Abstract
Current microarray assay technology predominately uses fluorescence as a detectable signal end-point. This study assessed real-time in situ surface hybridization capture kinetics for single printed DNA microspots on solid array surfaces using fluorescence. Influence of DNA target and probe cyanine dye position on oligo DNA duplex formation behavior was compared in solution versus surface-hybridized single DNA printed spots using fluorescence resonance energy transfer (FRET) analysis. Fluorophore Cy3/Cy5 fluorescent intensities were analyzed both through printed hybridized DNA spot thickness and radially across single spot surfaces. Confocal single spot imaging shows that real-time in situ hybridization kinetics with constant target concentrations change as a function of printed probe density. Target-specific imaging in single spots exhibits heterogeneous printed probe radial density that influences hybridization spatially and temporally via radial hemispherical diffusion of dye-labeled target from the outside edge of the spot to the interior. FRET of surface-captured target occurs irrespective of probe/target fluorophore position, resulting from excess printed probe density and spot thickness. Both heterogeneous probe density distributions in printed spots and fluorophore position on short DNA oligomers influence duplex formation kinetics, hybridization efficiencies and overall fluorescence intensity end-points in surface-capture formats. This analysis is important to understanding, controlling and quantifying array assay signal essential to reliable application of the surface-capture format.
Keywords: DNA microarray, printing, confocal imaging, duplex, real-time hybridization, fluorescence resonance energy transfer, FRET, spot heterogeneity
INTRODUCTION
DNA microarrays provide relative levels of expression for up to tens of thousands of genes in parallel on a single platform, and are widely employed in genotyping, drug discovery and gene expression profiling.1 Accurate analysis and signal interpretation presents immense challenges to quantitative analysis and clinical translation.2, 3 Multiple sources of microarray spot heterogeneity, reproducibility and repeatability are known, contributing to assay variability and inconsistency.4–7 The DNA oligomer surface hybridization process with printed probes exhibits a complex kinetic and thermodynamic dependence on local interfacial environmental factors including probe attachment modalities on solid supports8, surface probe density issues associated with steric and electrostatic interactions9, length and sequence specificities of probe with target,10 increased non-specific binding/cross-hybridization at higher probe densities causing target cross-binding to neighboring probes11 and non-specific influences.12, 13 Additional factors include surface chemistry to reduce non-specific background noise, binding interference and reliable probe immobilization and presentation.14 Strategies to improve target-surface hybridization to solid supports include microspot size to overcome mass transfer limitations15 and surface diluent spacers (e.g., mercaptohexanol,16 11-mercapto-1-undecanol,17 oligoethylene glycols17) that reduce non-specific probe-surface interactions, control electrostatic forces and steric issues between adjacent DNA probes, probe density, orientation and presentation to incoming target analyte. Furthermore, increasing ionic strength18 during oligonucleotide probe deposition, and electrostatic balance between immobilized oligonucleotide charge density and solution ionic strength19 can significantly increase assay hybridization efficiency.
DNA hybridization at the solid-liquid interphase is complex: a stochastic process with non-linear kinetics and analyte concentration dependence.20 The simplest two-state model involving hybridization of matched complementary oligomer sequences is represented as a one-step reversible reaction model with forward and reverse rate constants and a binding equilibrium constant. Capture is influenced by DNA surface transport, adsorption/desorption, probe/target length, ionic strength, extent of probe-target interactions, 2-D diffusion coefficient and immobilized probe density.13 Target binding is similar to ligand-receptor pairing: a hybrid of diffusion and reaction-controlled models with overall binding kinetics sensitive to both reaction rates and surface diffusion.21 Various modifications of Langmuirian binding isotherms have been proposed for DNA hybridization on solid surfaces. Vainrub and Pettitt22, 23 account for repulsion between target DNA and surface-immobilized oligonucleotide probes, producing a coulombic blockage of target binding. Chan et al.,11 incorporated heterogeneous hybridization isotherms considering both length and surface diffusion of target with changing probe surface coverage. Further modification24 considered competitive hybridization of surface vs. bulk hybridization, entropic and enthalpic contributions25 of probe immobilized on an impenetrable surface, polydispersity of probe and target, and electrostatic interactions within the probe layers. A Langmuir-based hybridization model predicts two different mechanisms: direct hybridization from solution and hybridization by targets that first adsorb nonspecifically and then surface-diffuse to probes, showing that DNA hybridization on solid supports may be extremely efficient for physically realistic 2-D diffusion coefficients, target concentrations and surface probe densities.11 Langmuir models deviate from ideality with increasingly crowded probes and duplexes. Hybridization yields become insensitive to target concentrations at high ionic strengths and within densely tethered films.26 Commercial microarray data under real-world conditions suggest that these non-idealities impact solid-phase hybridization versus identical sequences in solution.27, 28
End-tethered single-stranded DNA (ssDNA) at a surface is characterized as a flexible polyanionic chain while double-stranded (dsDNA) is compared to a rigid polyanionic rod. This plays an important role in the kinetics and thermodynamics of the hybridization process. dsDNA persistence length been shown to be ~50 nm in salt concentrations greater than 10 mM, while that for ssDNA is ~1 nm. Hence, ssDNA is not only much more flexible than dsDNA, but is also affected by conditions near a surface.29, 30 Moreover, DNA persistence length increases with strand length, increasing temperatures and ionic strength, attributed to exaggerated end effects from shorter strands.31 Increasing probe surface density creates a polyelectrolyte brush from collective crowding of nucleic acid tails. The ionic microenvironment within dense end-attached DNA oligonucleotides (i.e., DNA brushes) produces interfacial DNA brush electrostatics influencing target capture32: hybridization rates for longer oligomer lengths are reduced.33 Therefore, tethered DNA strand dynamics and interfacial properties (flexibility, elasticity, electrostatics all influencing steric contributions) affect ssDNA conformation at surfaces and alter DNA target hybridization from solution.
