury ( NCT00332254) Arthritis Res Ther. 2010;12:R229. doi: 10.1186/ar3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones AR, Chen S, Chai DH, Stevens AL, Gleghorn JP, Bonassar LJ, Grodzinsky AJ, Flannery CR. Modulation of lubricin biosynthesis and tissue surface properties following cartilage mechanical injury. Arthritis Rheum. 2009;60:133–142. doi: 10.1002/art.24143. [DOI] [PubMed] [Google Scholar]
- 45.Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, Sood R, Makalowska I, Baxevanis A, Johnstone B, et al. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat Genet. 1999;23:319–322. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- 46.Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, Jay GD, Stewart M, Wang H, Warman ML, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doss F, Menard J, Hauschild M, Kreutzer HJ, Mittlmeier T, Muller-Steinhardt M, Muller B. Elevated IL-6 levels in the synovial fluid of osteoarthritis patients stem from plasma cells. Scand J Rheumatol. 2007;36:136–139. doi: 10.1080/03009740701250785. [DOI] [PubMed] [Google Scholar]
- 48.Futani H, Okayama A, Matsui K, Kashiwamura S, Sasaki T, Hada T, Nakanishi K, Tateishi H, Maruo S, Okamura H. Relation between interleukin-18 and PGE2 in synovial fluid of osteoarthritis: a potential therapeutic target of cartilage degradation. J Immunother. 2002;25(1):S61–64. doi: 10.1097/00002371-200203001-00009. [DOI] [PubMed] [Google Scholar]
- 49.Malemud CJ. Anticytokine therapy for osteoarthritis: evidence to date. Drugs Aging. 2010;27:95–115. doi: 10.2165/11319950-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Elsaid KA, Machan JT, Waller K, Fleming BC, Jay GD. The impact of anterior cruciate ligament injury on lubricin metabolism and the effect of inhibiting tumor necrosis factor alpha on chondroprotection in an animal model. Arthritis Rheum. 2009;60:2997–3006. doi: 10.1002/art.24800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cameron M, Buchgraber A, Passler H, Vogt M, Thonar E, Fu F, Evans CH. The natural history of the anterior cruciate ligament-deficient knee. Changes in synovial fluid cytokine and keratan sulfate concentrations. Am J Sports Med. 1997;25:751–754. doi: 10.1177/036354659702500605. [DOI] [PubMed] [Google Scholar]
- 52.Marks PH, Donaldson ML. Inflammatory cytokine profiles associated with chondral damage in the anterior cruciate ligament-deficient knee. Arthroscopy. 2005;21:1342–1347. doi: 10.1016/j.arthro.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Cuellar VG, Cuellar JM, Golish SR, Yeomans DC, Scuderi GJ. Cytokine profiling in acute anterior cruciate ligament injury. Arthroscopy. 2010;26:1296–1301. doi: 10.1016/j.arthro.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Kahle P, Saal JG, Schaudt K, Zacher J, Fritz P, Pawelec G. Determination of cytokines in synovial fluids: correlation with diagnosis and histomorphological characteristics of synovial tissue. Ann Rheum Dis. 1992;51:731–734. doi: 10.1136/ard.51.6.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richette P, Francois M, Vicaut E, Fitting C, Bardin T, Corvol M, Savouret JF, Rannou F. A high interleukin 1 receptor antagonist/IL-1beta ratio occurs naturally in knee osteoarthritis. J Rheumatol. 2008;35:1650–1654. [PubMed] [Google Scholar]
- 56.Horiuchi T, Yoshida T, Koshihara Y, Sakamoto H, Kanai H, Yamamoto S, Ito H. The increase of parathyroid hormone-related peptide and cytokine levels in synovial fluid of elderly rheumatoid arthritis and osteoarthritis. Endocr J. 1999;46:643–649. doi: 10.1507/endocrj.46.643. [DOI] [PubMed] [Google Scholar]
- 57.Westacott CI, Whicher JT, Barnes IC, Thompson D, Swan AJ, Dieppe PA. Synovial fluid concentration of five different cytokines in rheumatic diseases. Ann Rheum Dis. 1990;49:676–681. doi: 10.1136/ard.49.9.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lettesjo H, Nordstrom E, Strom H, Nilsson B, Glinghammar B, Dahlstedt L, Moller E. Synovial fluid cytokines in patients with rheumatoid arthritis or other arthritic lesions. Scand J Immunol. 1998;48:286–292. doi: 10.1046/j.1365-3083.1998.00399.x. [DOI] [PubMed] [Google Scholar]
- 59.Nakanishi Y, Yamanaka H, Hakoda M, Nakazawa S, Saito S, Hara M, Kamatani N, Kashiwazaki S. Association between hypoxanthine concentration in synovial fluid and joint destruction in patients with rheumatoid arthritis. Modern Rheumatology. 1998;8:59–67. [Google Scholar]
- 60.Cameron ML, Fu FH, Paessler HH, Schneider M, Evans CH. Synovial fluid cytokine concentrations as possible prognostic indicators in the ACL-deficient knee. Knee Surg Sports Traumatol Arthrosc. 1994;2:38–44. doi: 10.1007/BF01552652. [DOI] [PubMed] [Google Scholar]
- 61.Irie K, Uchiyama E, Iwaso H. Intraarticular inflammatory cytokines in acute anterior cruciate ligament injured knee. Knee. 2003;10:93–96. doi: 10.1016/s0968-0160(02)00083-2. [DOI] [PubMed] [Google Scholar]
