Abstract
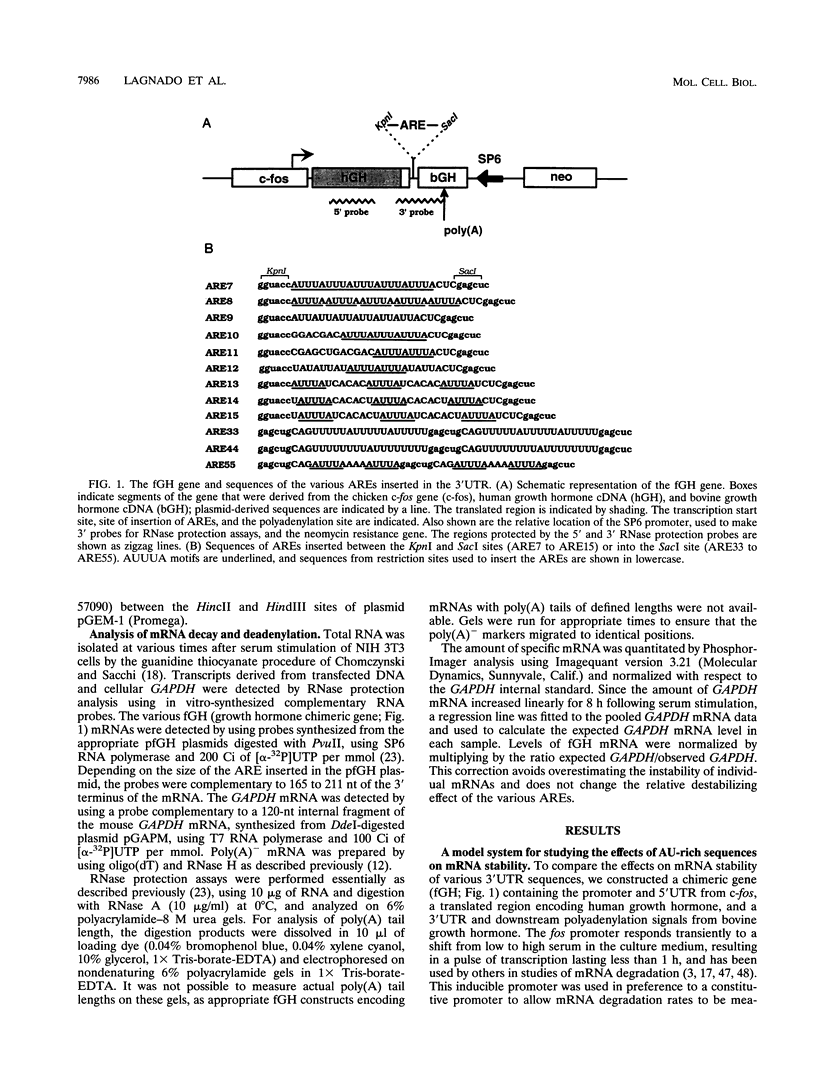
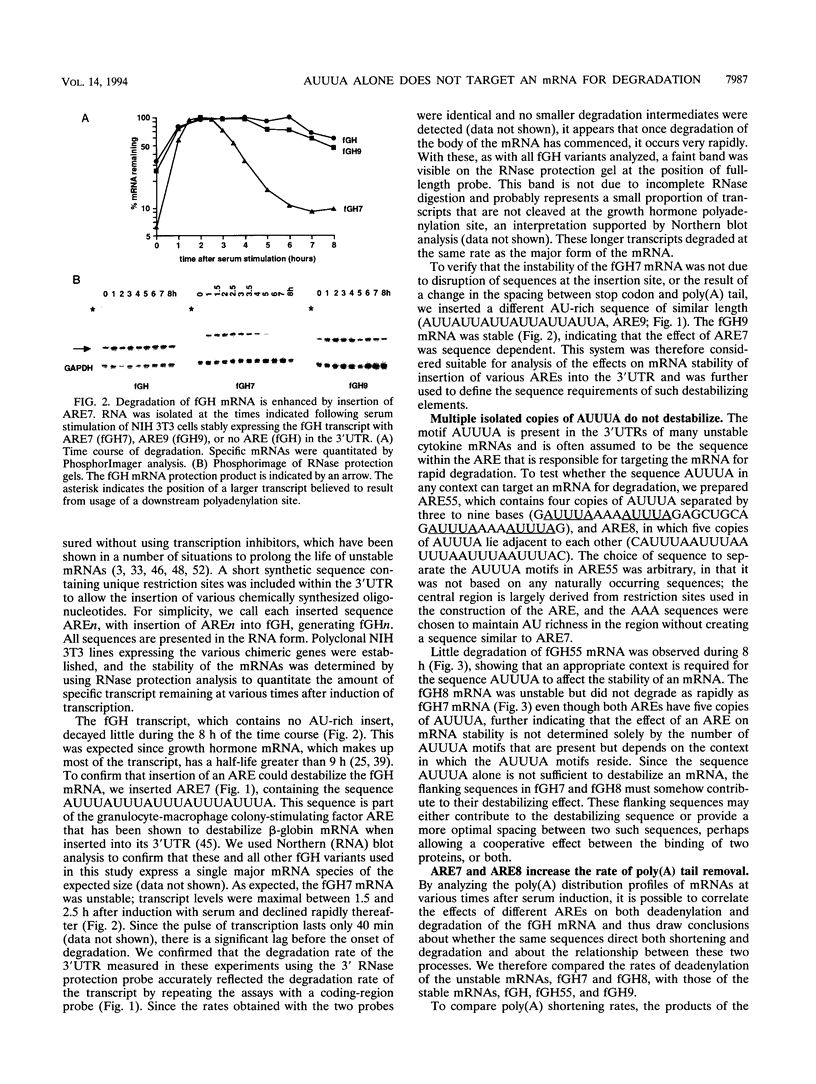
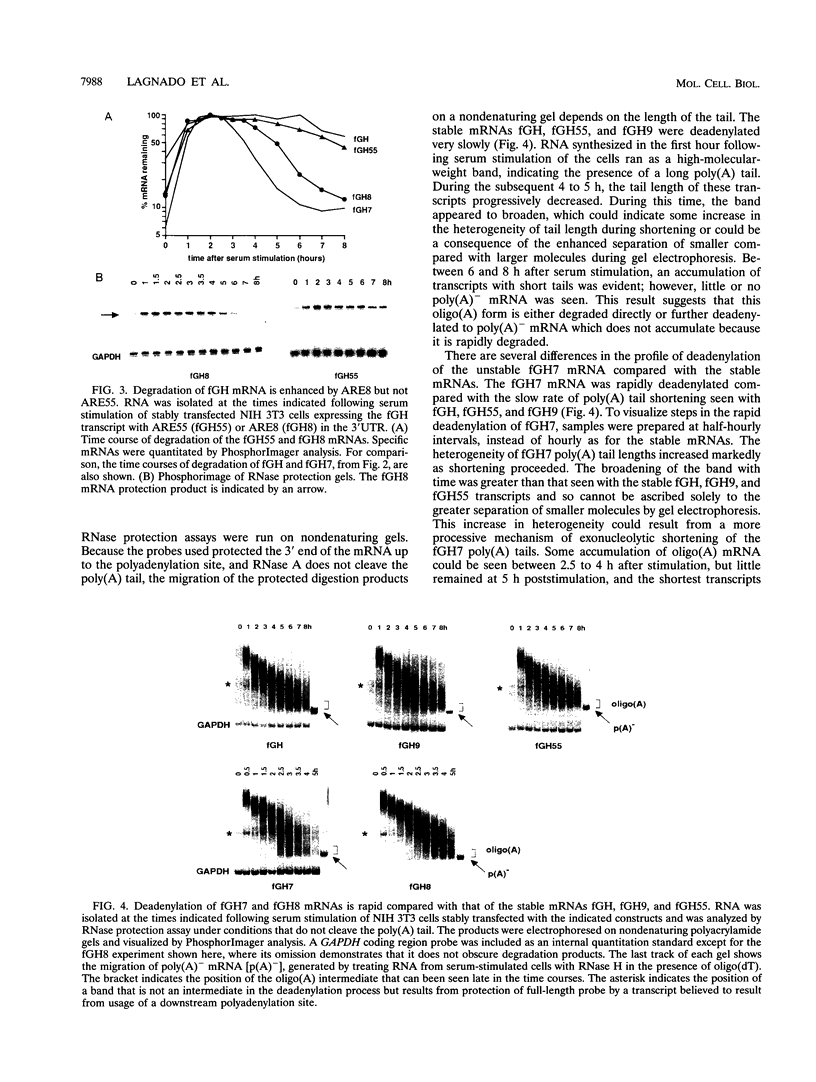
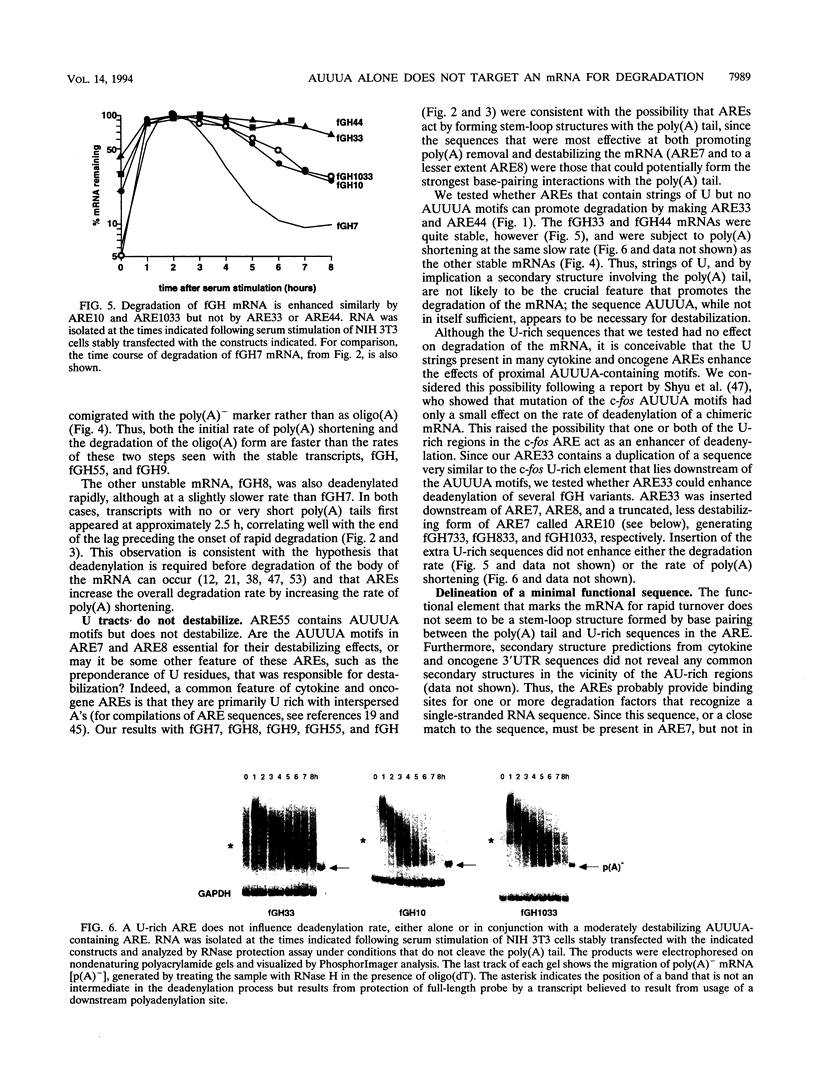
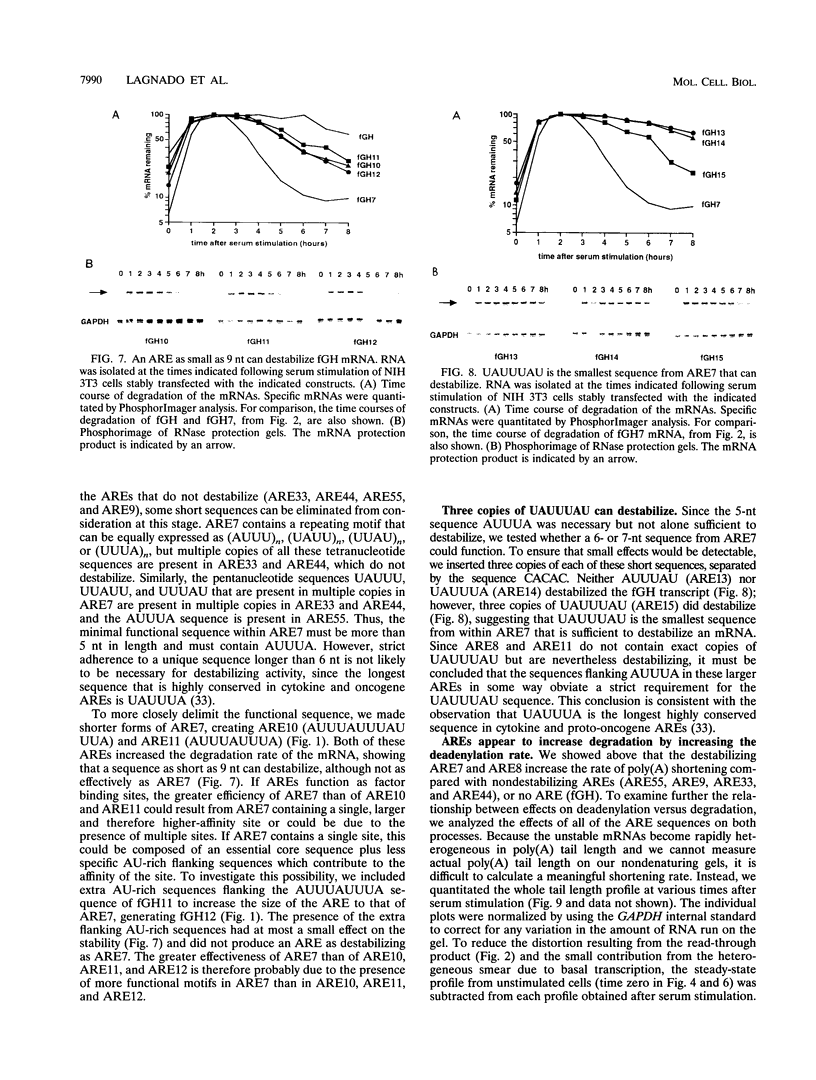
AU-rich elements (AREs) in the 3' untranslated regions of several cytokine and oncogene mRNAs have been shown to function as signals for rapid mRNA degradation, and it is assumed that the many other cytokine and oncogene mRNAs that contain AU-rich sequences in the 3' untranslated region are similarly targeted for rapid turnover. We have used a chimeric gene composed mostly of growth hormone sequences with expression driven by the c-fos promoter to investigate the minimal sequence required to act as a functional destabilizing element and to monitor the effect of these sequences on early steps in the degradation pathway. We find that neither AUUUA, UAUUUA, nor AUUUAU can function as a destabilizing element. However, the sequence UAUUUAU, when present in three copies, is sufficient to destabilize a chimeric mRNA. We propose that this sequence functions by virtue of being a sufficient portion of the larger sequence, UUAUUUA(U/A)(U/A), that we propose forms the optimal binding site for a destabilizing factor. The destabilizing effect depends on the number of copies of this proposed binding site and their degree of mismatch in the first two and last two positions, with mismatches in the AUUUA sequence not being tolerated. We found a strict correlation between the effect of an ARE on degradation rate and the effect on the rate of poly(A) shortening, consistent with deadenylation being the first and rate-limiting step in degradation, and the step stimulated by destabilizing AREs. Deadenylation was observed to occur in at least two phases, with