Abstract
Zymolyase (lyticase) is used for cell wall digestion in yeast experiments and is needed for incubation processes under moderate experimental conditions. This has been thought to cause unfavorable effects, and many researchers are aware that the enzyme method is unsuitable for RNA preparation following several reports of stress responses to the enzyme process. However, RNA preparation with enzyme digestion continues to be used. This may be because there have been insufficient data directly comparing RNA preparation conditions with previous studies. We investigated the influence of enzyme processes in RNA preparation using a DNA microarray, and compared superoxide dismutase (SOD) activities with a non-treated control and the results of previous research. Gene expressions were commonly changed by enzyme processes, and SOD activities increased only during short-term incubation. Meanwhile, both SOD gene expressions and SOD activity during RNA preparation indicated different results than gained under conditions of long-term incubation. These results suggest that zymolyase treatment surely influences gene expressions and enzyme activity, although the effect assumed by previous studies is not necessarily in agreement with that of RNA preparation.
Keywords: Zymolyase, Yeast, RNA preparation, DNA microarray, SOD activity
Introduction
Zymolyase (lyticase) is a very important enzyme in microbiology, isolated from Arthrobacter luteus. Zymolyase actively degrades yeast cell walls [1]. The principal ingredient in zymolyase is β-1,3-glucan laminaripentaohydrolase, which hydrolyzes glucose polymers linked by β-1,3-bonds, producing laminaripentaose [1–3]. Zymolyase also includes protease and mannase. These enzymes strongly degrade yeast cell walls. By changing pH or temperature, cell wall lysis and dehydration of glucan are easily achievable. For this reason, zymolyase is frequently used for the preparation of nucleic acids or soluble proteins from yeast cells, and many commercial kits involving such enzyme treatment have been released. However, digestion must be performed by incubating at moderate temperature for a given length of time, thereby having potential to change the yeast reaction. Forsburg [4] introduced the RNA preparation method, involving 6 U/ml of enzyme at 37 °C for 30 min, and Klassen [5] introduced 8 U/ml of enzyme over 1 h. These conditions may be considered moderate, and many commercial kits use higher concentrations (10–500 U/ml) and short-term incubation (15–30 min) at 37 °C. This is a strategy to avoid cell response resulting from enzymatic treatment.
Recently, there have been a number of studies on the influence of zymolyase. De Groot [6], for example, measured cell density under 0.1 U/ml concentration at 37 °C for 1–4 h, and also reported the induction of cell wall genes and mitogen-activated protein kinase (MAPK)-related genes under 0.26 U/ml of zymolyase at 28 °C for 1–4 h, or 0.8 U/ml at 24 °C for 3 h, or 5 U/ml 24 °C for 2 h [7–9]. Bermejo [10] also investigated the influence of mutation on the MAPK pathway receptor under 0.8 U/ml at 24 °C for 3 h. Since these studies utilize long-term incubation, from 1 to 4 h, at low concentrations of digestion enzyme, gene expression data show significant changes. However, the reaction times of digestion reported by these studies are relatively longer than under RNA preparation conditions. If gene expression changes also occur during short-term reaction times such as those used for RNA preparation, then RNA preparation using the digestion method is confronted with a critical problem. Because the concentrations of digestive enzyme in RNA preparations are generally higher than in enzymatic studies, it is thought that stress is generated.
Moreover, Bermejo [11] reported that SLT2, a protein kinase of the cell integrity pathway on the MAPK cascade, was induced by 0.4 U/ml of zymolyase treatment at 24 °C for 15 min, and induction was maximized for 2 h. Generally, the incubation time for zymolyase treatment is 15–30 min; therefore, this report suggests that unintended gene expressions will be caused by zymolyase treatment for RNA preparation. What then is the influence of such treatment? Many recent protocols do not use enzyme treatment because it is known to cause complications. However, the concentrations and incubation times used in previous research do not correspond to each other, and therefore information regarding the influence of the enzymatic process in RNA preparation is insufficient and complicates our understanding of this method. To address this problem, we investigated the true influence of the enzyme method on RNA preparation.
There are several methods of yeast RNA preparation. The hot phenol method, the glass beads method, and the enzyme digestion method are generally used. Since the treatment time is the shortest in the glass bead method, the possibility of changing the gene expression is thought to be very low. Phenol denatures cell proteins and enzymes, so the hot phenol method is expected to suspend cell activity promptly. We used a commercial kit able to exploit the advantages of both methods. The influence of enzyme treatment was detected as gene expression changes using a DNA microarray.