Monitoring real-time DNA hybridization kinetics with different optical (ellipsometry34, 35, evanescent wave methods36 and surface plasmon resonance37), physical or electrochemical (potentiometric stripping analysis38, amperometry39) and fluorescence techniques (FRET40, 41, fluorescence correlation spectroscopy42) analytical tools is common. Confocal fluorescence microscopy has been used in real-time detection of single oligonucleotides in solution.43 FRET-based analysis for DNA in situ hybridization with donor-acceptor fluorophore pairs (i.e., Cy3-probe and Cy5-target) is extensively used44, 45 to report intermolecular fluorophore distances, conformations, molecular dynamics and kinetics.46 FRET efficiency between Cy3 and Cy5 fluorophores attached to the different locations in DNA duplexes demonstrates a power law dependence on separation distance and relative fluorophore dipole orientations.47–49 FRET’s orientational dependence is likely most significant when the fluorophores are constrained, such as for DNA bound to solid substrates where fluorophore motion is limited.50 Acceptor photobleaching is also used to validate FRET, quantitatively determined by a “de-quenching” method comparing the intensities of Cy3-donor emission before and after photobleaching of the Cy5-acceptor.51, 52 Notably, cyanine dyes are predominately used for single molecule FRET measurement and for DNA microarray analysis due to their favorable fluorescence quantum yields and photostabilities, low pH sensitivities and ability to employ inexpensive diode lasers.53 Additionally, repeated microarray optical scanning and other intense laser exposure can promote non-intentional dye photobleaching.54
In this study, we monitored real-time hybridization of target to surface–tethered, dye-labeled DNA probes using confocal microscopy of individual printed DNA spots on array substrates. This analysis follows our recent comparisons of single printed probe microspots using epifluorescence and scanner-based imaging compared to mass spectrometry chemical imaging analysis of identical printed spots.55 Real-time direct visualization of target hybridization within single spots follows fluorescent intensities for both cyanine probe and target dyes in situ over time. Pronounced differences in hybridization kinetics to achieve spot target saturation were observed, although spot-to-spot hybridization kinetics for identical probe DNA concentrations and fractional probe DNA-fluorophore spot densities were comparable. Time-lapse images for spot hybridization show heterogeneous probe density distributions within printed DNA spots, resulting in time-dependent hybridization controlled by radial hemispherical diffusive flux of target across the spot from the edge (less dense probe) to the center (denser probe). Also, spot hybridization is time-dependent with homogenous probe densities due to transport limitations.15 Moreover, Cy3-probe/Cy5-target FRET in printed spots using acceptor photobleaching over time and with spot depth showed little observable differences from the two distinct duplex-induced dye-dye orientations, possibly reflecting dense probe molecular environments and multiple nearest-neighbor donor-acceptor interactions in printed, micron-thick printed spots. By contrast, bulk solution-phase FRET readily distinguished the two different dye-dye duplex orientations, emphasizing distinct microenvironments for duplex formation in solution vs. surface-bound assays. Furthermore, single spot heterogeneity was probed by locally photobleaching the target Cy5 dye over the spot thickness comprising densely printed probe. Photobleaching studies also revealed target-Cy5 signal at different depths in DNA microspots as shown by optical Z slices and additionally indicated that the photostability of Cy3/Cy5 pairings is critical to their analysis under multiple laser scans in microarray assays.
MATERIALS AND METHODS
Materials
HPLC-purified (≥90% purity) DNA oligomers with several different modifications (Table S-1) were purchased from TriLink Biotechnologies (San Diego, USA). Ultrapure water (18.2 MΩ-cm) was used for all solutions and protocols. Cyanine Cy3 (probe) and Cy5 (target) fluorophores are incorporated at 5’ or 3’ oligonucleotide termini during synthesis using phosphoramidite reagents through a C6-linker to ssDNA.56 All chemicals were used as received. Buffer salts, sarcosine, sodium dodecyl sulfate (SDS), and ethanolamine were ACS grade (Sigma-Aldrich, St. Louis, USA). Microarray slides (Slide H, amine-reactive 3D polymer-coated slides, Schott Nexterion, Louisville, USA) were used for DNA oligomer printing. New slides taken from the manufacturers packaging were used without pre-treatment. Unused slides were stored under nitrogen at 4°C and used within a month to preserve the slide’s ester reactivity.57
DNA immobilization on microarray substrates using contact printing
Microarrays were generated by printing multiple DNA probe concentrations (10, 20, and 40 micromolar) and different bulk solution ratios of Cy3-labeled (Cy3-Oligo1-NH2) to unlabeled (Oligo1-NH2) sequence-identical oligo DNA probes (0:1, 1:3, 3:1, and 1:0 ratios) to provide printed rows containing no Cy3-labeled probes(0:1) with rows containing Cy3 gradient contents up to 100% Cy3-labeled probe (1:0). DNA probes solutions were prepared in print buffers containing 150mM sodium phosphate, 0.001% Tween20 and 0.001% sarcosine at pH 8.5. Spotted DNA arrays (5×4 spots of each probe concentration) with varying amounts of Cy3-DNA were printed in triplicate on separate slides to compare probe-only and hybridized duplex arrays. Contact printing was performed using a SpotBot®2 contact printer and Stealth™ pins from Arrayit (Sunnyvale, USA). Further details of printing DNA spots and post-printing protocols are discussed in Supporting Information.
Fluorescence scanner images of DNA microarrays
Printed DNA microarray slides were scanned after blocking but prior to hybridization steps (GenePix Microarray scanner 4100A, v.6 software) to record Cy3-DNA probe and Cy5-target DNA spot intensities. DNA printed/hybridized slides were excited using two optical channels (535 and 635 nm for Cy3 and Cy5, respectively, laser power 100%, brightness and contrast 92%, PMT gain 400, pixel size 10 micron and the line average set at 3).
Real-time confocal imaging of hybridization in single DNA spots
A fully-automated Nikon A1R confocal microscope capable of capturing high-quality images was used to image real-time target hybridization, FRET and photobleaching for single DNA microarray spots. Ultra high-speed high quality imaging scanned 512 × 512 pixels at 30 frames/second with a 40XELWD NA 0.80 air lens. Depth-dependent Z slice imaging in single printed spots was recorded with a 0.5 micron step-size at 15-second intervals for 30 minute time lapses. Slides were imaged with printed side facing up and away from the objective to allow addition of hybridization buffer and target while imaging.
Real-time hybridization and FRET with confocal microscopy
To study real-time hybridization kinetics and FRET of DNA target oligomers with 5’-Cy3-labeled immobilized probe (i.e., 5’-Cy3 dye position on oligonucleotide probes) and Cy5-labeled target (either 3’ or 5’ termini) in single spots, hybridization was performed in situ on the confocal microscope stage. Hybridization conditions typical of commercial microarray generation were adapted to study real-time imaging under confocal microscopy. A stage-mounted temperature controlled-humidity chamber (set to 23°C and 60% humidity, Pathology Devices) was utilized, and 20µl of 1µM target DNA in hybridization buffer (4X saline sodium citrate (SSC) containing 0.1% SDS at pH 7) was added over the probe-printed surface slide and a cover slip was overlaid to control evaporation. Following confocal imaging, slides were washed with hybridization buffer solutions (4X, 2X, 1X and 0.2X dilutions) for 5 minutes followed by water wash. Samples were then blown dry with nitrogen and immediately scanned. Further details are discussed in Supporting Information.
For Cy3–Cy5 FRET analysis, target Cy5 emission signal was recorded under Cy3-specific excitation over time in single printed spots. To confirm FRET, acceptor photobleaching of Cy5 with 633nm laser excitation was performed and then both Cy3 and Cy5 emission signals were recorded. In areas where FRET transfer between Cy3 and Cy5 is facilitated by duplex formation and sufficient dye proximity, photobleaching of the acceptor (Cy5) should result in an increase in donor (Cy3) emission signal relative to that prior to bleaching. For FRET analysis, additional controls were performed to confirm FRET by monitoring changes in intensities for different excitation and emission of Cy3-probe and Cy5-target with unlabeled target DNA hybridized to a single spot.
DNA duplex-induced Cy3–Cy5 FRET in solution
FRET produced from DNA duplex formation in solution was assessed. Fluorescence emission spectra from 545–750 nm were obtained using a Cary Eclipse fluorescence spectrophotometer (Varian, Walnut Creek, USA). Excitation wavelengths for Cy3-DNA and Cy5-DNA were 547nm and 644nm, respectively, with emission wavelengths of 569nm and 655nm (λmax), respectively. Further details are provided in Supporting Information.