- 62.Pagura SM, Thomas SG, Woodhouse LJ, Ezzat S, Marks P. Circulating and synovial levels of IGF-I, cytokines, physical function and anthropometry differ in women awaiting total knee arthroplasty when compared to men. J Orthop Res. 2005;23:397–405. doi: 10.1016/j.orthres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 63.Guerne PA, Zuraw BL, Vaughan JH, Carson DA, Lotz M. Synovium as a source of interleukin 6 in vitro. Contribution to local and systemic manifestations of arthritis. J Clin Invest. 1989;83:585–592. doi: 10.1172/JCI113921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao XT, Feng L, Gu LJ, Wu LJ, Feng TT, Yang YM, Wu NP, Yao HP. Expression of interleukin-18, IL-18BP, and IL-18R in serum, synovial fluid, and synovial tissue in patients with rheumatoid arthritis. Clin Exp Med. 2009;9:215–221. doi: 10.1007/s10238-009-0036-2. [DOI] [PubMed] [Google Scholar]
- 65.Tchetverikov I, Lohmander LS, Verzijl N, Huizinga TW, TeKoppele JM, Hanemaaijer R, DeGroot J. MMP protein and activity levels in synovial fluid from patients with joint injury, inflammatory arthritis, and osteoarthritis. Ann Rheum Dis. 2005;64:694–698. doi: 10.1136/ard.2004.022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishiguro N, Ito T, Ito H, Iwata H, Jugessur H, Ionescu M, Poole AR. Relationship of matrix metalloproteinases and their inhibitors to cartilage proteoglycan and collagen turnover: analyses of synovial fluid from patients with osteoarthritis. Arthritis Rheum. 1999;42:129–136. doi: 10.1002/1529-0131(199901)42:1<129::AID-ANR16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 67.Dahlberg L, Friden T, Roos H, Lark MW, Lohmander LS. A longitudinal study of cartilage matrix metabolism in patients with cruciate ligament rupture-synovial fluid concentrations of aggrecan fragments, stromelysin-1 and tissue inhibitor of metalloproteinase-1. Br J Rheumatol. 1994;33:1107–1111. doi: 10.1093/rheumatology/33.12.1107. [DOI] [PubMed] [Google Scholar]
- 68.Lohmander LS, Roos H, Dahlberg L, Hoerrner LA, Lark MW. Temporal patterns of stromelysin-1, tissue inhibitor, and proteoglycan fragments in human knee joint fluid after injury to the cruciate ligament or meniscus. J Orthop Res. 1994;12:21–28. doi: 10.1002/jor.1100120104. [DOI] [PubMed] [Google Scholar]
- 69.Lohmander LS, Atley LM, Pietka TA, Eyre DR. The release of crosslinked peptides from type II collagen into human synovial fluid is increased soon after joint injury and in osteoarthritis. Arthritis Rheum. 2003;48:3130–3139. doi: 10.1002/art.11326. [DOI] [PubMed] [Google Scholar]
- 70.Levick JR, McDonald JN. Fluid movement across synovium in healthy joints: role of synovial fluid macromolecules. Ann Rheum Dis. 1995;54:417–423. doi: 10.1136/ard.54.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Binette JP, Schmid K. The Proteins of Synovial Fluid: A Study of the Alpha 1-Alpha 2 Globulin Ratio. Arthritis Rheum. 1965;8:14–28. doi: 10.1002/art.1780080103. [DOI] [PubMed] [Google Scholar]
- 72.Steiner G, Studnicka-Benke A, Witzmann G, Hofler E, Smolen J. Soluble receptors for tumor necrosis factor and interleukin-2 in serum and synovial fluid of patients with rheumatoid arthritis, reactive arthritis and osteoarthritis. J Rheumatol. 1995;22:406–412. [PubMed] [Google Scholar]
- 73.Belcher C, Yaqub R, Fawthrop F, Bayliss M, Doherty M. Synovial fluid chondroitin and keratan sulphate epitopes, glycosaminoglycans, and hyaluronan in arthritic and normal knees. Ann Rheum Dis. 1997;56:299–307. doi: 10.1136/ard.56.5.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsson S, Lohmander LS, Struglics A. Synovial fluid level of aggrecan ARGS fragments is a more sensitive marker of joint disease than glycosaminoglycan or aggrecan levels: a cross-sectional study. Arthritis Res Ther. 2009;11:R92. doi: 10.1186/ar2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lohmander LS, Saxne T, Heinegard DK. Release of cartilage oligomeric matrix protein (COMP) into joint fluid after knee injury and in osteoarthritis. Ann Rheum Dis. 1994;53:8–13. doi: 10.1136/ard.53.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poole AR. Cartilage in health and disease. In: Koopman WJ, editor. Arthritis and Allied Conditions. 15. Vol. 1. Philadelphia: Lippincott, Williams and Wilkins; 2005. pp. 223–269. [Google Scholar]
- 77.Dahlberg L, Friden T, Roos H, Lark MW, Lohmander LS. Longitudinal-Study of Cartilage Matrix Metabolism in Patients with Cruciate Ligament Rupture - Synovial-Fluid Concentrations of Aggrecan Fragments, Stromelysin-1 and Tissue Inhibitor of Metalloproteinase-1. British Journal of Rheumatology. 1994;33:1107–1111. doi: 10.1093/rheumatology/33.12.1107. [DOI] [PubMed] [Google Scholar]
- 78.Coggeshall HC, Warren CF, Bauer W. The cytology of normal human synovial fluid. Anat Rec. 1940;77:129–144. [Google Scholar]
- 79.McEwen C. Cytologic Studies on Rheumatic Fever: II. Cells of Rheumatic Exudates. J Clin Invest. 1935;14:190–201. doi: 10.1172/JCI100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fye KH. Arthrocentesis, Synovial Fluid Analysis, and Synovial Biopsy. In: Klippel JH, Stone JH, Crofford LJ, White PH, editors. Primer on the Rheumatic Diseases. 13. Atlanta,GA: Arthritis Foundation; 2008. pp. 21–27. [Google Scholar]
- 81.Kratz A, Pesce MA, Fink DJ. “Appendix: Laboratory Values of Clinical Importance” (Chapter) Fauci A, Braunwald E, Kasper D, Hauser S, Longo D, Jameson J, Loscalzo J, editors. Harrison's Principles of Internal Medicine. (17e) http://www.accessmedicine.com/content.aspx?aID=2904600.