Materials and Methods
Yeast Strain and RNA Preparation
Cells were prepared from yeast (Saccharomyces cerevisiae) strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0; Open Biosystems, Huntsville, AL, USA). The frozen (−80 °C) glycerol stocks of yeast cells were transferred with an inoculating needle into 10 ml of YPD medium (1 % yeast extract, 2 % peptone, and 2 % glucose) in glass tubes, and incubated on a rotary shaker at 150 rpm and 25 °C for 2 days until the stationary phase. The cells were transferred to new flasks, and cell density was adjusted to 0.2–0.3 at an absorbance of 650 nm (A 650). Yeast cells in the flasks were incubated with 150-rpm rotation at 25 °C for a few hours, until the A 650 became about 1.0, and then a cell pellet was obtained by centrifuging at 3,000 rpm for 5 min. Total RNA was prepared using a commercial kit (FastRNA Pro Red kit; MP Biomedicals, Irvine, CA, USA). A non-treated control sample was prepared through following the manufacturer’s instructions. To prepare a zymolyase-treated sample, zymolyase 20T (Seikagaku, Tokyo, Japan) was dissolved in buffer solution [0.9 M sorbitol, 0.1 M EDTA, 50 mM DTT (pH 7.5); stock solution concentration 600 U/ml], and this was added to the cell pellet (final concentration 300 U/ml). This was incubated at 37 °C for 10 min before following the manufacturer's instructions. For a low concentration zymolyase sample, zymolyase solution was added to the diluted cells (A 650 = 0.2–0.3, final concentration 4 U/ml), and this was incubated with 150 rpm rotation at 25 °C for 2 h. RNA preparation was performed in the same way as for the non-treated control.
DNA Microarray Analysis
Total RNA samples of the non-treated control and the zymolyase (300 U/ml) treatment were prepared. To avoid contamination by genomic DNA, RNA samples were treated with DNase (DNase I; Takara, Shiga, Japan) before reverse transcription. The quality of total RNA was evaluated using a nucleic acid analyzer (Experion; Bio-Rad, Hercules, CA, USA). RNA samples were used for synthesizing labeled RNA (amplified RNA) using a 3′ IVT Express kit (Affymetrix, Santa Clara, CA, USA). DNA microarrays (GeneChip Yeast Genome 2.0 array; Affymetrix) were processed following the manufacturer’s instructions. Array data sets were transferred into GeneSpring analysis software (ver. 10; Agilent Technologies, Santa Clara, CA, USA), and cluster analyses were performed for each treatment. After using the MAS5 algorithm to obtain summarized probe-level expression data, the average expression of three replicates was normalized to the control. The T test algorism was used for statistical analysis, and differences in gene expression with a p value <0.05 (p < 0.05) and a fold change >2.0 (FC > 2.0) were considered significant. However, all extracted genes (p < 0.05) were checked to find the expression trend within each category. To avoid the detection of false positives, a multiple testing correction (Benjamini–Hochberg FDR) was used to obtain corrected p values. Selected genes were categorized following the annotation used by the Munich Information Center for Protein Sequences (MIPS [12]) and the Saccharomyces Genome Database (SGD [13]). Several clusters were visualized using spreadsheet software (Excel 2007; Microsoft, Redmond, WA, USA). The microarray dataset has been assigned accession number GSE36955 in the Gene Expression Omnibus Database (GEO [14]).
Intracellular SOD Activity
To avoid the inhibition of measurement of SOD activity, zymolyase was dissolved into another buffer solution (0.9 M sorbitol, 0.1 M EDTA) not containing DTT. The non-treated control and the zymolyase treatment cells were collected in microcentrifuge tubes, and the cell pellets were stored on ice. Samples were washed three times with PBS containing protease inhibitor (1 pellet/10 ml, Complete mini EDTA-free; Roche, Basel, Switzerland), and finally, 50 μl of PBS was left in each microtube. A small volume of 0.5 mm sterilized glass beads was added to the microtube, and cell samples were ground by pestle for 3 min on ice. Then 450 μl of PBS was added to each microtube, and these were centrifuged at 10,000×g and 4 °C for 15 min. The supernatant was used as each sample. Samples were used in the SOD Assay Kit (Dojindo, Kumamoto, Japan), and measurements followed the manufacturer’s instructions. Because SOD inhibits the transformation from WST-1 (2-(4-lodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium) to WST-1 formazan, we observed the inhibition rate (%) and conversion into SOD activity (U/mg).