RESULTS AND DISCUSSION
Single spot real-time DNA oligomer hybridization kinetics
Hybridization of immobilized 5’-Cy3-oligo1-probe with the 5’-Cy5-target-oligo2 and Oligo2–3’-Cy5-target was analyzed in single printed probe spots in real-time. In order to evaluate dye-dye distance and steric influences on duplex formation, experiments were performed in different hybridization orientations shown in Figure S-1. Individual spots (40µM probe DNA containing 50% Cy3-DNA duplexed) with Cy5-target (1µM containing 1% Cy5-DNA) were analyzed in different dye-duplex relative orientations depicted in Figure S-1(also depicted in schematics in Figure 1A and 2A).
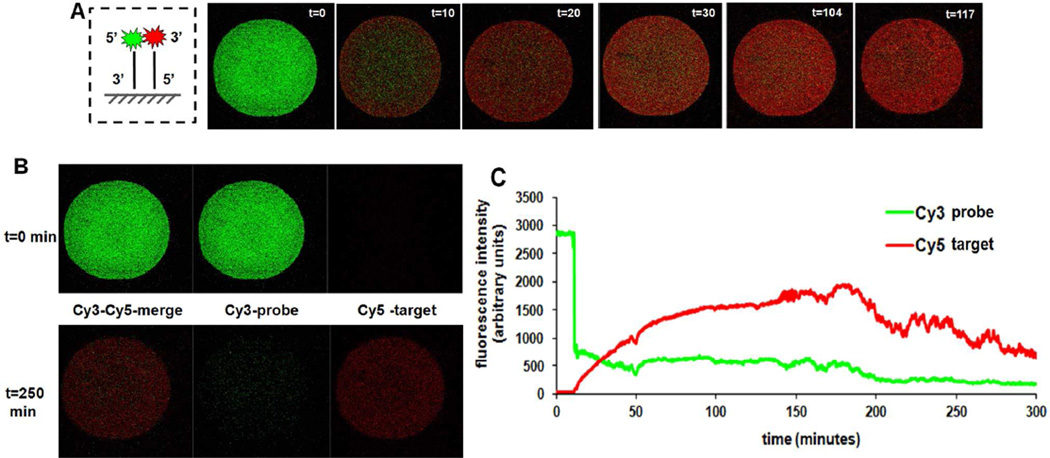
Figure 1.
Real-time hybridization of single DNA spot with dyes on opposite ends of the probe-target duplex. A) Schematic representation of the duplex: 5’-Cy3-oligo1-probe with 5’-Cy5-oligo2-target; images of DNA spot with merged Cy3 and Cy5 emission intensity at various time points. B) Images of spots in Cy3 or Cy5 or both filter channels during real-time imaging of single array spots before (t=0) and after (t=90 min) hybridization. C) Real-time hybridization of Cy3/Cy5 fluorescence intensity changes in single spots versus time. Duplex formation kinetics under this unstirred transport-limited regime follows pseudo-first order kinetics (see supplementary information and Figure S-7).
Note: microscopy image contrast has been digitally enhanced for the figures to better facilitate direct observation of the optical changes observed over time in situ.
Figure 2.
Real-time hybridization of single DNA spot with dyes on same end of the probe-target duplex. A) Schematic representation of the duplex: 5’-Cy3-Oligo1-probe: Oligo2–3’-Cy5-target with; images of DNA spot with merged Cy3 and Cy5 emission intensity at various time points. B) Images of spots in Cy3 or Cy5 or both filter channels during real-time imaging of single array spots before (t=0) and after (t=250 min) hybridization. C) Real-time hybridization of Cy3/Cy5 fluorescence intensity changes in single spots versus time. Duplex formation kinetics under this unstirred transport-limited regime follows pseudo-first order kinetics (see supplementary information and Figure S-7).
Note: microscopy image contrast has been digitally enhanced for the figures to better facilitate direct observation of the optical changes observed over time in situ.
Each single printed spot image in Figure 1A depicts an overlay of immobilized probe-Cy3 (green) and target-Cy5 (red) intensities versus time to assess distributions of Cy5 target captured in the same Cy3-DNA spot over time. Confocal spot images corresponding to start (t=0) and end (t=90 min) of hybridization in different imaging channels are depicted in Figure 1B. However, Cy3/Cy5 intensities were recorded for up to 140 total minutes as indicated in Figure 1C. Figure 1A reveals a bright Cy3 emission (green) color at t=0, and its intensity decreases after addition of Cy5-target-DNA with a simultaneous increase in Cy5 emission (red) intensity. With increasing time, Cy5 intensity (red) increases (e.g., t=25 min.), producing bright red spots at t=90 min, indicating the expected increased target hybridization over time. Figure 1A also reveals a brighter Cy3 green intensity at the spot center compared to its peripheral intensity, indicating the non-uniformity of Cy3-DNA immobilized probe distribution. Previous work has detailed the causes of this micro-spot heterogeneity from printed spot drying dynamics.55 Probe DNA density is high at spot centers as evidenced by centralized green intensity, decreasing radially to spot edges. This central green Cy3 emission intensity prevails throughout the Cy5-target hybridization process (although a relative decrease in intensity is observed); indicating limited duplex formation at the spot center.
Figure 1B compares Cy3-probe and Cy5-target only intensities of the same single DNA spot along with its Cy3–Cy5 merged intensity before and after hybridization (i.e., at t=0 and t=90 min). Contrasting intensities in DNA spots after hybridization are shown by mixing Cy3- and Cy5 signal intensities in the merged image for spots. Increasing Cy5 intensity versus Cy3 indicates probe-target duplex formation in the DNA spot after 90 minutes. Furthermore, hybridization completeness is confirmed by fluorescence scanner data. Figure 1C tracks relative spot integrated emission intensities from each fluorophore over time, showing rapid decreases in Cy3 fluorescence signal intensity immediately upon addition of Cy5-target, attributed to altered Cy3 environment from the duplex formation, including quenching. This is confirmed using confocal imaging by corresponding decreases in spot integrated Cy3-probe emission intensity upon addition of unlabeled target DNA in hybridization buffer (4XSSC) (see Figure S-2). Simultaneous increases in Cy5 target intensity from the spot characterizes the onset of surface hybridization, occurring less than 2 minutes after addition of (unstirred) target solution over each spot. The in situ hybridization time-lapse imaging of single spots was performed over 30–60 minutes, and shows that target emission intensity replaces probe emission intensity within the spot at ~6 minutes, seen both in Figures 1A and 1B spot images, and in the intensity-specific time plot in Figure 1C. At 45 minutes, a crossover of probe and target emission intensities (i.e., decreasing Cy3 intensity and increasing Cy5 intensity) is observed, attributed to increasing spot-based FRET (further confirmed in the following sections) from increasing densities of dye-labeled probe-target duplexes at high hybridization efficiencies. Figure 1C also indicates a higher intensity of Cy5 emission compared to Cy3 emission at the later time points, either due to enhanced FRET, quenched Cy3, or greater quantum yield from Cy5 vs. Cy3 emitter on-spot. Steady signal intensities of Cy3 and Cy5 at longer times indicate saturation of probes and completeness of hybridization for a 100-micron diameter spot at approximately 60 minutes. Similar experiments performed with 40µM total DNA probe and 100%, 75% or 25% fractional ratios of 5’-Cy3-Oligo-1-probe with 5’-Cy5-Oligo2-target showed similar trends, though differences in time required to attain stability of final Cy3 and Cy5 emission intensities were observed.
