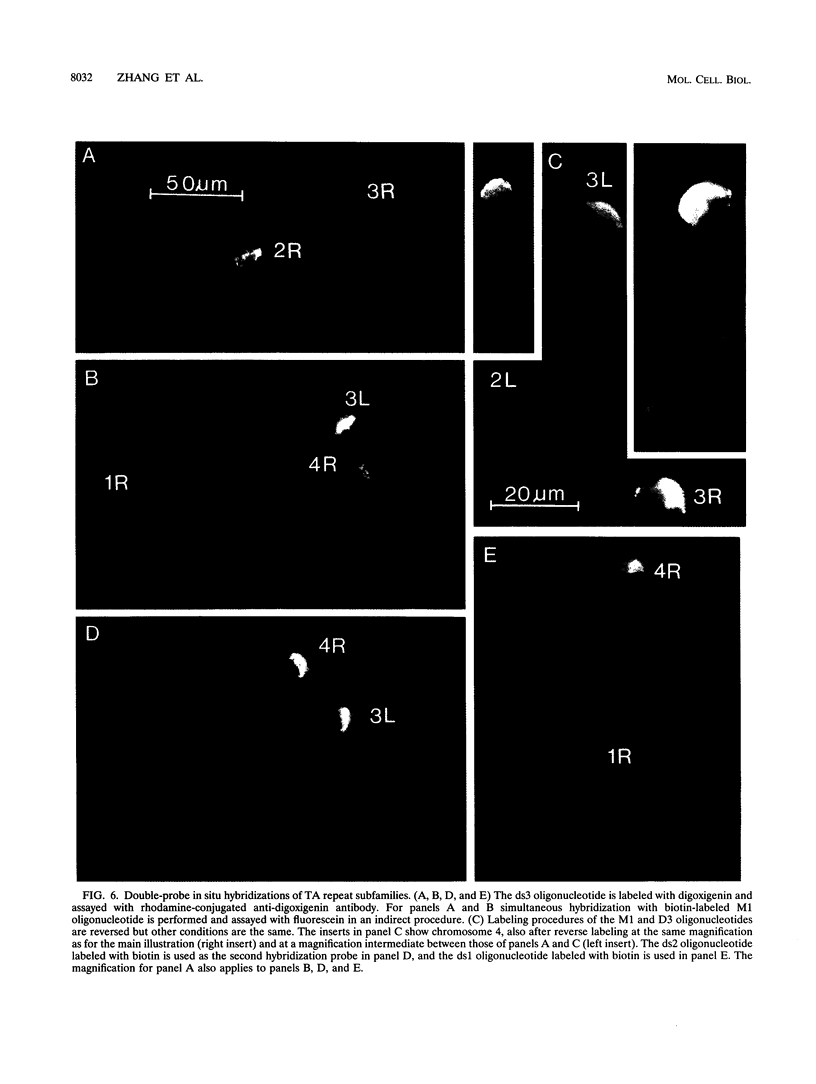
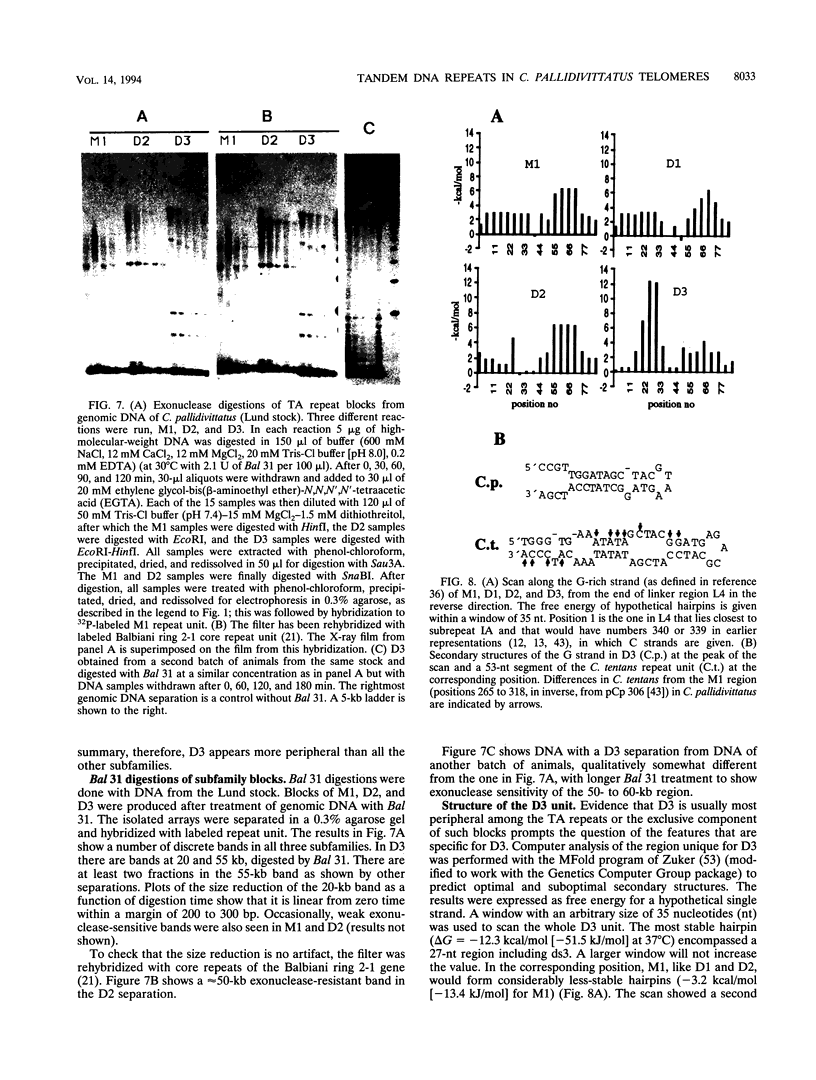
Abstract
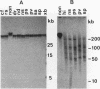
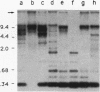
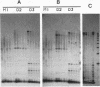
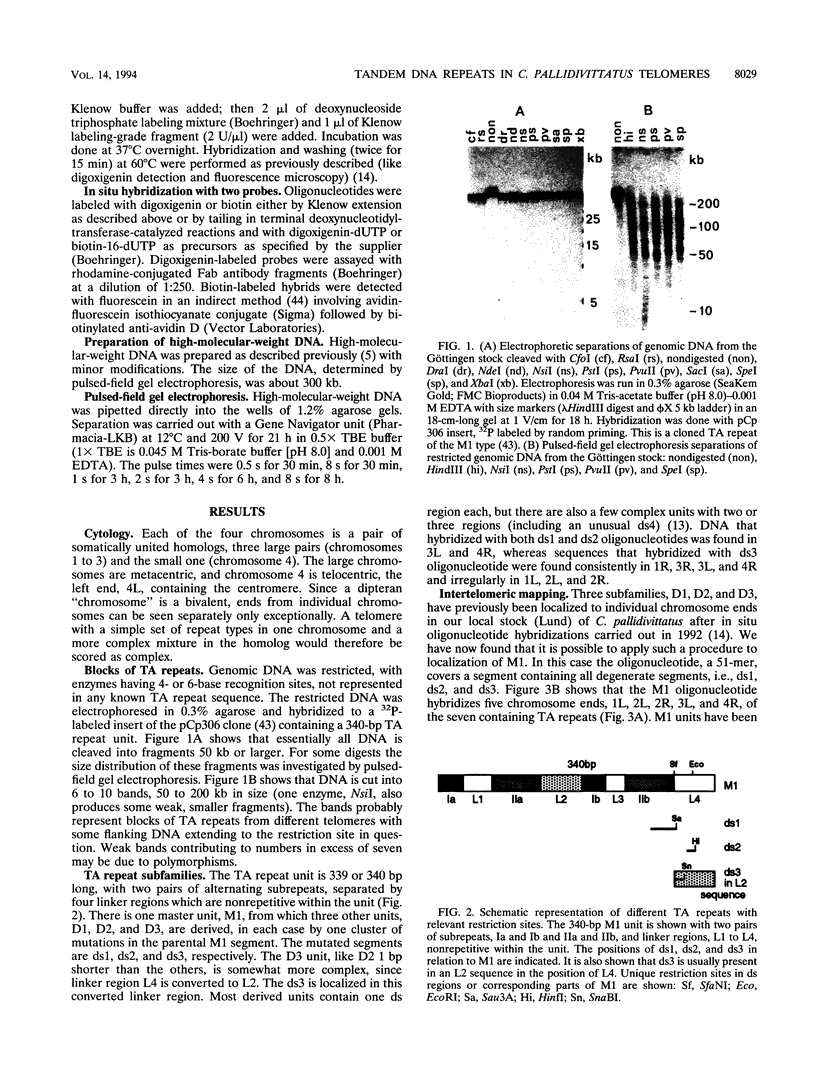
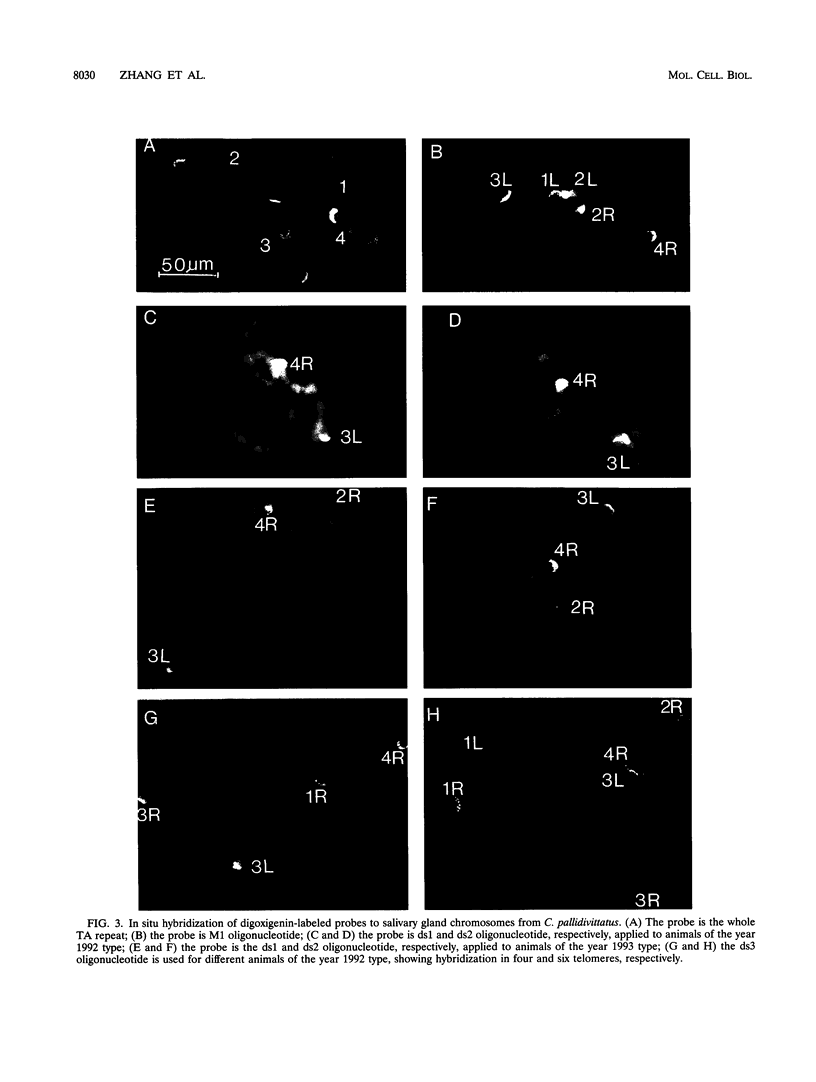
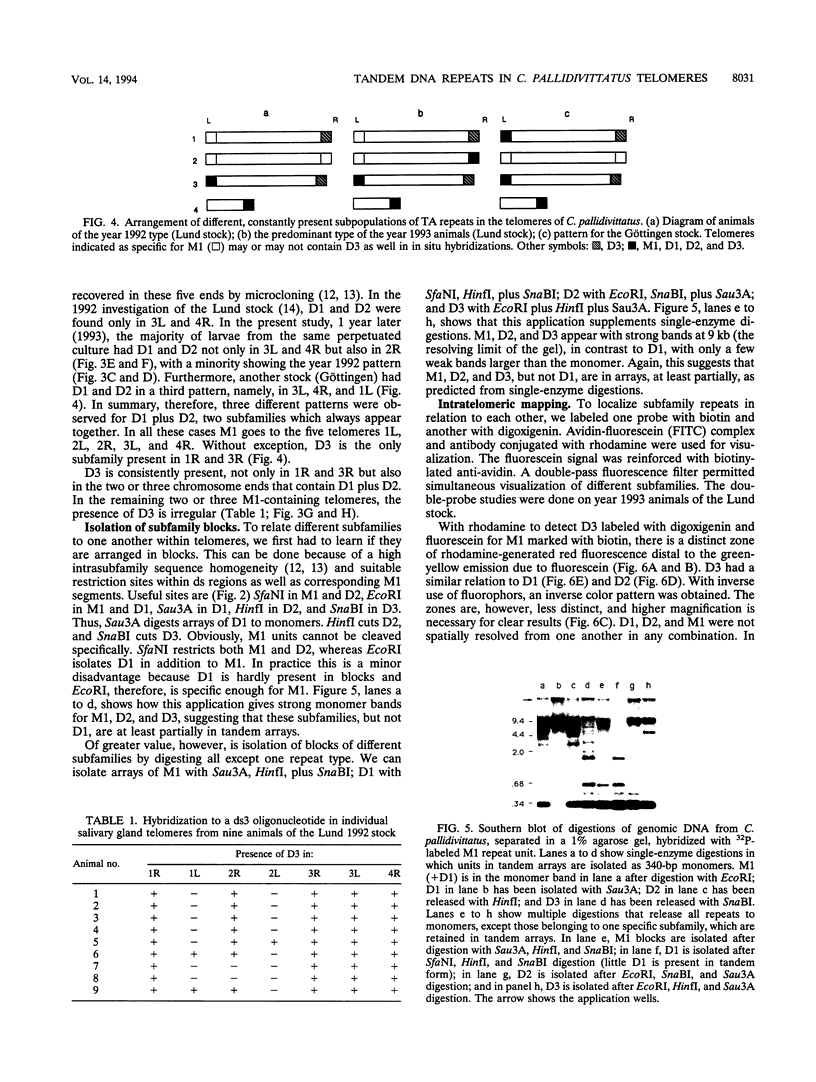
A family of 340-bp tandem telomere-associated DNA repeats is present in 50- to 200-kb blocks in seven of the eight paired chromosome ends in Chironomus pallidivittatus. It consists of four main subfamilies, differing from each other by small clusters of mutations. This differentiation may reflect different functional roles for the repeats. Here we find that one subfamily, D3, is consistently localized most peripherally and extends close to the ends of the chromosomes, as shown by its sensitivity to the exonuclease Bal 31. The amounts of D3 are highly variable between individuals. The repeat characteristic for D3 forms a segment with pronounced dyad symmetry, which in single-strand form would give rise to a hairpin. Evidence from an interspecies comparison suggests that a similar structure is the result of selective forces. Another subfamily, M1, is present more proximally in a subgroup of telomeres characterized by a special kind of repeat variability. Thus, a complex block with three kinds of subfamilies may occupy different M1 telomeres depending on the stock of animals. We conclude that subfamilies are differentially distributed between and within telomeres and are likely to serve different functions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biessmann H., Champion L. E., O'Hair M., Ikenaga K., Kasravi B., Mason J. M. Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J. 1992 Dec;11(12):4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H., Mason J. M., Ferry K., d'Hulst M., Valgeirsdottir K., Traverse K. L., Pardue M. L. Addition of telomere-associated HeT DNA sequences "heals" broken chromosome ends in Drosophila. Cell. 1990 May 18;61(4):663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Mason J. M. Genetics and molecular biology of telomeres. Adv Genet. 1992;30:185–249. doi: 10.1016/s0065-2660(08)60321-1. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Valgeirsdottir K., Lofsky A., Chin C., Ginther B., Levis R. W., Pardue M. L. HeT-A, a transposable element specifically involved in "healing" broken chromosome ends in Drosophila melanogaster. Mol Cell Biol. 1992 Sep;12(9):3910–3918. doi: 10.1128/mcb.12.9.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Challoner P. B. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell. 1984 Feb;36(2):447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Gall J. G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978 Mar 25;120(1):33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. Structure and function of telomeres. Nature. 1991 Apr 18;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- Brown W. R., MacKinnon P. J., Villasanté A., Spurr N., Buckle V. J., Dobson M. J. Structure and polymorphism of human telomere-associated DNA. Cell. 1990 Oct 5;63(1):119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- Choi K. H., Choi B. S. Formation of a hairpin structure by telomere 3' overhang. Biochim Biophys Acta. 1994 Apr 6;1217(3):341–344. doi: 10.1016/0167-4781(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Cohn M., Edström J. E. Chromosome ends in Chironomus pallidivittatus contain different subfamilies of telomere-associated repeats. Chromosoma. 1992 Oct;101(10):634–640. doi: 10.1007/BF00360541. [DOI] [PubMed] [Google Scholar]