Semi-quantitative PCR
cDNA templates were synthesized from total RNA samples using a commercial reverse transcription kit (Transcriptor First Strand cDNA Synthesis Kit; Roche). Sequence information was obtained from SGD for the target genes (SLT2, SOD1, and SOD2), and primer sets that corresponded to the position around the 3′-end sequences were designed using the Primer 3 web site [15]. The following primer sets, SLT2 (forward, 5′-CTCAAGAGAGTATGATGGAGATGAGA-3′; reverse, 5′-GTGATTCTATACTTCCCCGGTTACT-3′), SOD1 (forward, 5′-ATCAAGCTTATCGGTCCTACCTC-3′; reverse, 5′-GAATGAATTGATGCGCTTACTACTT-3′), and SOD2 (forward, 5′-TTGTTCAAACCTACAACCAGGATAC-3′; reverse, 5′-TTCTTTCTTTCTTCAGATCTTGC-3′) corresponded to each target sequence. ACT1 was used as an internal control. The ACT1 primer set (forward, 5′-ATTGCCGAAAGAATGCAAAAGG-3′; reverse, 5′-CGCACAAAAGCAGAG ATTAGAAACA-3′), which has the same sequence as in a previous report [16], was prepared. For quantification, positive control templates for constructing standard curves were prepared using standard PCR. We prepared 20 μl of reaction solutions [1 μl of each 10 μM primer, 4 μl 5× buffer, 1.4 μl 25 mM MgCl2, 0.6 μl 10 mM dNTPs, 1 μl DNA template, 11 μl distilled water, and 0.1 μl taq polymerase (KAPA taq Extra; Kapa Biosystems, Woburn, MA, USA)] and subjected this to 95 °C for 2 min, followed by 30 cycles of 95 °C for 25 s, 55 °C for 15 s, and 72 °C for 45 s, in a thermal cycler (C1000; Bio-Rad). For each probe set, a template dilution series was prepared from these PCR products, and 1 μl of the samples was dispensed into a 96-well PCR plate. The same volume of control or zymolyase-treated templates was also dispensed into the plate. A total of 19 μl of reaction mix [0.4 μl of each 10 μM primer, 8.2 μl distilled water, and 10 μl 2× Master mix (KAPA SYBR FAST qPCR kit; Kapa Biosystems)] was added to the PCR plate. The sample plate was subjected to 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s and 62 °C for 20 s, in a thermal cycler (MX3000P; Agilent Technologies). The amplified ACT1 product was used as an internal control, and triplicates were averaged.
Results and Discussion
Gene Expression Changes from Zymolyase Treatment
Zymolyase reaction conditions are variously optimized by commercial kits. Moreover, various concentrations of zymolyase (approximately 10 to 1,000 U/ml) are used. Conditions that involve low concentrations (approximately 0.1 to 5 U/ml) have been thought not able to be evaluated using the results of former studies. Hence, we prepared a 300 U/ml concentration of zymolyase solution, and 100 μl of the solution was added to a yeast cell pellet. The sample was incubated for 10 min and then compared with a non-treated control sample using gene expression analysis. The microarray result extracted 3,218 gene expression changes from a total of 5,714 probes mounted on the DNA microarray (p < 0.05). Moreover, 1,529 induction genes and 1,689 repression genes were also counted from the total extracted genes, and a significant deviation to one side was not observed (Fig. 1). Meanwhile, a total of 537 genes indicated a more than 2-fold gene expression change; 435 of these genes indicated induction, and 112 indicated repression. Since much research has focused on gene induction, incorrect signals may be obtained by this expression analysis.
Fig. 1.
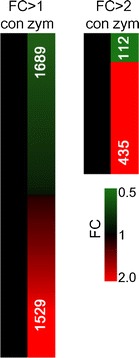
Gene expression profiles following zymolyase treatment. FC fold change, con non-treated control, zym 300 U/ml for 10 min
Functional classification was performed on extracted genes, indicating a more than 2-fold expression change (Table 1). The ratio of detection in the secondary metabolism category was the highest. However, whole categories indicated a certain level of detection ratio, and therefore a significant change was not observed in almost all functional categories except for protein synthesis. A total of 469 protein synthesis genes were contained in the yeast genome DNA microarray, and considering this, the detection number within this category seemed comparatively low. This implies that protein synthesis processes were not strongly induced because of short-term digestion. Meanwhile, localization classification indicated significant changes in cell wall synthesis-related categories (Table 2). However, a large number of detection genes were observed in the cytoplasm, nucleus, and mitochondria rather than in the cell wall-related genes. Taken together, the results of both functional and localization classifications suggested that the expression of many functional genes was changed by zymolyase treatment.