an oligo(A) intermediate transiently accumulating, consistent with the suggestion that the degradation processes may be similar in yeast and mammalian cells. AREs that are especially U rich and contain no UUAUUUA(U/A)(U/A) motifs failed to influence the degradation rate or the deadenylation rate, either when downstream of suboptimal destabilizing AREs or when alone.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aghib D. F., Bishop J. M., Ottolenghi S., Guerrasio A., Serra A., Saglio G. A 3' truncation of MYC caused by chromosomal translocation in a human T-cell leukemia increases mRNA stability. Oncogene. 1990 May;5(5):707–711. [PubMed] [Google Scholar]
- Aharon T., Schneider R. J. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3' noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol. 1993 Mar;13(3):1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern S. M., Miyata T., Sadler J. E. Regulation of human tissue factor expression by mRNA turnover. J Biol Chem. 1993 Jan 25;268(3):2154–2159. [PubMed] [Google Scholar]
- Akashi M., Loussararian A. H., Adelman D. C., Saito M., Koeffler H. P. Role of lymphotoxin in expression of interleukin 6 in human fibroblasts. Stimulation and regulation. J Clin Invest. 1990 Jan;85(1):121–129. doi: 10.1172/JCI114401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aström J., Aström A., Virtanen A. Properties of a HeLa cell 3' exonuclease specific for degrading poly(A) tails of mammalian mRNA. J Biol Chem. 1992 Sep 5;267(25):18154–18159. [PubMed] [Google Scholar]