- Cohn M., Edström J. E. Evolutionary relations between subtypes of telomere-associated repeats in Chironomus. J Mol Evol. 1991 Jun;32(6):463–468. doi: 10.1007/BF02102648. [DOI] [PubMed] [Google Scholar]
- Cohn M., Edström J. E. Telomere-associated repeats in Chironomus form discrete subfamilies generated by gene conversion. J Mol Evol. 1992 Aug;35(2):114–122. doi: 10.1007/BF00183222. [DOI] [PubMed] [Google Scholar]
- Corcoran L. M., Thompson J. K., Walliker D., Kemp D. J. Homologous recombination within subtelomeric repeat sequences generates chromosome size polymorphisms in P. falciparum. Cell. 1988 Jun 3;53(5):807–813. doi: 10.1016/0092-8674(88)90097-9. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O. N., Petrov D. A., Pavlova M. N., Koga A., Kurenova E. V., Hartl D. L. A repetitive DNA element, associated with telomeric sequences in Drosophila melanogaster, contains open reading frames. Chromosoma. 1992 Dec;102(1):32–40. doi: 10.1007/BF00352288. [DOI] [PubMed] [Google Scholar]
- Dore E., Pace T., Ponzi M., Picci L., Frontali C. Organization of subtelomeric repeats in Plasmodium berghei. Mol Cell Biol. 1990 May;10(5):2423–2427. doi: 10.1128/mcb.10.5.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Cech T. R. The beta subunit of Oxytricha telomere-binding protein promotes G-quartet formation by telomeric DNA. Cell. 1993 Sep 10;74(5):875–885. doi: 10.1016/0092-8674(93)90467-5. [DOI] [PubMed] [Google Scholar]
- Foote S. J., Kemp D. J. Chromosomes of malaria parasites. Trends Genet. 1989 Oct;5(10):337–342. doi: 10.1016/0168-9525(89)90139-x. [DOI] [PubMed] [Google Scholar]
- Galler R., Saiga H., Widmer R. M., Lezzi M., Edström J. E. Two genes in Balbiani ring 2 with metabolically different 75S transcripts. EMBO J. 1985 Nov;4(11):2977–2982. doi: 10.1002/j.1460-2075.1985.tb04032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989 Jan 26;337(6205):331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985 Dec;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Horowitz H., Thorburn P., Haber J. E. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger D., Philippsen P. Many yeast chromosomes lack the telomere-specific Y' sequence. Mol Cell Biol. 1989 Dec;9(12):5754–5757. doi: 10.1128/mcb.9.12.5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. M. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993 Dec 17;75(6):1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- Louis E. J., Haber J. E. Mitotic recombination among subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics. 1990 Mar;124(3):547–559. doi: 10.1093/genetics/124.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Haber J. E. The structure and evolution of subtelomeric Y' repeats in Saccharomyces cerevisiae. Genetics. 1992 Jul;131(3):559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Haber J. E. The subtelomeric Y' repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990 Mar;124(3):533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994 Mar;136(3):789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E. H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993 Apr 23;73(2):347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- McEachern M. J., Blackburn E. H. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin G. B., Cech T. R. Mitochondrial telomeres: surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell. 1988 Feb 12;52(3):367–374. doi: 10.1016/s0092-8674(88)80029-1. [DOI] [PubMed] [Google Scholar]
- Morin G. B., Cech T. R. The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell. 1986 Sep 12;46(6):873–883. doi: 10.1016/0092-8674(86)90069-3. [DOI] [PubMed] [Google Scholar]
- Moyzis R. K., Buckingham J. M., Cram L. S., Dani M., Deaven L. L., Jones M. D., Meyne J., Ratliff R. L., Wu J. R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L., Edström J. E. Complex telomere-associated repeat units in members of the genus Chironomus evolve from sequences similar to simple telomeric repeats. Mol Cell Biol. 1993 Mar;13(3):1583–1589. doi: 10.1128/mcb.13.3.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L., Schmidt E. R., Edström J. E. Subrepeats result from regional DNA sequence conservation in tandem repeats in Chironomus telomeres. J Mol Biol. 1990 Dec 5;216(3):577–584. doi: 10.1016/0022-2836(90)90385-Y. [DOI] [PubMed] [Google Scholar]
- Okazaki S., Tsuchida K., Maekawa H., Ishikawa H., Fujiwara H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol Cell Biol. 1993 Mar;13(3):1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A. F., Zakian V. A. Recombination occurs during telomere formation in yeast. Nature. 1989 Feb 2;337(6206):429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- Raghuraman M. K., Cech T. R. Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 1990 Aug 11;18(15):4543–4552. doi: 10.1093/nar/18.15.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz-Pohl R., Bialojan S. A DNA sequence of Drosophila melanogaster with a differential telomeric distribution. Chromosoma. 1984;89(3):206–211. doi: 10.1007/BF00295001. [DOI] [PubMed] [Google Scholar]
- Rovira C., Beermann W., Edström J. E. A repetitive DNA sequence associated with the centromeres of Chironomus pallidivittatus. Nucleic Acids Res. 1993 Apr 25;21(8):1775–1781. doi: 10.1093/nar/21.8.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga H., Edström J. E. Long tandem arrays of complex repeat units in Chironomus telomeres. EMBO J. 1985 Mar;4(3):799–804. doi: 10.1002/j.1460-2075.1985.tb03700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Köhler M., Vogt P., von Malsch K., Schweizer D. Chromosomal in situ hybridization with double-labeled DNA: signal amplification at the probe level. Cytogenet Cell Genet. 1992;60(1):4–7. doi: 10.1159/000133282. [DOI] [PubMed] [Google Scholar]
- Traverse K. L., Pardue M. L. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8116–8120. doi: 10.1073/pnas.85.21.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgeirsdóttir K., Traverse K. L., Pardue M. L. HeT DNA: a family of mosaic repeated sequences specific for heterochromatin in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7998–8002. doi: 10.1073/pnas.87.20.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselkov A. G., Malkov V. A., Frank-Kamenetskll M. D., Dobrynin V. N. Triplex model of chromosome ends. Nature. 1993 Aug 5;364(6437):496–496. doi: 10.1038/364496a0. [DOI] [PubMed] [Google Scholar]
- Walmsley R. W., Chan C. S., Tye B. K., Petes T. D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984 Jul 12;310(5973):157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- Young B. S., Pession A., Traverse K. L., French C., Pardue M. L. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983 Aug;34(1):85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- Zahler A. M., Williamson J. R., Cech T. R., Prescott D. M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991 Apr 25;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Blanton H. M. Distribution of telomere-associated sequences on natural chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2257–2260. doi: 10.1128/mcb.8.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- de Lange T., Shiue L., Myers R. M., Cox D. R., Naylor S. L., Killery A. M., Varmus H. E. Structure and variability of human chromosome ends. Mol Cell Biol. 1990 Feb;10(2):518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]