Table 1.
Functional classifications of detected genes
| Descriptionsa | Detection number | Detection ratio (%) |
|---|---|---|
| Metabolism | ||
| Amino acid metabolism (171)b | 25 | 10.2 |
| Nitrogen and sulfur metabolism (93) | 8 | 8.2 |
| Nucleotide metabolism (221) | 17 | 7.5 |
| Phosphate metabolism (418) | 36 | 8.6 |
| C-compound and carbohydrate metabolism (495) | 59 | 11.7 |
| Lipid, fatty acid, and isoprenoid metabolism (287) | 35 | 12.5 |
| Metabolism of vitamins, cofactors (158) | 17 | 10.4 |
| Secondary metabolism (76) | 17 | 21.8 |
| Energy (339) | 38 | 10.6 |
| Cell cycle and DNA processing (969) | 85 | 8.5 |
| Transcription (1,012) | 95 | 9.3 |
| Protein synthesis (469) | 22 | 4.6 |
| Protein fate (1,133) | 98 | 8.5 |
| Protein with binding function (1,026) | 110 | 10.6 |
| Protein activity regulation (245) | 18 | 7.4 |
| Cellular transport (1,010) | 111 | 10.7 |
| Cellular communication (231) | 20 | 8.5 |
| Cell rescue (521) | 63 | 11.4 |
| Cellular interaction (445) | 47 | 10.3 |
| Development (67) | 7 | 10.0 |
| Biogenesis of cellular components (844) | 83 | 9.7 |
| Cell type differentiation (449) | 47 | 10.5 |
| Unclassified proteins (879) | 85 | 4.3 |
aFunctional categories referred to MIPS [12]
bProbe set number mounted on array chip
Table 2.
Localization of detected genes
| Localizationa | Detection number | Detection ratio (%) |
|---|---|---|
| Cell wall (40)b | 12 | 30.0 |
| Extracellular (49) | 12 | 24.5 |
| Cytoskeleton (202) | 34 | 16.8 |
| Integral membrane (158) | 21 | 13.3 |
| Cell periphery (211) | 26 | 12.3 |
| Punctate composite (131) | 16 | 12.2 |
| Bud (145) | 17 | 11.7 |
| ER (521) | 56 | 10.7 |
| Plasma membrane (181) | 19 | 10.5 |
| Cytoplasm (2,645) | 275 | 10.4 |
| Nucleus (2,036) | 191 | 9.4 |
| Vacuole (269) | 25 | 9.3 |
| Endosome (54) | 5 | 9.3 |
| Transport vesicle (137) | 12 | 8.8 |
| Mitochondria (993) | 84 | 8.5 |
| Ambiguous (225) | 19 | 8.4 |
| Golgi (154) | 12 | 7.8 |
| Golgi membrane (94) | 4 | 4.3 |
| Peroxisome (52) | 1 | 1.9 |
aCellular localization referred to MIPS [12]
bProbe set number mounted on array chip
Activation of RNA Polymerase
Because the transcription process is essential for gene expression, we observed significantly changed (FC > 2.0) RNA polymerase-related genes. In Table 1, the detection ratio of transcription-related genes (9.3 %) corresponds to the average (around 10 %) in all functional categories; this category did not therefore show significant characteristics. However, the class of RNA polymerase was biased; a number of RNA polymerase II-related genes that work on transcription of mRNA indicated more than 2-fold changes, while only a small number of genes in classes I and III showed the same changes (Fig. 2). This finding suggests that short-term zymolyase reaction causes the induction of mRNA transcription.
Fig. 2.