- Bernstein P., Peltz S. W., Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989 Feb;9(2):659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel M., Iwai Y., Pluznik D. H., Cohen R. B. Binding of sequence-specific proteins to the adenosine- plus uridine-rich sequences of the murine granulocyte/macrophage colony-stimulating factor mRNA. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10001–10005. doi: 10.1073/pnas.89.21.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder R., Hwang S. P., Ratnasabapathy R., Williams D. L. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5'-AAU-3'/5'-UAA-3' elements in single-stranded loop domains of the 3'-noncoding region. J Biol Chem. 1989 Oct 5;264(28):16910–16918. [PubMed] [Google Scholar]
- Bohjanen P. R., Petryniak B., June C. H., Thompson C. B., Lindsten T. AU RNA-binding factors differ in their binding specificities and affinities. J Biol Chem. 1992 Mar 25;267(9):6302–6309. [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Brewer G., Ross J. Poly(A) shortening and degradation of the 3' A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988 Apr;8(4):1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. D., Zipkin I. D., Harland R. M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993 Aug;7(8):1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J. L., Koeller D. M., Ramin V. C., Klausner R. D., Harford J. B. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 1989 Dec 1;8(12):3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Chen T. M., Shyu A. B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994 Jan;14(1):416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Coutts M., Brawerman G. A 5' exoribonuclease from cytoplasmic extracts of mouse sarcoma 180 ascites cells. Biochim Biophys Acta. 1993 Apr 29;1173(1):57–62. doi: 10.1016/0167-4781(93)90242-6. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993 Aug;7(8):1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Gillis P., Malter J. S. The adenosine-uridine binding factor recognizes the AU-rich elements of cytokine, lymphokine, and oncogene mRNAs. J Biol Chem. 1991 Feb 15;266(5):3172–3177. [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Hamilton B. J., Nagy E., Malter J. S., Arrick B. A., Rigby W. F. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993 Apr 25;268(12):8881–8887. [PubMed] [Google Scholar]
- Helms S. R., Rottman F. M. Characterization of an inducible promoter system to investigate decay of stable mRNA molecules. Nucleic Acids Res. 1990 Jan 25;18(2):255–259. doi: 10.1093/nar/18.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis G. F., Gazdar A. F., Bertness V., Kirsch I. R. Complex translocation disrupts c-myc regulation in a human plasma cell myeloma. Mol Cell Biol. 1988 Jan;8(1):124–129. doi: 10.1128/mcb.8.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. Y., Tholanikunnel B. G., Vakalopoulou E., Malbon C. C. The M(r) 35,000 beta-adrenergic receptor mRNA-binding protein induced by agonists requires both an AUUUA pentamer and U-rich domains for RNA recognition. J Biol Chem. 1993 Dec 5;268(34):25769–25775. [PubMed] [Google Scholar]
- Jackson R. J., Standart N. Do the poly(A) tail and 3' untranslated region control mRNA translation? Cell. 1990 Jul 13;62(1):15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- Jones T. R., Cole M. D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987 Dec;7(12):4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabnick K. S., Housman D. E. Determinants that contribute to cytoplasmic stability of human c-fos and beta-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988 Aug;8(8):3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeffler H. P., Gasson J., Tobler A. Transcriptional and posttranscriptional modulation of myeloid colony-stimulating factor expression by tumor necrosis factor and other agents. Mol Cell Biol. 1988 Aug;8(8):3432–3438. doi: 10.1128/mcb.8.8.