- 82.Krey PR, Bailen DA. Synovial fluid leukocytosis. A study of extremes. Am J Med. 1979;67:436–442. doi: 10.1016/0002-9343(79)90790-3. [DOI] [PubMed] [Google Scholar]
- 83.Firestein GS. Etiology and pathogenesis of rheumatoid arthritis. In: Firestein GS, editor. Kelley's Textbook of Rheumatology. 8th. Philadelphia, PA: Saunders/Elsevier; 2009. pp. 1035–1086. [Google Scholar]
- 84.Schumacher BL, Hughes CE, Kuettner KE, Caterson B, Aydelotte MB. Immunodetection and partial cDNA sequence of the proteoglycan, superficial zone protein, synthesized by cells lining synovial joints. J Orthop Res. 1999;17:110–120. doi: 10.1002/jor.1100170117. [DOI] [PubMed] [Google Scholar]
- 85.Schumacher BL, Schmidt TA, Voegtline MS, Chen AC, Sah RL. Proteoglycan 4 (PRG4) synthesis and immunolocalization in bovine meniscus. J Orthop Res. 2005;23:562–568. doi: 10.1016/j.orthres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 86.Sabaratnam S, Arunan V, Coleman PJ, Mason RM, Levick JR. Size selectivity of hyaluronan molecular sieving by extracellular matrix in rabbit synovial joints. J Physiol. 2005;567:569–581. doi: 10.1113/jphysiol.2005.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simkin PA. Synovial permeability in rheumatoid arthritis. Arthritis Rheum. 1979;22:689–696. doi: 10.1002/art.1780220701. [DOI] [PubMed] [Google Scholar]
- 88.Levick JR, McDonald JN. Ultrastructure of transport pathways in stressed synovium of the knee in anaesthetized rabbits. J Physiol. 1989;419:493–508. doi: 10.1113/jphysiol.1989.sp017882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Granger HJ, Taylor AE. Permeability of connective tissue linings isolated from implanted capsules; implications for interstitial pressure measurements. Circ Res. 1975;36:222–228. doi: 10.1161/01.res.36.1.222. [DOI] [PubMed] [Google Scholar]
- 90.Sabaratnam S, Coleman PJ, Mason RM, Levick JR. Interstitial matrix proteins determine hyaluronan reflection and fluid retention in rabbit joints: effect of protease. J Physiol. 2007;578:291–299. doi: 10.1113/jphysiol.2006.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kushner I, Somerville JA. Permeability of human synovial membrane to plasma proteins. Relationship to molecular size and inflammation. Arthritis Rheum. 1971;14:560–570. doi: 10.1002/art.1780140503. [DOI] [PubMed] [Google Scholar]
- 92.Pejovic M, Stankovic A, Mitrovic DR. Determination of the apparent synovial permeability in the knee joint of patients suffering from osteoarthritis and rheumatoid arthritis. Br J Rheumatol. 1995;34:520–524. doi: 10.1093/rheumatology/34.6.520. [DOI] [PubMed] [Google Scholar]
- 93.Safari M, Bjelle A, Gudmundsson M, Hogfors C, Granhed H. Clinical assessment of rheumatic diseases using viscoelastic parameters for synovial fluid. Biorheology. 1990;27:659–674. doi: 10.3233/bir-1990-27504. [DOI] [PubMed] [Google Scholar]
- 94.Schurz J, Ribitsch V. Rheology of synovial fluid. Biorheology. 1987;24:385–399. doi: 10.3233/bir-1987-24404. [DOI] [PubMed] [Google Scholar]
- 95.Fam H, Bryant JT, Kontopoulou M. Rheological properties of synovial fluids. Biorheology. 2007;44:59–74. [PubMed] [Google Scholar]
- 96.Jay GD, Torres JR, Warman ML, Laderer MC, Breuer KS. The role of lubricin in the mechanical behavior of synovial fluid. Proc Natl Acad Sci U S A. 2007;104:6194–6199. doi: 10.1073/pnas.0608558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmidt TA, Gastelum NS, Nguyen QT, Schumacher BL, Sah RL. Boundary lubrication of articular cartilage: role of synovial fluid constituents. Arthritis Rheum. 2007;56:882–891. doi: 10.1002/art.22446. [DOI] [PubMed] [Google Scholar]
- 98.Schmidt TA, Sah RL. Effect of synovial fluid on boundary lubrication of articular cartilage. Osteoarthritis Cartilage. 2007;15:35–47. doi: 10.1016/j.joca.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 99.Gleghorn JP, Jones AR, Flannery CR, Bonassar LJ. Boundary mode lubrication of articular cartilage by recombinant human lubricin. J Orthop Res. 2009;27:771–777. doi: 10.1002/jor.20798. [DOI] [PubMed] [Google Scholar]
- 100.Teeple E, Elsaid KA, Fleming BC, Jay GD, Aslani K, Crisco JJ, Mechrefe AP. Coefficients of friction, lubricin, and cartilage damage in the anterior cruciate ligament-deficient guinea pig knee. J Orthop Res. 2007 doi: 10.1002/jor.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jay GD, Elsaid KA, Zack J, Robinson K, Trespalacios F, Cha CJ, Chichester CO. Lubricating ability of aspirated synovial fluid from emergency department patients with knee joint synovitis. J Rheumatol. 2004;31:557–564. [PubMed] [Google Scholar]