Figure 2 shows the Case 2 (see Figure S-1 and schematics in Figure 2A) situation for DNA duplexes forming with both fluorophores located at the same dsDNA terminus. These experiments show a similar emission kinetics plot and characteristic intensity-time changes for hybridization as seen in Figure 1 for the Case 1 anti-parallel fluorophore duplex. Although hybridization begins within 2 minutes (similar to Figure 1), the spot exhibits Cy3–Cy5 emission intensity “crossover” more rapidly at 25 minutes. Cy5-target emission intensity is higher compared to Cy3 and also plateaus due to completion of hybridization (assay saturation). Both signals then decrease beyond 160 minutes with eventual endpoint stabilization of both Cy3-probe and Cy5-target emission intensities, with final Cy5 signal again greater than that for Cy3 (Figure 2C).
The general trend in real-time kinetics of target binding to surface-tethered probe with dye label (shown in Figure S-3) shows the initial rapid reduction in Cy3-probe emission intensity as target hybridization buffer (Cy5 DNA, 4X SSC) is added over the spot. Emission intensity reduction (over 50% reduction) is observed for both surface array phase (single spot confocal image) and solution phase (fluorescence spectrometry) hybridization involving the Cy3 probe. Previous reports indicate that Cy3 fluorescence quantum yield is highest when attached to the 5’ terminus of single-stranded DNA (i.e., Cy3–5’-ssDNA) and decreases by a factor of 2.4 when the (unlabeled) complementary strand is annealed to form duplex DNA (Cy3–5’-dsDNA).58 Control experiments for Cy3-ssDNA-oligo1 hybridized with target oligo2 (i.e., no Cy5 dye label) confirm this reduction in Cy3 emission intensity from duplex formation (see supplementary Figures S-2 and S-4 for spot and bulk fluorophore behaviors). The observed fluorophore intensity crossover point where Cy5 target signal intensity increases and Cy3 probe signal decreases (i.e., shown in Figures 1 and 2) is speculated to result from FRET at higher degrees of hybridization when fluorophores are duplexed in close proximity.59 Also, final signal saturation reflecting hybridization end points are further confirmed with fluorescence scanning data of spots with homogeneous Cy5 signal (Figure S5). Late time-point reduced Cy3 and Cy5 emission is attributed to FRET, plus dye stacking due to overcrowding, dye-DNA and dye-DNA-dye induced self-quenching and changes in the local fluorophore environments as a function of spot hybridization density.60
Similar experiments performed with 10, 20, and 40µM DNA with Cy3-DNA content (50%) for both Case 1 and 2 targets show the trend depicted in Figure S-6, with no remarkable differences in the crossover and saturation time points for hybridization using Case 1 (antiparallel) and Case 2 (parallel) dye positions on probes vs. targets. Results are discussed in Supporting Information. The assay incubation time (i.e., time to hybridization endpoint stabilization) can be very long (i.e., up to 14h reported9, 20, 61) depending on bulk composition, target concentration, ionic strength and probe grafting density. Incubation times often vary, different even for similar concentrations of probe and target, but depending on study conditions25, 62 and largely influenced by probe density distributions.
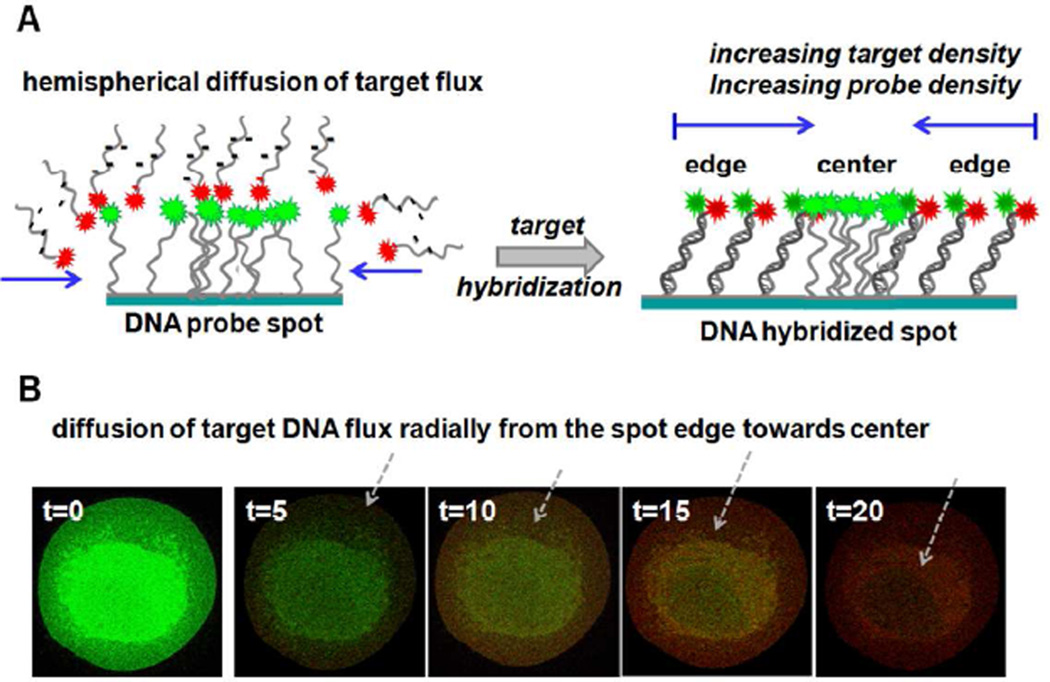
Real-time hybridization imaging of single DNA probe spots evidences for spot fluorescence probe immobilization and density heterogeneities resulting from the probe printing and drying processes.55 Printed dried probe spots often exhibit increased DNA density at spot centers compared to peripheries. This probe heterogeneity has implications for target uptake and resulting spot duplex heterogeneity as a consequence due to local differences in interfacial steric and coulombic properties that affect target-probe pairing efficiencies and kinetics.12, 15 Figures 1 and 2 indicate probe heterogeneity in each hybridized spot upon target introduction. Initially, increased probe DNA signal is evident at spot centers compared to the outer perimeter. Target hybridization therefore occurs from spot edges radially inwards as indicated by increasing Cy5 emission spot intensity profiles (Figure 3). Both non-uniform radial probe density from spot drying55 and target radial transport issues15 produce non-homogeneous target hybridization with time. Target mass transport depends on array format (static vs. flow), target concentration, and its diffusion coefficient.15, 63 Near spot centers, target is delivered to the probe surface solely through linear normal diffusion whereas toward the outer edge, a lateral (radial) component of diffusive flux augments the normal linear transport component, producing hemispherical diffusion.15 This results in spot outer edges accumulating more target through greater proximate fluid volume transport versus spot centers, depicted in Figure 3A schematics. Figure 3B shows the observed increasing target Cy5 emission intensity at the spot periphery initially, increasing radially towards the spot center over time as depicted from the time lapse. This validates the radial planar diffusion mechanism kinetically. It also shows that improving the uniformity of probe density across the spot feature could improve assay signal consistency at any given assay endpoint, even prior to saturation. We recognize this duplex signal imbalance across the spot cannot be mitigated under target transport limitations in the assay, but might be addressed at long assay times approaching equilibrium by producing more uniform spot probe densities sufficiently low so that steric and coulombic duplex hindrances are minimized and only target transport then remains kinetically limiting to signal generation. This is impossible currently in printed/dried spot formats and also could result in assay signal read-out thresholds below those for common scanners.