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski J., Denhardt D. T. Regulation of the mRNA for monocyte-derived neutrophil-activating peptide in differentiating HL60 promyelocytes. Mol Cell Biol. 1989 May;9(5):1946–1957. doi: 10.1128/mcb.9.5.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa I. A., de Wit C. L., Elfferich P., van der Eb A. J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990 Dec;10(12):6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman A. P., Pitha P. M., Shin M. L. Poly(A) removal is the kinase-regulated step in tumor necrosis factor mRNA decay. J Biol Chem. 1992 Feb 5;267(4):2123–2126. [PubMed] [Google Scholar]
- Lowell J. E., Rudner D. Z., Sachs A. B. 3'-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992 Nov;6(11):2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Meijlink F., Curran T., Miller A. D., Verma I. M. Removal of a 67-base-pair sequence in the noncoding region of protooncogene fos converts it to a transforming gene. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4987–4991. doi: 10.1073/pnas.82.15.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992 Nov;6(11):2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Paek I., Axel R. Glucocorticoids enhance stability of human growth hormone mRNA. Mol Cell Biol. 1987 Apr;7(4):1496–1507. doi: 10.1128/mcb.7.4.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppel K., Baglioni C. Deadenylation and turnover of interferon-beta mRNA. J Biol Chem. 1991 Apr 15;266(11):6663–6666. [PubMed] [Google Scholar]