- 102.Momberger TS, Levick JR, Mason RM. Hyaluronan secretion by synoviocytes is mechanosensitive. Matrix Biol. 2005;24:510–519. doi: 10.1016/j.matbio.2005.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ingram KR, Wann AK, Angel CK, Coleman PJ, Levick JR. Cyclic movement stimulates hyaluronan secretion into the synovial cavity of rabbit joints. J Physiol. 2008;586:1715–1729. doi: 10.1113/jphysiol.2007.146753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grad S, Lee CR, Gorna K, Gogolewski S, Wimmer MA, Alini M. Surface motion upregulates superficial zone protein and hyaluronan production in chondrocyte-seeded three-dimensional scaffolds. Tissue Eng. 2005;11:249–256. doi: 10.1089/ten.2005.11.249. [DOI] [PubMed] [Google Scholar]
- 105.Nugent GE, Aneloski NM, Schmidt TA, Schumacher BL, Voegtline MS, Sah RL. Dynamic shear stimulation of bovine cartilage biosynthesis of proteoglycan 4 (PRG4) Arthritis Rheum. 2006;54:1888–1896. doi: 10.1002/art.21831. [DOI] [PubMed] [Google Scholar]
- 106.Grad S, Gogolewski S, Alini M, Wimmer MA. Effects of simple and complex motion patterns on gene expression of chondrocytes seeded in 3D scaffolds. Tissue Eng. 2006;12:3171–3179. doi: 10.1089/ten.2006.12.3171. [DOI] [PubMed] [Google Scholar]
- 107.Wimmer MA, Alini M, Grad S. The effect of sliding velocity on chondrocytes activity in 3D scaffolds. J Biomech. 2009;42:424–429. doi: 10.1016/j.jbiomech.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 108.Neu CP, Khalafi A, Komvopoulos K, Schmid TM, Reddi AH. Mechanotransduction of bovine articular cartilage superficial zone protein by transforming growth factor beta signaling. Arthritis Rheum. 2007;56:3706–3714. doi: 10.1002/art.23024. [DOI] [PubMed] [Google Scholar]
- 109.Nugent-Derfus GE, Takara T, O'Neill JK, Cahill SB, Gortz S, Pong T, Inoue H, Aneloski NM, Wang WW, Vega KI, et al. Continuous passive motion applied to whole joints stimulates chondrocyte biosynthesis of PRG4. Osteoarthritis Cartilage. 2007;15:566–574. doi: 10.1016/j.joca.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCarty WJ, Nguyen QT, Hui AY, Chen AC, Sah RL. Cartilage Tissue Engineering. In: Ducheyne P, Healy KE, Hutmacher DE, Grainger DE, Kirkpatrick CJ, editors. Comprehensive Biomaterials. 1. Vol. 5. 2011. pp. 199–212. [Google Scholar]
- 111.Sellam J, Berenbaum F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol. 2010;6:625–635. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- 112.Badot V, Galant C, Nzeusseu Toukap A, Theate I, Maudoux AL, Van den Eynde BJ, Durez P, Houssiau FA, Lauwerys BR. Gene expression profiling in the synovium identifies a predictive signature of absence of response to adalimumab therapy in rheumatoid arthritis. Arthritis Res Ther. 2009;11:R57. doi: 10.1186/ar2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dehne T, Karlsson C, Ringe J, Sittinger M, Lindahl A. Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix-associated autologous chondrocyte transplantation. Arthritis Res Ther. 2009;11:R133. doi: 10.1186/ar2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pohlers D, Beyer A, Koczan D, Wilhelm T, Thiesen HJ, Kinne RW. Constitutive upregulation of the transforming growth factor-beta pathway in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2007;9:R59. doi: 10.1186/ar2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rockel JS, Bernier SM, Leask A. Egr-1 inhibits the expression of extracellular matrix genes in chondrocytes by TNFalpha-induced MEK/ERK signalling. Arthritis Res Ther. 2009;11:R8. doi: 10.1186/ar2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sandell LJ, Xing X, Franz C, Davies S, Chang LW, Patra D. Exuberant expression of chemokine genes by adult human articular chondrocytes in response to IL-1beta. Osteoarthritis Cartilage. 2008;16:1560–1571. doi: 10.1016/j.joca.2008.