Figure 3.
Mass transfer-limited diffusion of target binding to immobilized probe in a highly densed heterogeneous DNA spot.15 A) Schematic representation of target DNA radial hybridization progress from the outside edge of the spot to the interior for a single DNA immobilized probe spot. B) Confocal images of real-time hybridization of a single printed DNA spot showing the Cy3–Cy5 overlay emission signal. Target Cy5 intensity increases from the spot edge initially and moves gradually radially into the spot center supporting outside-to-inside mass transport-mediated and probe density-controlled spot hybridization.
Note: microscopy image contrast has been digitally enhanced for the figures to better facilitate direct observation of the optical changes observed over time in situ.
Kinetic modeling analysis was performed for DNA hybridization for both (Case 1) antiparallel and Case 2 (parallel) dye position of the duplex reactions from the confocal spot imaging data for Cy5-shown in Figures 1 and 2 (Figure S-7). Both cases provide reliable fits for first-order reaction surface-capture kinetics (R2>0.996). Probe-printed spots slide containing target in hybridization chambers are fixed and stationary, and analyte fluxes are not large enough to induce bulk flow (i.e., convection flow) and also sealed to prevent evaporative losses. The rate of target diffusion strongly affects hybridization rates and in an unstirred chamber is transport limiting. The hybridization reaction shows pseudo-first-order reaction kinetics when the concentration of bound probe strands is much greater than the concentration of corresponding target mobile strands in solution, similar to experimental conditions adapted in this work and in other previous published models. Previous observations demonstrate first-order and pseudo-first order rate kinetics in time64, 65 and second-order in volume15, 66 for very similar target capture in probe spots.
Real-time in situ results for both Case 1 and 2 hybridizations (see Figures 1 and 2) indicate that irrespective of the relative Cy3–Cy5 duplex orientation, Cy3 emission intensity decreases. The simultaneous increase in Cy5 emission intensity reflects FRET effects. Typically, FRET is most efficient when energy-matched fluorophores are within the Forster radius, typically several nanometers. Additionally, FRET efficiency depends on the energy overlap between donor emission and acceptor excitation spectra, relative orientation of the fluorophore transition dipoles, donor quantum yield, extinction coefficient of the acceptor and the (sixth power of the) distance between fluorophores.67 The 20-mer DNA oligomers used have substantial Cy3–Cy5 FRET potential41, 47 with a theoretical probe and target duplex with dyes in antiparallel position with an end-end distance of 6.8nm. In contrast to model thiolated DNA monolayers on gold surfaces,17,18 DNA microarray spots printed on most commercial array surfaces dry under rapid non-equilibrium conditions, producing a dense, concentrated DNA film on the slide surface with buffer salts and additives and random orientations of many DNA probe layers, sometimes microns in thickness.68 Rinsing however only partially removes contents from this thick film, leaving multilayer DNA spots with random probe orientations. Target DNA analyte then hybridizes to this thick printed film with many differently oriented probes in uncontrolled densities. Irrespective of the labeling position of target Cy5 during hybridization, the abundant duplex opportunities with dense, unoriented probe spots should facilitate FRET. In unstirred solutions, hybridization is observed to occur immediately (within seconds of target addition) and concludes in ~60 minutes. Each single spot scanned using a fluorescence scanner after 60 minutes indicates peak Cy5 intensity and endpoint for hybridization in this format (see Supplementary Information; Figure S-5).
Fluorescence Resonance Energy Transfer (FRET) in DNA microarrays
The printed surface capture array format should produce FRET when Cy3 and Cy5 dyes satisfy Forster conditions as depicted in Figure S-8. For dye-dye distances greater than 10nm, FRET is unlikely to occur (Figure S-8). To confirm the assertion for FRET in the observed Cy3–Cy5 intensity crossover and further reduction in Cy3 emission intensity (see Figures 1C and 2C), FRET signal was compared in both bulk solution phase (i.e., DNA-dye duplexes in solution) using spectrophotometry and surface-capture (i.e., on printed microarrays) with confocal microscopy.
In solution, 5’-Cy3-oligo1-probe duplexes with 5’-Cy5-oligo2-target DNA (Figure 1, Case 1, dyes antiparallel) does not exhibit observable shifts in respective emission wavelengths (λem): duplexed dyes are theoretically approximately 7nm apart). However, when both fluorophores are proximal in parallel duplexes (Figure S-1, Case 2: 5’-Cy3-Oligo-1-probe: Oligo2–3’-Cy5-target, approximate theoretical separation of 2nm, shown in Figure 4A), Cy5 emission intensity increases, characteristic of FRET. Supplementary Figure S-4 shows respective fluorescence excitation and emission spectra for all probe and target oligonucleotides in bulk solution duplex experiments.
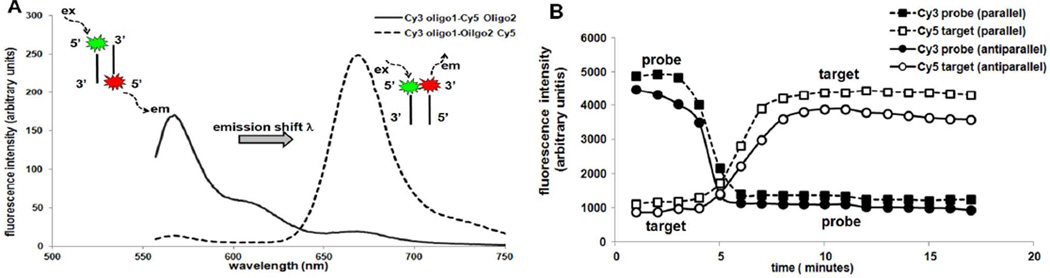
Figure 4.
Comparison of FRET analysis in bulk (solution) and printed array spots on solid substrate for two different orientation of dye (as described in Figure S-1). A) Oligo DNA target Cy5 emission shift indicating FRET in bulk solution duplexes for 5’-Cy3-oligo1-probe:Oligo2–3’-Cy5-target (Case 2 dye on the same side of duplexes indicated here by dotted line) and not for 5’-Cy3-oligo1-probe:5’-Cy5-oligo2-target (Case 1 with anti-parallel dye duplexes indicated by solid line). B) Single spot analysis of fluorescence signals for Cy3 probe and Cy5 target emissions from surface capture assays monitored in real-time by confocal microscopy for 5’-Cy3 oligo1-probe:5’-Cy5-Oligo2-target parallel duplex (dotted lines) and for 5’-Cy3 oligo1-probe:Oligo2–3’-Cy5-target antiparallel duplex (solid lines) indicating FRET with increasing Cy5 emission intensity and simultaneously decrease in Cy3 probe intensity. Solution-phase FRET occurs in (A) for two fluorophores in close proximity (i.e., on same duplex ends) whereas single spot array assays (B) indicated FRET for both duplex dye orientations upon hybridization in printed spots.