- Raj N. B., Pitha P. M. 65-kDa protein binds to destabilizing sequences in the IFN-beta mRNA coding and 3' UTR. FASEB J. 1993 May;7(8):702–710. doi: 10.1096/fasebj.7.8.8500695. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Schiavi S. C., Belasco J. G., Greenberg M. E. Regulation of proto-oncogene mRNA stability. Biochim Biophys Acta. 1992 Dec 16;1114(2-3):95–106. doi: 10.1016/0304-419x(92)90009-n. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shaw J., Meerovitch K., Bleackley R. C., Paetkau V. Mechanisms regulating the level of IL-2 mRNA in T lymphocytes. J Immunol. 1988 Apr 1;140(7):2243–2248. [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Greenberg M. E., Belasco J. G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989 Jan;3(1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- Stoeckle M. Y., Guan L. High-resolution analysis of gro alpha mRNA poly(A) shortening: regulation by interleukin-1 beta. Nucleic Acids Res. 1993 Apr 11;21(7):1613–1617. doi: 10.1093/nar/21.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P., Raué H. A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3'-terminal part of the coding region. Mol Cell Biol. 1992 Jul;12(7):2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore L. A., Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990 Apr;10(4):1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3' AU-rich sequences. Nature. 1988 Nov 24;336(6197):396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- Wisdom R., Lee W. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991 Feb;5(2):232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wagner B. J., Ehrenman K., Schaefer A. W., DeMaria C. T., Crater D., DeHaven K., Long L., Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993 Dec;13(12):7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]