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andreas K, Haupl T, Lubke C, Ringe J, Morawietz L, Wachtel A, Sittinger M, Kaps C. Antirheumatic drug response signatures in human chondrocytes: potential molecular targets to stimulate cartilage regeneration. Arthritis Res Ther. 2009;11:R15. doi: 10.1186/ar2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andreas K, Lubke C, Haupl T, Dehne T, Morawietz L, Ringe J, Kaps C, Sittinger M. Key regulatory molecules of cartilage destruction in rheumatoid arthritis: an in vitro study. Arthritis Res Ther. 2008;10:R9. doi: 10.1186/ar2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lequerre T, Bansard C, Vittecoq O, Derambure C, Hiron M, Daveau M, Tron F, Ayral X, Biga N, Auquit-Auckbur I, et al. Early and long-standing rheumatoid arthritis: distinct molecular signatures identified by gene-expression profiling in synovia. Arthritis Res Ther. 2009;11:R99. doi: 10.1186/ar2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huber R, Hummert C, Gausmann U, Pohlers D, Koczan D, Guthke R, Kinne RW. Identification of intra-group, inter-individual, and gene-specific variances in mRNA expression profiles in the rheumatoid arthritis synovial membrane. Arthritis Res Ther. 2008;10:R98. doi: 10.1186/ar2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Arora PS, Yamagiwa H, Srivastava A, Bolander ME, Sarkar G. Comparative evaluation of two two-dimensional gel electrophoresis image analysis software applications using synovial fluids from patients with joint disease. J Orthop Sci. 2005;10:160–166. doi: 10.1007/s00776-004-0878-0. [DOI] [PubMed] [Google Scholar]
- 122.Firestein GS, Pisetsky DS. DNA microarrays: Boundless technology or bound by technology? Guidelines for studies using microarray technology. Arthritis Rheum. 2002;46:859–861. doi: 10.1002/art.10236. [DOI] [PubMed] [Google Scholar]
- 123.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van der Pouw Kraan TC, van Gaalen FA, Huizinga TW, Pieterman E, Breedveld FC, Verweij CL. Discovery of distinctive gene expression profiles in rheumatoid synovium using cDNA microarray technology: evidence for the existence of multiple pathways of tissue destruction and repair. Genes Immun. 2003;4:187–196. doi: 10.1038/sj.gene.6363975. [DOI] [PubMed] [Google Scholar]
- 125.van der Pouw Kraan TC, van Gaalen FA, Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, Fero M, Tak PP, Huizinga TW, Pieterman E, et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003;48:2132–2145. doi: 10.1002/art.11096. [DOI] [PubMed] [Google Scholar]
- 126.Kasperkovitz PV, Timmer TC, Smeets TJ, Verbeet NL, Tak PP, van Baarsen LG, Baltus B, Huizinga TW, Pieterman E, Fero M, et al. Fibroblast-like synoviocytes derived from patients with rheumatoid arthritis show the imprint of synovial tissue heterogeneity: evidence of a link between an increased myofibroblast-like phenotype and high-inflammation synovitis. Arthritis Rheum. 2005;52:430–441. doi: 10.1002/art.20811. [DOI] [PubMed] [Google Scholar]
- 127.Fitzgerald JB, Jin M, Dean D, Wood DJ, Zheng MH, Grodzinsky AJ. Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem. 2004;279:19502–19511. doi: 10.1074/jbc.M400437200. [DOI] [PubMed] [Google Scholar]
- 128.Fitzgerald JB, Jin M, Grodzinsky AJ. Shear and compression differentially regulate clusters of functionally related temporal transcription patterns in cartilage tissue. J Biol Chem. 2006;281:24095–24103. doi: 10.1074/jbc.M510858200. [DOI] [PubMed] [Google Scholar]
- 129.Lee JH, Fitzgerald JB, Dimicco MA, Grodzinsky AJ. Mechanical injury of cartilage explants causes specific time-dependent changes in chondrocyte gene expression. Arthritis Rheum. 2005;52:2386–2395. doi: 10.1002/art.21215. [DOI] [PubMed] [Google Scholar]
- 130.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Karlsson C, Dehne T, Lindahl A, Brittberg M, Pruss A, Sittinger M, Ringe J. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18:581–592. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 132.Appleton CT, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum. 2007;56:1854–1868. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 133.Gebauer M, Saas J, Haag J, Dietz U, Takigawa M, Bartnik E, Aigner T. Repression of anti-proliferative factor Tob1 in osteoarthritic cartilage. Arthritis Res Ther. 2005;7:R274–284. doi: 10.1186/ar1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hermansson M, Sawaji Y, Bolton M, Alexander S, Wallace A, Begum S, Wait R, Saklatvala J. Proteomic analysis of articular cartilage shows increased type II collagen synthesis in osteoarthritis and expression of inhibin betaA (activin A), a regulatory molecule for chondrocytes. J Biol Chem. 2004;279:43514–43521. doi: 10.1074/jbc.M407041200. [DOI] [PubMed] [Google Scholar]
- 135.Wu J, Liu W, Bemis A, Wang E, Qiu Y, Morris EA, Flannery CR, Yang Z. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheum. 2007;56:3675–3684. doi: 10.1002/art.22876. [DOI] [PubMed] [Google Scholar]
- 136.Kabuyama Y, Kitamura T, Yamaki J, Homma MK, Kikuchi S, Homma Y. Involvement of thioredoxin reductase 1 in the regulation of redox balance and viability of rheumatoid synovial cells. Biochem Biophys Res Commun. 2008;367:491–496. doi: 10.1016/j.bbrc.2007.12.178. [DOI] [PubMed] [Google Scholar]