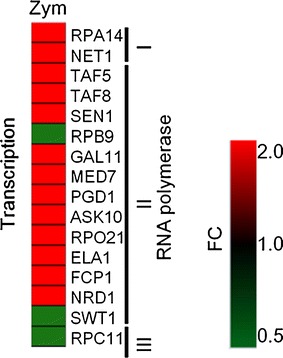
Gene expression changes induced by RNA polymerase. RNA polymerase-coding genes, which indicated significant changes (FC > 2.0), were extracted. The gene expressions of non-treated control samples were adjusted 1-fold. FC fold change
Regulation of Cell Wall Genes
Changes in cell wall synthesis-related genes following zymolyase treatment have been well reported [6–9, 17]. Moreover, detection ratios in cell wall synthesis categories at localization classification were significantly large, so we investigated the genes in this category. For instance, both FIT1 and FIT3, required for smooth metabolism with iron conservation in periplasmic space [18], were induced by the enzyme treatment. This might have been used against cell lysis by cell wall repair and the activation of cellular metabolism. However, not many genes indicated greater than 2-fold change (Fig. 3a), and expression of SLT2, which codes mitogen-activated protein kinase (MAPK) in the cell integration pathway and is related to cell wall remodeling [19], was a little less than 2-fold (data not shown). A former study reported that 15 min of zymolyase treatment did not cause significant change to SLT2 compared with 2 h of treatment [11]. Meanwhile, RNA preparation uses high concentrations (300 U/ml) of zymolyase, and this does not correspond with former studies. We therefore prepared a sample with low zymolyase concentration (4 U/ml) and long-term incubation (2 h) to enable comparison with former studies (Table 3). Our semi-quantitative PCR indicated that zymolyase treatment for RNA preparation resulted in a small induction shown in the DNA microarray results (Fig. 3b). Likewise, the sample that was under long-term incubation indicated relatively high induction though it was less than 2-fold. The induction level of SLT2 was proportional to incubation time, but not to enzyme concentration. Taken together, we thought that RNA preparation with short-term incubation (10–15 min) only caused slight changes to cell wall genes.
Fig. 3.
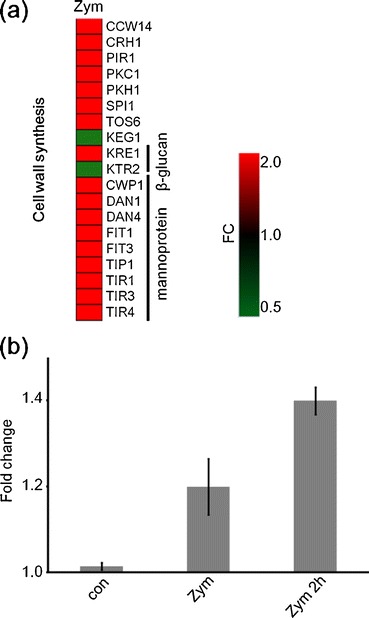
Gene expression changes in cell wall genes. Cell wall protein-coding genes which indicated significant changes (FC > 2.0) were extracted (a). To enable comparisons with previous research, the gene expression of SLT2 was measured by semi-quantitative PCR (b). FC fold change, con non-treated control, Zym 300 U/ml for 10 min, Zym 2h 4 U/ml for 2 h. Bars = SE. n = 3
Table 3.
Zymolyase treatment conditions
| Con | Zym | Zym 2 h | |
|---|---|---|---|
| Time | 0 | 10 min | 2 h |
| Zymolyase (U/ml) | – | 300 | 4 |
However, DAN genes indicated large expression changes, and also TIR1, 3, and 4 were induced more than 3-fold. DAN1 and TIR1–4 each code homologous mannoproteins [20]. The expression of cell wall genes and, in particular, the expression of many mannoprotein genes was induced. These changes suggest that yeast cells respond to enzyme treatment.