On printed arrays, Cy5 target emission intensity increases accompany decreasing Cy3 emission intensity upon addition of complementary Cy5-labeled target analyte to printed Cy3-probes (see Figure 1 and 2). In contrast to bulk duplex FRET studies, altered Cy5 and Cy3 emission is evident for both parallel and anti-parallel dye orientations upon duplexing in printed spots (Figure 4A and 4B). FRET is observed with both antiparallel and parallel dye-dye duplex orientations on arrayed single spot studies (Figure 4B). We attribute this to high probe density in dried multilayer printed spots, with many possible probe arrangements capable of duplexing Cy5-target. This produces high possibilities for energy transfer with multiple nearest neighbors or in duplexes between strands regardless of relative Cy3–Cy5 orientations in the two duplex designs.
Fluorescence resonance energy transfer (FRET) is widely used to report nucleotide structures and dynamics transitions using the Cy3/Cy5 FRET pairing.47, 69–71 Optical complications involving multiple dye relaxation pathways are minimized since the absolute inter-dye distance is well within the FRET range (Forster radius: 1–8nm).51 Moreover, orientational FRET dependence is significant when fluorophores are constrained on solid substrates.50, 71 Previous NMR evidence for Cy3 and Cy5 bound to DNA indicates dye stacking on the DNA helix terminus act as an additional base pair signifying dependence on fluorophore orientation for FRET efficiency. Various factors affect the fluorescence intensity in addition to complexity in analyzing FRET of dye-DNA bound to the surface of high-density probes. With parallel and antiparallel dye positions from probe-target hybridization (seen in real-time hybridization and FRET of DNA spots) we expect a high FRET probability, irrespective of distances between dyes, due to heterogeneity in probe density distribution facilitating abundant duplex opportunities with dense unoriented probes.
Duplex FRET Cy5 acceptor photobleaching
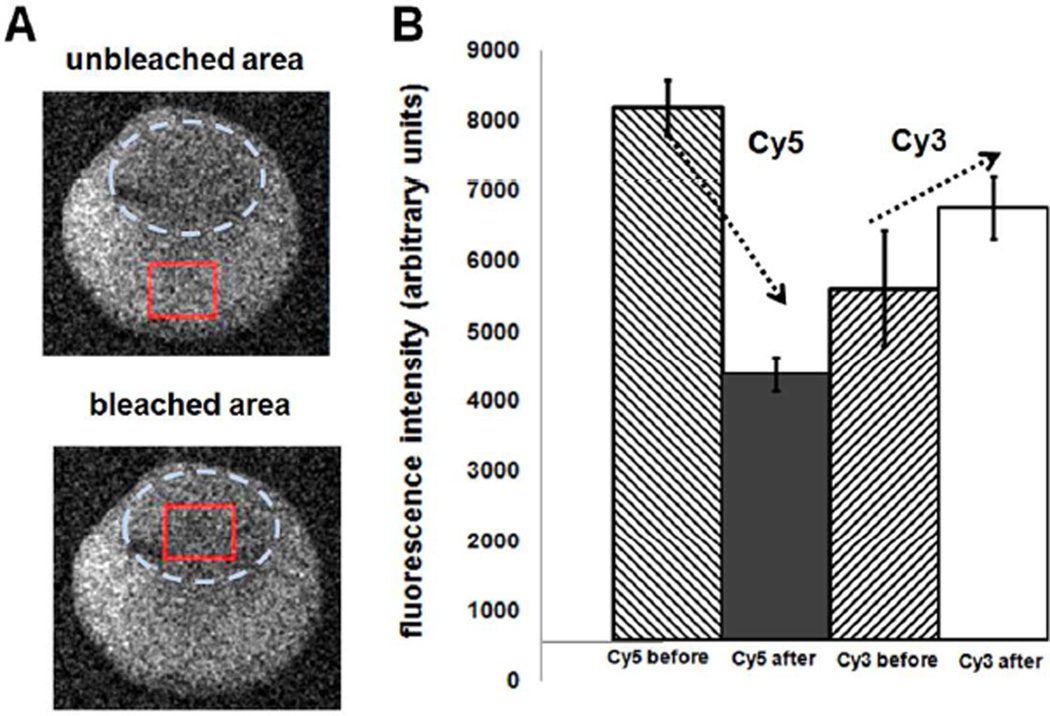
Cy5 acceptor photobleaching in hybridized DNA single spots was used to investigate FRET in spots. This was achieved by comparing donor fluorescence emission intensity in the same printed spot before and after deliberately photolysing the Cy5-photoacceptor on duplexed target. FRET was validated by analyzing initial attenuation of Cy3 emission by energy transfer to Cy5, increasing Cy5 emission. Upon selective Cy5 photobleaching, the Cy5-acceptor no longer accepts Cy3 emission energy. Cy3 emission therefore increases and Cy5 emission is attenuated. FRET efficiency is quantified by FRETeff = 1−(ID/I´D) where ID is donor emission intensity during FRET and I´D is the emission intensity after acceptor photobleaching.51 Photobleaching experiments with DNA spots hybridized with Cy5-oligo2-target in a region of a DNA spot (indicated by circular dotted line, Figure 5A). Cy3 and Cy5 emission intensities in spots were recorded over time and compared with unbleached regions in spots (indicated by the square zone, Figure 5A). FRET efficiency in single hybridized DNA spots, depicted in Figure 5B, is about 18% before Cy5 photobleaching. Target Cy5 emission intensity decreases to approximately 30% with a significant increase in the Cy3 probe emission attributed to Cy5 acceptor photobleaching. These results confirmed FRET in single hybridized DNA spots.
Figure 5.
Photobleaching analysis of FRET signal of a single hybridized DNA spot. Comparison of Cy3 and Cy5 emission intensities in unbleached and bleached regions of a single hybridized DNA microspot. A) Images of both unbleached and bleached areas (dotted oval) of the same DNA spot after confocal local photobleaching. B) Quantification of both Cy3 probe and Cy5 target emission intensities before and after photobleaching (solid squares) showing reduction of Cy5 target signal and increase in Cy3 probe signal once Cy5 fluorophores are photolyzed, consistent with FRET effects.
DNA spot photobleaching depth-dependent studies
To assess Cy5-DNA target penetration into printed probe microspots, photobleaching through the spot thickness in printed DNA spots was performed and both Cy3 and Cy5 emission intensities were collected at each depth (each Z slice ~ 0.5 microns) in single DNA spots. Intensities of Cy3 and Cy5 emission in localized bleached areas in single spots were measured in each Z step (depth). Emission intensities for both Cy3 and Cy5 across a small bleached vs. unbleached area in a single spot are shown in Figure S-9. Regions for analysis were chosen at spot corners since hybridization efficiencies are greater at edges than centrally, with more Cy5-DNA duplexed on spot peripheries as seen in Figure 3. Photobleaching of immobilized Cy5 target fluorophore is indicated by decreasing emission intensity through the Z-series within bleached areas compared to unbleached areas in the same single DNA spot (Figure S-9B). FRET phenomenon is observed in unbleached regions as the Cy5 emission intensity is higher compared to that from Cy3. In the different depth photobleached regions for DNA spots, the overlaid intensities of Cy3 and Cy5 emissions overlap, indicating decreasing Cy5 intensity compared to unbleached Cy3 intensity. Previous reports have shown that Cy5 is more sensitive to photobleaching than Cy3.72 Cy3 dye has remarkable stability against photobleaching; however it is much more sensitive than Cy5 to environmental factors such as temperature and local rigidity.58, 73 To improve assay signal detection and spot sensitivity, higher excitation laser power is preferred. However, increased energy on-array leads to dye bleaching and reduced spot fluorescent signal emission during successive scans. Photobleaching studies reveal specific photolysing of the dyes, penetration of target oligo DNA into the printed probe spot during the hybridization process and FRET in unbleached spots with comparable signal intensities to unbleached spots.