- 137.Gallagher J, Howlin J, McCarthy C, Murphy EP, Bresnihan B, FitzGerald O, Godson C, Brady HR, Martin F. Identification of Naf1/ABIN-1 among TNF-alpha-induced expressed genes in human synoviocytes using oligonucleotide microarrays. FEBS Lett. 2003;551:8–12. doi: 10.1016/s0014-5793(03)00823-8. [DOI] [PubMed] [Google Scholar]
- 138.Honda T, Segi-Nishida E, Miyachi Y, Narumiya S. Prostacyclin-IP signaling and prostaglandin E2-EP2/EP4 signaling both mediate joint inflammation in mouse collagen-induced arthritis. J Exp Med. 2006;203:325–335. doi: 10.1084/jem.20051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haag J, Gebhard PM, Aigner T. SOX gene expression in human osteoarthritic cartilage. Pathobiology. 2008;75:195–199. doi: 10.1159/000124980. [DOI] [PubMed] [Google Scholar]
- 140.Aigner T, Zien A, Hanisch D, Zimmer R. Gene expression in chondrocytes assessed with use of microarrays. J Bone Joint Surg Am. 2003;85-A(2):117–123. doi: 10.2106/00004623-200300002-00016. [DOI] [PubMed] [Google Scholar]
- 141.Sato T, Konomi K, Yamasaki S, Aratani S, Tsuchimochi K, Yokouchi M, Masuko-Hongo K, Yagishita N, Nakamura H, Komiya S, et al. Comparative analysis of gene expression profiles in intact and damaged regions of human osteoarthritic cartilage. Arthritis Rheum. 2006;54:808–817. doi: 10.1002/art.21638. [DOI] [PubMed] [Google Scholar]
- 142.Dell'accio F, De Bari C, Eltawil NM, Vanhummelen P, Pitzalis C. Identification of the molecular response of articular cartilage to injury, by microarray screening: Wnt-16 expression and signaling after injury and in osteoarthritis. Arthritis Rheum. 2008;58:1410–1421. doi: 10.1002/art.23444. [DOI] [PubMed] [Google Scholar]
- 143.Kamphorst JJ, van der Heijden R, DeGroot J, Lafeber FP, Reijmers TH, van El B, Tjaden UR, van der Greef J, Hankemeier T. Profiling of endogenous peptides in human synovial fluid by NanoLC-MS: method validation and peptide identification. J Proteome Res. 2007;6:4388–4396. doi: 10.1021/pr0704534. [DOI] [PubMed] [Google Scholar]
- 144.Sinz A, Bantscheff M, Mikkat S, Ringel B, Drynda S, Kekow J, Thiesen HJ, Glocker MO. Mass spectrometric proteome analyses of synovial fluids and plasmas from patients suffering from rheumatoid arthritis and comparison to reactive arthritis or osteoarthritis. Electrophoresis. 2002;23:3445–3456. doi: 10.1002/1522-2683(200210)23:19<3445::AID-ELPS3445>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 145.Gobezie R, Kho A, Krastins B, Sarracino DA, Thornhill TS, Chase M, Millett PJ, Lee DM. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liao H, Wu J, Kuhn E, Chin W, Chang B, Jones MD, O'Neil S, Clauser KR, Karl J, Hasler F, et al. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 147.Drynda S, Ringel B, Kekow M, Kuhne C, Drynda A, Glocker MO, Thiesen HJ, Kekow J. Proteome analysis reveals disease-associated marker proteins to differentiate RA patients from other inflammatory joint diseases with the potential to monitor anti-TNFalpha therapy. Pathol Res Pract. 2004;200:165–171. doi: 10.1016/j.prp.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 148.Ruiz-Romero C, Lopez-Armada MJ, Blanco FJ. Proteomic characterization of human normal articular chondrocytes: a novel tool for the study of osteoarthritis and other rheumatic diseases. Proteomics. 2005;5:3048–3059. doi: 10.1002/pmic.200402106. [DOI] [PubMed] [Google Scholar]
- 149.Cillero-Pastor B, Ruiz-Romero C, Carames B, Lopez-Armada MJ, Blanco FJ. Proteomic analysis by two-dimensional electrophoresis to identify the normal human chondrocyte proteome stimulated by tumor necrosis factor alpha and interleukin-1beta. Arthritis Rheum. 2010;62:802–814. doi: 10.1002/art.27265. [DOI] [PubMed] [Google Scholar]
- 150.De Ceuninck F, Marcheteau E, Berger S, Caliez A, Dumont V, Raes M, Anract P, Leclerc G, Boutin JA, Ferry G. Assessment of some tools for the characterization of the human osteoarthritic cartilage proteome. J Biomol Tech. 2005;16:256–265. [PMC free article] [PubMed] [Google Scholar]
- 151.Stevens AL, Wishnok JS, Chai DH, Grodzinsky AJ, Tannenbaum SR. A sodium dodecyl sulfate-polyacrylamide gel electrophoresis-liquid chromatography tandem mass spectrometry analysis of bovine cartilage tissue response to mechanical compression injury and the inflammatory cytokines tumor necrosis factor alpha and interleukin-1beta. Arthritis Rheum. 2008;58:489–500. doi: 10.1002/art.23120. [DOI] [PubMed] [Google Scholar]