Induction of MAPK Pathway Genes
Slt2 is a component of the MAPK pathway, and therefore we believe that up-stream and down-stream genes were also influenced by zymolyase treatment. We extracted MAPK-related genes, and these were categorized based on the Kyoto Encyclopedia of Genes and Genomes (KEGG [21]). According to our previous studies, MAPK genes did not show significant expression changes (p < 0.05; fold change, FC > 2.0) even under highly toxic conditions [22, 23]. Hence, we extracted all MAPK genes that indicated changes (p < 0.05, FC > 1.0) (Fig. 4). The extracted genes belonged to various MAPK pathways, and they included GTPase-coding genes such as CDC42 and RHO1 [24, 25], and the cellular membrane receptor genes, SHO1, WSC2, and STE3 [26–29]. Those receptor and GTPase genes indicated repression trends. Conversely, the down-stream genes of MAPKKK in each MAPK pathway pointed to a trend in gene induction. While the reason for repression of up-stream genes was uncertain, this result suggested that these MAPK pathways were induced by zymolyase treatment, regardless of functional category. Moreover, DIG1 and 2 coding proteins, Dig1 and 2, bind Ste12, and function as inhibition regulators down-stream of the MAPK Filamentation or Mating pathway [30]; these indicated a different manner of expression. Because it has been reported that Dig1 and 2 regulate Ste12 differently from each other [31], it was thought that the differences in gene expression reflected the regulatory manner of Ste12. STE12 expression was not observed, whereas the genes of both TEC1 and MCM1, which bind Ste12 to form a complex [32, 33], were also induced. Expression of TEC1 and MCM1, and also of MSN2 and RLM1, was induced, and these changes corresponded to the expression patterns of up-stream genes such as FUS3, SLT2, and HOG1. MAPK pathways were thought to be induced by zymolyase treatment regardless of differences in functional cascade.
Fig. 4.
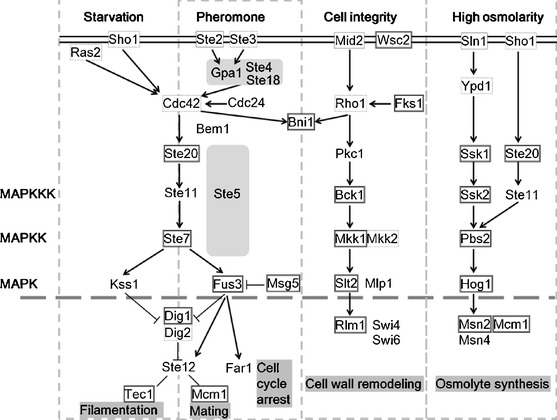
Gene expression changes in the MAPK pathway. MAPK pathway genes (FC > 1.0, p < 0.05) were extracted and classified to each pathway. The root structure of MAPK was constructed by referring to KEGG [20]. Lined box, induced gene; dashed line box, repressed gene. MAPKKK MAP kinase kinase kinase, MAPKK MAP kinase kinase, MAPK MAP kinase
Oxidative Stress Responses and Intracellular SOD Activity
Since enzymatic digestion is thought to cause oxidative stress conditions in cells, we also extracted the oxidative stress response genes. Only a small number of genes were observed to indicate significant change (FC > 2.0), although we extracted all oxidative response genes (p < 0.05, FC > 1.0) (Fig. 5a). The result of extraction number did not indicate bias between induction and repression. One third of the oxidative stress response genes coded for mitochondria-located proteins, and the others coded for cytoplasm- or nucleus-located proteins. Previous studies have not to this extent reported oxidative stress response genes during cell digestion [8, 9]. Our study indicated small expression changes. However, oxidative stress is the most likely stressor causing damage to cells, and it is possible that zymolyase treatment causes oxidative, stress-derived damage [34]. Hence, we tried to observe intracellular oxidative stress responses through the comparison of SOD activity. SOD1 and 2, encoding superoxide dismutase [35, 36], were also detected and slightly induced. SOD is a major anti-stressor of cells, so it was expected that SOD activity would accurately reflect the intracellular circumstances. The method we used measured SOD activity but not enzyme volume because it monitored the inhibition reaction of free SOD (see “Materials and Methods” section). Sod1 is localized in the cytoplasm, nucleus, and mitochondria, while Sod2 is localized in the mitochondrial membrane surfaces. However, we did not make allowances for the localization of SOD because the sample preparation for this monitoring was not suitable for separating the levels of SOD activity. We also used a low concentration (4 U/ml) zymolyase-conditioned sample with long-term incubation (2 h), so that this could be easily compared with previous studies. The non-treated control indicated 175 U/mg (SE = 4), 300 U/ml of zymolyase-conditioned sample indicated 220 U/mg (SE = 12), and the low concentration (4 U/ml) zymolyase-conditioned sample indicated 117 U/mg (SE = 18) (Fig. 5b). Compared with the non-treated control, SOD activity in the high concentration zymolyase sample increased 26 %, while that in the low concentration sample decreased 33 %, and each zymolyase sample indicated statistical significance (p < 0.01). The decrease in SOD activity in the long-term incubated sample complicated the interpretation of the effect of zymolyase treatment. We cannot be sure of the cause. However, regardless of time course or enzyme concentration, SOD activities changed in enzyme-treated samples. Considering the gene expression changes in SOD genes obtained by semi-quantitative PCR, the change in SOD activity corresponds to the gene expressions of SOD (Fig. 5b). These findings suggest that zymolyase treatment in RNA preparation causes changes in gene expression and enzymatic activity, which, although small, should not be ignored.