CONCLUSIONS
Fluorescence analysis of individual array-printed DNA microspots using fluorophore-labeled DNA probe-target pairings with confocal microscopy provides important new information regarding the spot molecular environment of immobilized probes, target hybridization processes to substrate-bound probe and aspects of incubation time periods for duplex-forming assays. Single spot image analysis and intensity measurements for immobilized Cy3-oligo DNA probe and Cy5-oligo DNA target compared for two different dye positions on the duplex showed few differences in real-time hybridization time points. The detailed image analysis of real-time hybridization for target DNA confirmed the mass transfer-limited diffusion of target binding to immobilized probe in highly dense but heterogeneous DNA spots. Though real-time hybridization (duplex formation) detection of probe-target is challenging for surface-bound assays, FRET enables real-time monitoring of duplex formation and hybridization rates by monitoring Cy3/Cy5 signal intensities during hybridization processes. Hybridization in solution produced different FRET effects for the 2 different duplexed dye positions in contrast to DNA surface-capture assays that facilitate FRET in both positions in single DNA hybridized spots, attributed to high probe densities. In situ photobleaching of Cy3 and Cy5 within the spot’s cross-section supports heterogeneous probe-target distribution in multilayered-thick printed spots. Overall, new confocal DNA hybridization data provides important new information supporting spot fluorescence signal intensity variations resulting from spot probe printing and drying. Real-time in situ fluorescence image analysis of hybridization supports non-uniform probe distribution density within each spot as a key influence on target binding kinetics and fluorescence signal ambiguities. Microarray assays based on spot printing and target capture should consider these sources of intrinsic variability, and their influence on assay answer reliability.
Supplementary Material
ACKNOWLEDGMENTS
Support from NIH grant EB001473 and assistance with imaging/intensity and data analysis from Dr. K.R. Carney (University of Utah) are both gratefully acknowledged.
REFERENCES
- 1.Stears RL, Martinsky T, Schena M. Nat. Med. 2003;9(1):140–145. doi: 10.1038/nm0103-140. [DOI] [PubMed] [Google Scholar]
- 2.Šášik R, Woelk CH, Corbeil J. J. Mol. Endocrinol. 2004;33(1):1–9. doi: 10.1677/jme.0.0330001. [DOI] [PubMed] [Google Scholar]
- 3.Simon R, Radmacher MD, Dobbin K, McShane LM. J. Natl. Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 4.Wu P, Castner DG, Grainger DW. J. Biomater. Sci. Polym. Ed. 2008;19(6):725–753. doi: 10.1163/156856208784522092. [DOI] [PubMed] [Google Scholar]
- 5.Zou S, He H-J, Zong Y, Shi L, Wang L. DNA Microarrays: Applications, Future Trends, and the Need for Standardization. Standardization and Quality Assurance in Fluorescence Measurements II. 2008;Vol. 6:215–237. Springer Series on Fluorescence. [Google Scholar]
- 6.Shi L, Campbell G, Jones WD, Campagne F, Wen Z. Nat. Biotechnol. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L. Nat. Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zammatteo N, Jeanmart L, Hamels S, Courtois S, Louette P, Hevesi L, Remacle J. Anal. Biochem. 2000;280(1):143–150. doi: 10.1006/abio.2000.4515. [DOI] [PubMed] [Google Scholar]
- 9.Peterson AW, Heaton RJ, Georgiadis RM. Nucleic Acids Res. 2001;29(24):5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou CC, Chen CH, Lee TT, Peck K. Nucleic Acids Res. 2004;32(12) doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan V, Graves DJ, McKenzie SE. Biophys. J. 1995;69(6):2243–2255. doi: 10.1016/S0006-3495(95)80095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai H, Meyer M, Stepaniants S, Ziman M, Stoughton R. Nucleic Acids Res. 2002;30(16 e86):1–8. doi: 10.1093/nar/gnf085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaton RJ, Peterson AW, Georgiadis RM. Proc. Natl. Acad. Sci. U. S. A. 2001;98(7):3701–3704. doi: 10.1073/pnas.071623998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steven W, Metzger MJL, David W. Grainger. Surface chemistries enable application of protein microarray technologies to diagnostics. Manufacturing & Processing Technologies. 2002:10. [Online], http://www.ivdtechnology.com/article/improving-performance-protein-based-microarrays. [Google Scholar]
- 15.Dandy DS, Wu P, Grainger DW. Proc. Natl. Acad. Sci. U. S. A. 2007;104(20):8223–8228. doi: 10.1073/pnas.0606054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levicky R, Herne TM, Tarlov MJ, Satija SK. J. Am. Chem. Soc. 1998;120(38):9787–9792. [Google Scholar]
- 17.Lee CY, Gong P, Harbers GM, Grainger DW, Castner DG, Gamble LJ. Anal. Chem. 2006;78(10):3316–3325. doi: 10.1021/ac052137j. [DOI] [PubMed] [Google Scholar]
- 18.Demers LM, Mirkin CA, Mucic RC, Reynolds Iii RA, Letsinger RL, Elghanian R, Viswanadham G. Anal. Chem. 2000;72(22):5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 19.Gong P, Levicky R. Proc. Natl. Acad. Sci. U. S. A. 2008;105(14):5301–5306. doi: 10.1073/pnas.0709416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassibi A, Vikalo H, Riechmann JL, Hassibi B. Nucleic Acids Res. 2009;37(20 e132):1–12. doi: 10.1093/nar/gkp675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Axelrod D, Wang MD. Biophys. J. 1994;66(3):588–600. doi: 10.1016/s0006-3495(94)80834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vainrub A, Pettitt BM. J. Am. Chem. Soc. 2003;125(26):7798–7799. doi: 10.1021/ja035020q. [DOI] [PubMed] [Google Scholar]
- 23.Vainrub A, Pettitt BM. Phys. Rev. E. Stat. Nonlin. Soft. Matter. Phys. 2002;66(4):041905/1–041905/4. doi: 10.1103/PhysRevE.66.041905. [DOI] [PubMed] [Google Scholar]
- 24.Halperin A, Buhot A, Zhulina EB. J. Phys.: Condens. Matter. 2006;18(18):S463–S490. [Google Scholar]
- 25.Halperin AS, Buhot A, Zhulina EB. Biophys. J. 2004;86(2):718–730. doi: 10.1016/S0006-3495(04)74150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irving D, Gong P, Levicky R. J. Phys. Chem. B. 2010;114(22):7631–7640. doi: 10.1021/jp100860z. [DOI] [PubMed] [Google Scholar]