- 152.Rosenthal AK, Gohr CM, Ninomiya J, Wakim BT. Proteomic analysis of articular cartilage vesicles from normal and osteoarthritic cartilage. Arthritis Rheum. 2011;63:401–411. doi: 10.1002/art.30120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ruiz-Romero C, Lopez-Armada MJ, Blanco FJ. Mitochondrial proteomic characterization of human normal articular chondrocytes. Osteoarthritis Cartilage. 2006;14:507–518. doi: 10.1016/j.joca.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 154.Vincourt JB, Lionneton F, Kratassiouk G, Guillemin F, Netter P, Mainard D, Magdalou J. Establishment of a reliable method for direct proteome characterization of human articular cartilage. Mol Cell Proteomics. 2006;5:1984–1995. doi: 10.1074/mcp.T600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 155.Dasuri K, Antonovici M, Chen K, Wong K, Standing K, Ens W, El-Gabalawy H, Wilkins JA. The synovial proteome: analysis of fibroblast-like synoviocytes. Arthritis Res Ther. 2004;6:R161–168. doi: 10.1186/ar1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bo GP, Zhou LN, He WF, Luo GX, Jia XF, Gan CJ, Chen GX, Fang YF, Larsen PM, Wu J. Analyses of differential proteome of human synovial fibroblasts obtained from arthritis. Clin Rheumatol. 2009;28:191–199. doi: 10.1007/s10067-008-1013-y. [DOI] [PubMed] [Google Scholar]
- 157.Tilleman K, Van Beneden K, Dhondt A, Hoffman I, De Keyser F, Veys E, Elewaut D, Deforce D. Chronically inflamed synovium from spondyloarthropathy and rheumatoid arthritis investigated by protein expression profiling followed by tandem mass spectrometry. Proteomics. 2005;5:2247–2257. doi: 10.1002/pmic.200401109. [DOI] [PubMed] [Google Scholar]
- 158.Lorenz P, Ruschpler P, Koczan D, Stiehl P, Thiesen HJ. From transcriptome to proteome: differentially expressed proteins identified in synovial tissue of patients suffering from rheumatoid arthritis and osteoarthritis by an initial screen with a panel of 791 antibodies. Proteomics. 2003;3:991–1002. doi: 10.1002/pmic.200300412. [DOI] [PubMed] [Google Scholar]
- 159.Fell HB. Synoviocytes. J Clin Pathol Suppl (R Coll Pathol) 1978;12:14–24. [PMC free article] [PubMed] [Google Scholar]
- 160.Dingle JT, Saklatvala J, Hembry R, Tyler J, Fell HB, Jubb R. A cartilage catabolic factor from synovium. Biochem J. 1979;184:177–180. doi: 10.1042/bj1840177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Haubeck HD, Kock R, Fischer DC, Van de Leur E, Hoffmeister K, Greiling H. Transforming growth factor beta 1, a major stimulator of hyaluronan synthesis in human synovial lining cells. Arthritis Rheum. 1995;38:669–677. doi: 10.1002/art.1780380515. [DOI] [PubMed] [Google Scholar]
- 162.Recklies AD, White C, Melching L, Roughley PJ. Differential regulation and expression of hyaluronan synthases in human articular chondrocytes, synovial cells and osteosarcoma cells. Biochem J. 2001;354:17–24. doi: 10.1042/0264-6021:3540017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kawakami M, Suzuki K, Matsuki Y, Ishizuka T, Hidaka T, Konishi T, Matsumoto M, Kataharada K, Nakamura H. Hyaluronan production in human rheumatoid fibroblastic synovial lining cells is increased by interleukin 1 beta but inhibited by transforming growth factor beta 1. Ann Rheum Dis. 1998;57:602–605. doi: 10.1136/ard.57.10.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stuhlmeier KM, Pollaschek C. Differential effect of transforming growth factor beta (TGF-beta) on the genes encoding hyaluronan synthases and utilization of the p38 MAPK pathway in TGF-beta-induced hyaluronan synthase 1 activation. J Biol Chem. 2004;279:8753–8760. doi: 10.1074/jbc.M303945200. [DOI] [PubMed] [Google Scholar]
- 165.Lee SY, Niikura T, Reddi AH. Superficial zone protein (lubricin) in the different tissue compartments of the knee joint: modulation by transforming growth factor beta 1 and interleukin-1 beta. Tissue Eng Part A. 2008 doi: 10.1089/ten.tea.2007.0367. [DOI] [PubMed] [Google Scholar]
- 166.Hardingham TE, Bayliss MT, Rayan V, Noble DP. Effects of growth factors and cytokines on proteoglycan turnover in articular cartilage. Br J Rheumatol. 1992;31S1:1–6. [PubMed] [Google Scholar]
- 167.Chiroiu V, Mosnegutu V, Munteanu L, Ioan R. On a micromorphic model for the synovial fluid in the human knee. Mech Res Commun. 2010;37:246–255. [Google Scholar]
- 168.Nigam KM, Manohar K, Chaudhry KK. Viscoelastic fluid film lubrication with reference to human joints. Wear. 1981;70:63–75. [Google Scholar]