Fig. 5.
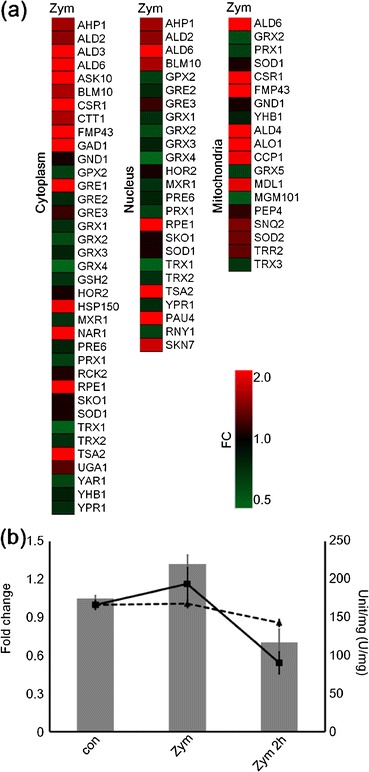
Oxidative stress responses. Oxidative stress response genes (FC > 1.0, p < 0.05) were extracted and classified to each localization (a). FC fold change. SOD expressions and SOD activity (b). Dashed line graph, SOD1 expression; line graph, SOD2 expression; gray bar graph, SOD activity. con non-treated control, Zym 300 U/ml for 10 min, Zym 2h 4 U/ml for 2 h. Bars = SE. n = 3
Conclusion
RNA preparation from yeast cells is an important and basic technique in molecular biology. A number of RNA preparation methods have been proposed and adopted. However, this has included great variation in technique following large differences in equipment between laboratories. The development of zymolyase treatment has been welcomed to overcome this problem. Northern hybridization analysis, in particular, needs large quantities of RNA. Therefore, enzymatic digestion suitable for bulk preparation of RNA was hailed as an appropriate technique. In recent years, new techniques such as real-time PCR have been developed, for which large quantities of RNA are not necessary. Recent reports have recommended avoiding enzymatic digestion [37], and this method has already been ignored by many other reports describing RNA preparation. It is thus almost time to stop using enzymatic digestion. Our study and other previous studies suggest that yeast cells respond to stress during the incubation process. Only small changes are indicated; however, mRNA levels will not reflect the original data that we want to obtain. In particular, the MAPK pathway genes and the RNA polymerase genes influence many of the other gene categories, producing moderate changes. It is possible to miss the changes produced by zymolyase treatment, meaning a great loss in research understanding. To avoid this situation, scientists should pay attention to careful interpretation of the effects of enzymatic reactions.
Abbreviations
- MAPK
Mitogen-activated protein kinase
- PCR
Polymerase chain reaction
- FC
Fold change
- SE
Standard error
Contributor Information
Tadahiro Suzuki, Email: suzut@affrc.go.jp.
Yumiko Iwahashi, Phone: +81-29-8388103, FAX: +81-29-8387996, Email: yumiko@affrc.go.jp.