- 27.Held GA, Grinstein G, Tu Y. Proc. Natl. Acad. Sci. U. S. A. 2003;100(13):7575–7580. doi: 10.1073/pnas.0832500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ota N, Hirano K, Warashina M, Andrus A, Mullah B, Hatanaka K, Taira K. Nucleic Acids Res. 1998;26(3):735–743. doi: 10.1093/nar/26.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mearns FJ, Wong ELS, Short K, Hibbert DB, Gooding JJ. Electroanalysis. 2006;18(19–20):1971–1981. [Google Scholar]
- 30.Ambia-Garrido J, Vainrub A, Pettitt BM. Comput. Phys. Commun. 2010;181(12):2001–2007. doi: 10.1016/j.cpc.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morriss-Andrews A, Rottler J, Plotkin SS. J. Chem. Phys. 2010;132(3) doi: 10.1063/1.3269994. [DOI] [PubMed] [Google Scholar]
- 32.Shen G, Tercero N, Gaspard MA, Varughese B, Shepard K, Levicky R. J. Am. Chem. Soc. 2006;128(26):8427–8433. doi: 10.1021/ja0571500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halperin A, Buhot A, Zhulina EB. Biophys. J. 2005;89(2):796–811. doi: 10.1529/biophysj.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassalle N, Vieil E, Correia JP, Abrantes LM. Biosens. Bioelectron. 2001;16(4–5):295–303. doi: 10.1016/s0956-5663(01)00133-6. [DOI] [PubMed] [Google Scholar]
- 35.Fritz J, Cooper EB, Gaudet S, Sorger PK, Manalis SR. Proc. Natl. Acad. Sci. U. S. A. 2002;99(22):14142–14146. doi: 10.1073/pnas.232276699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abel AP, Weller MG, Duveneck GL, Ehrat M, Widmer HM. Anal. Chem. 1996;68(17):2905–2912. doi: 10.1021/ac960071+. [DOI] [PubMed] [Google Scholar]
- 37.Peterlinz KA, Georgiadis RM, Herne TM, Tarlov MJ. J. Am. Chem. Soc. 1997;119(14):3401–3402. [Google Scholar]
- 38.Wang J, Cai X, Tian B, Shiraishi H. Analyst. 1996;121(7):965–970. [Google Scholar]
- 39.Siontorou CG, Nikolelis DP, Miernik A, Krull UJ. Electrochim. Acta. 1998;43(23):3611–3617. [Google Scholar]
- 40.Gaylord BS, Heeger AJ, Bazan GC. J. Am. Chem. Soc. 2003;125(4):896–900. doi: 10.1021/ja027152+. [DOI] [PubMed] [Google Scholar]
- 41.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM. J. Proc. Natl. Acad. Sci. U. S. A. 2008;105(32):11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwille P, Oehlenschlager F, Walter NG. Biochemistry (Mosc) 1996;35(31):10182–10193. doi: 10.1021/bi960517g. [DOI] [PubMed] [Google Scholar]
- 43.Nie S, Chiu DT, Zare RN. Anal. Chem. 1995;67(17):2849–2857. [Google Scholar]
- 44.Clegg RM. Rev. Mol. Biotechnol. 2002;82(3):177–179. [PubMed] [Google Scholar]
- 45.Sapsford KE, Berti L, Medintz IL. Angewandte Chemie - International Edition. 2006;45(28):4562–4588. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 46.Clegg RM. Curr. Opin. Biotechnol. 1995;6(1):103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 47.Clegg RM, Murchie AIH, Zechel A, Lilley DM. J. Proc. Natl. Acad. Sci. U. S. A. 1993;90(7):2994–2998. doi: 10.1073/pnas.90.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dale RE, Eisinger J, Blumberg WE. Biophys. J. 1979;26(2):161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stryer L. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 50.VanBeek DB, Zwier MC, Shorb JM, Krueger BP. Biophys. J. 2007;92(12):4168–4178. doi: 10.1529/biophysj.106.092650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabanayagam CR, Eid JS, Meller A. J. Chem. Phys. 2005;122(6):061103-1–061103-5. doi: 10.1063/1.1854120. [DOI] [PubMed] [Google Scholar]
- 52.Dinant C, Van Royen ME, Vermeulen W, Houtsmuller AB. J. Microsc. 2008;231(1):97–104. doi: 10.1111/j.1365-2818.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 53.Widengren J, Schwille P. J. Phys. Chem. A. 2000;104(27):6416–6428. [Google Scholar]
- 54.Leung YF, Cavalieri D. Trends Genet. 2003;19(11):649–659. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 55.Rao AN, Vandencasteele N, Gamble LJ, Grainger DW. Anal. Chem. 2012 doi: 10.1021/ac3019334. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22(20):1859–1862. [Google Scholar]
- 57.Gong P, Grainger DW. Surf. Sci. 2004;570(1–2):67–77. [Google Scholar]
- 58.Sanborn ME, Connolly BK, Gurunathan K, Levitus M. J. Phys. Chem. B. 2007;111(37):11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- 59.Didenko VV. Biotechniques. 2001;31(5):1106–1121. doi: 10.2144/01315rv02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregory Cox W, Beaudet MP, Agnew JY, Ruth JL. Anal. Biochem. 2004;331(2):243–254. doi: 10.1016/j.ab.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Toegl A, Kirchner R, Gauer C, Wixforth A. J. Biomol. Tech. 2003;14(3):197–204. [PMC free article] [PubMed] [Google Scholar]
- 62.Carletti E, Guerra E, Alberti S. Trends Biotechnol. 2006;24(10):443–448. doi: 10.1016/j.tibtech.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Erickson D, Liu X, Krull U, Li D. Anal. Chem. 2004;76(24):7269–7277. doi: 10.1021/ac049396d. [DOI] [PubMed] [Google Scholar]
- 64.Stillman BA, Tonkinson JL. Anal. Biochem. 2001;295(2):149–157. doi: 10.1006/abio.2001.5212. [DOI] [PubMed] [Google Scholar]
- 65.Gadgil C, Yeckel A, Derby JJ, Hu WS. J. Biotechnol. 2004;114(1–2):31–45. doi: 10.1016/j.jbiotec.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Bishop J, Blair S, Chagovetz AM. Biophys. J. 2006;90(3):831–840. doi: 10.1529/biophysj.105.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Piston DW, Kremers GJ. Trends Biochem. Sci. 2007;32(9):407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Dugas V, Broutin J, Souteyrand E. Langmuir. 2005;21(20):9130–9136. doi: 10.1021/la050764y. [DOI] [PubMed] [Google Scholar]
- 69.Ha T. Methods. 2001;25(1):78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 70.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Biophys. J. 2004;86(4):2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Selvin PR. Nat. Struct. Biol. 2000;7(9):730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 72.Bilban M, Buehler LK, Head S, Desoye G, Quaranta V. Curr. Issues Mol. Biol. 2002;4(2):57–64. [PubMed] [Google Scholar]
- 73.Aramendia PF, Martin Negri R, San Roman E. J. Phys. Chem. 1994;98(12):3165–3173. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.