- 169.Ogston AG, Stanier JE, Toms BA, Strawbridge DJ. Elastic properties of ox synovial fluid. Nature. 1950;165:571. [Google Scholar]
- 170.Rainer F, Ribitsch V, Ulreich A. Viscosity of synovial fluid and possible artificial lubricants (author's transl) Acta Med Austriaca. 1980;7:92–95. [PubMed] [Google Scholar]
- 171.Tandon PN, Bong NH, Kushwaha K. A new model for synovial joint lubrication. Int J Biomed Comput. 1994;35:125–140. doi: 10.1016/0020-7101(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 172.Charnley J. The lubrication of animal joints in relation to surgical reconstruction by arthroplasty. Ann Rheum Dis. 1960;19:10–19. doi: 10.1136/ard.19.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.MacConaill MA. The function of intna-anticular fibrocartilages, with special reference to the knee and inferior radio-ulnar joints. J Anat. 1932;66:210–227. [PMC free article] [PubMed] [Google Scholar]
- 174.Walker PS, Dowson D, Longfield MD, Wright V. “Boosted lubrication” in synovial joints by fluid entrapment and enrichment. Ann Rheum Dis. 1968;27:512–520. doi: 10.1136/ard.27.6.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.DiMicco MA, Sah RL. Dependence of cartilage matrix composition on biosynthesis, diffusion, and reaction. Transport Porous Media. 2003;50:57–73. [Google Scholar]
- 176.Hascall VC, Luyten FP, Plaas AHK, Sandy JD. Steady-state metabolism of proteoglycans in bovine articular cartilage. In: Maroudas A, Kuettner K, editors. Methods in Cartilage Research. San Diego: Academic Press; 1990. pp. 108–112. [Google Scholar]
- 177.Klein TJ, Sah RL. Modulation of depth-dependent properties in tissue-engineered cartilage with a semi-permeable membrane and perfusion: a continuum model of matrix metabolism and transport. Biomech Model Mechanobiol. 2007;6:21–32. doi: 10.1007/s10237-006-0045-y. [DOI] [PubMed] [Google Scholar]
- 178.Maroudas A. Physicochemical properties of cartilage in the light of ion exchange theory. Biophys J. 1968;8:575–595. doi: 10.1016/S0006-3495(68)86509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Maroudas A. Distribution and diffusion of solutes in articular cartilage. Biophys J. 1970;10:365–379. doi: 10.1016/S0006-3495(70)86307-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Torzilli PA, Mow VC. On the fundamental fluid transport mechanisms through normal and pathological articular cartilage during function--I. The formulation J Biomech. 1976;9:541–552. doi: 10.1016/0021-9290(76)90071-3. [DOI] [PubMed] [Google Scholar]
- 181.Torzilli PA, Mow VC. On the fundamental fluid transport mechanisms through normal and pathological articular cartilage during function - II. The analysis, solution, and conclusions. J Biomech. 1976;9:587–606. doi: 10.1016/0021-9290(76)90100-7. [DOI] [PubMed] [Google Scholar]
- 182.Zhang L, Gardiner BS, Smith DW, Pivonka P, Grodzinsky A. The effect of cyclic deformation and solute binding on solute transport in cartilage. Arch Biochem Biophys. 2007;457:47–56. doi: 10.1016/j.abb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 183.Levick JR. An analysis of the effect of synovial capillary distribution upon trans-synovial concentration profiles and exchange. Q J Exp Physiol. 1984;69:289–300. doi: 10.1113/expphysiol.1984.sp002806. [DOI] [PubMed] [Google Scholar]
- 184.Levick JR. Blood flow and mass transport in synovial joints. In: Renkin M, Michel C, editors. Handbook of Physiology, Section 2, The Cardiovascular System, Volume IV, The Microcirculation. The American Physiological Society; 1984. pp. 917–947. [Google Scholar]
- 185.Levick JR. An analysis of the interaction between interstitial plasma protein, interstitial flow, and fenestral filtration and its application to synovium. Microvasc Res. 1994;47:90–125. doi: 10.1006/mvre.1994.1007. [DOI] [PubMed] [Google Scholar]
- 186.Levick JR. A method for estimating macromolecular reflection by human synovium, using measurements of intra-articular half-lives. Ann Rheum Dis. 1998;57:339–344. doi: 10.1136/ard.57.6.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Blewis ME, Nugent-Derfus GE, Schmidt TA, Schumacher BL, Sah RL. A model of synovial fluid lubricant composition in normal and injured joints. Eur Cell Mater. 2007;13:26–39. doi: 10.22203/ecm.v013a03. [DOI] [PubMed] [Google Scholar]
- 188.Rullmann JA, Struemper H, Defranoux NA, Ramanujan S, Meeuwisse CM, van Elsas A. Systems biology for battling rheumatoid arthritis: application of the Entelos PhysioLab platform. Syst Biol (Stevenage) 2005;152:256–262. doi: 10.1049/ip-syb:20050053. [DOI] [PubMed] [Google Scholar]