References
- 1.Kitamura K, Kaneko T, Yamamoto Y. Archives of Biochemistry and Biophysics. 1971;145:402–404. doi: 10.1016/0003-9861(71)90053-1. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura K, Yamamoto Y. Archives of Biochemistry and Biophysics. 1972;153:403–406. doi: 10.1016/0003-9861(72)90461-4. [DOI] [PubMed] [Google Scholar]
- 3.Kaneko T, Kitamura K, Yamamoto Y. Agricultural and Biological Chemistry. 1973;37:2295–2302. doi: 10.1271/bbb1961.37.2295. [DOI] [Google Scholar]
- 4.Forsburg SL, Rhind N. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klassen R, Fricke J, Pfeiffer A, Meinhardt F. Biotechnology Letters. 2008;30:1041–1044. doi: 10.1007/s10529-008-9648-y. [DOI] [PubMed] [Google Scholar]
- 6.de Groot PW, Ruiz C, de Vázquez de Aldana CR, Duenas E, Cid VJ, Del Rey F, et al. Comparative and Functional Genomics. 2001;2:124–142. doi: 10.1002/cfg.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boorsma A, de Nobel H, ter Riet B, Bargmann B, Brul S, Hellingwerf K, et al. Yeast. 2004;21:413–427. doi: 10.1002/yea.1109. [DOI] [PubMed] [Google Scholar]
- 8.Garcia R, Bermejo C, Grau C, Perez R, Rodriguez-Pena JM, Francois J, et al. The Journal of Biological Chemistry. 2004;279:15183–15195. doi: 10.1074/jbc.M312954200. [DOI] [PubMed] [Google Scholar]
- 9.García R, Rodríguez-Peña J, Bermejo C, Nombela C, Arroyo J. The Journal of Biological Chemistry. 2009;284:10901–10911. doi: 10.1074/jbc.M808693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bermejo C, García R, Straede A, Rodríguez-Peña JM, Nombela C, Heinisch JJ, et al. OMICS. 2010;14:679–688. doi: 10.1089/omi.2010.0060. [DOI] [PubMed] [Google Scholar]
- 11.Bermejo C, Rodríguez E, García R, Rodríguez-Peña J, Rodríguez de la Concepción M, Rivas C, et al. Molecular Biology of the Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munich Information Center for Protein Sequences. Available from: http://mips.gsf.de/. Accessed December 10, 2011.
- 13.Saccharomyces Genome Database. Available from: http://www.yeastgenome.org/. Accessed October 15, 2011.
- 14.Gene Expression Omnibus Database. Available from: http://www.ncbi.nlm.nih.gov/geo/. Accessed March 20, 2012.
- 15.Primer3. Available from: http://frodo.wi.mit.edu/. Accessed December 20, 2011.
- 16.Iwahashi H, Odani M, Ishidou E, Kitagawa E. FEBS Letters. 2005;579:2847–2852. doi: 10.1016/j.febslet.2005.03.100. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Du W, Zhao J, Zhang L, Zhu Z, Jiang L. FEMS Yeast Research. 2010;10:441–451. doi: 10.1111/j.1567-1364.2010.00626.x. [DOI] [PubMed] [Google Scholar]
- 18.Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, Botstein D, et al. The Journal of Biological Chemistry. 2001;276:49244–49250. doi: 10.1074/jbc.M109220200. [DOI] [PubMed] [Google Scholar]
- 19.Lee KS, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, et al. Molecular and Cellular Biology. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abramova NE, Cohen BD, Sertil O, Kapoor R, Davies KJ, Lowry CV. Genetics. 2001;157:1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyoto Encyclopedia of Genes and Genomes. Available from: http://www.genome.jp/kegg/. Accessed May 31, 2012.
- 22.Suzuki T, Iwahashi Y. Chem-Bio Information Journal. 2009;9:94–107. doi: 10.1273/cbij.9.94. [DOI] [Google Scholar]
- 23.Suzuki T, Iwahashi Y. Journal of Agricultural and Food Chemistry. 2011;59:7145–7154. doi: 10.1021/jf104938p. [DOI] [PubMed] [Google Scholar]
- 24.Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, et al. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 25.Johnson DI. Microbiology and Molecular Biology Reviews. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustin M, Albertyn J, Alexander M, Davenport K. Microbiology and Molecular Biology Reviews. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagen DC, McCaffrey G, Sprague GF. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarrinpar A, Park S-H, Lim WA. Nature. 2003;426:676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 29.Verna J, Lodder A, Lee K, Vagts A, Ballester R. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCullagh E, Seshan A, El-Samad H, Madhani HD. Nature Cell Biology. 2010;12:954–962. doi: 10.1038/ncb2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou S, Lane S, Liu H. Molecular and Cellular Biology. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elble R, Tye BK. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilella F, Herrero E, Torres J, de la Torre-Ruiz MA. The Journal of Biological Chemistry. 2005;280:9149–9159. doi: 10.1074/jbc.M411062200. [DOI] [PubMed] [Google Scholar]
- 35.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. The Journal of Biological Chemistry. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 36.Strålin P, Jacobsson H, Marklund SL. Biochimica et Biophysica Acta. 2003;1619:1–8. doi: 10.1016/S0304-4165(02)00419-1. [DOI] [PubMed] [Google Scholar]
- 37.Amin-ul Mannan M, Sharma S, Ganesan K. Analytical Biochemistry. 2009;389:77–79. doi: 10.1016/j.ab.2009.03.014. [DOI] [PubMed] [Google Scholar]


