Abstract
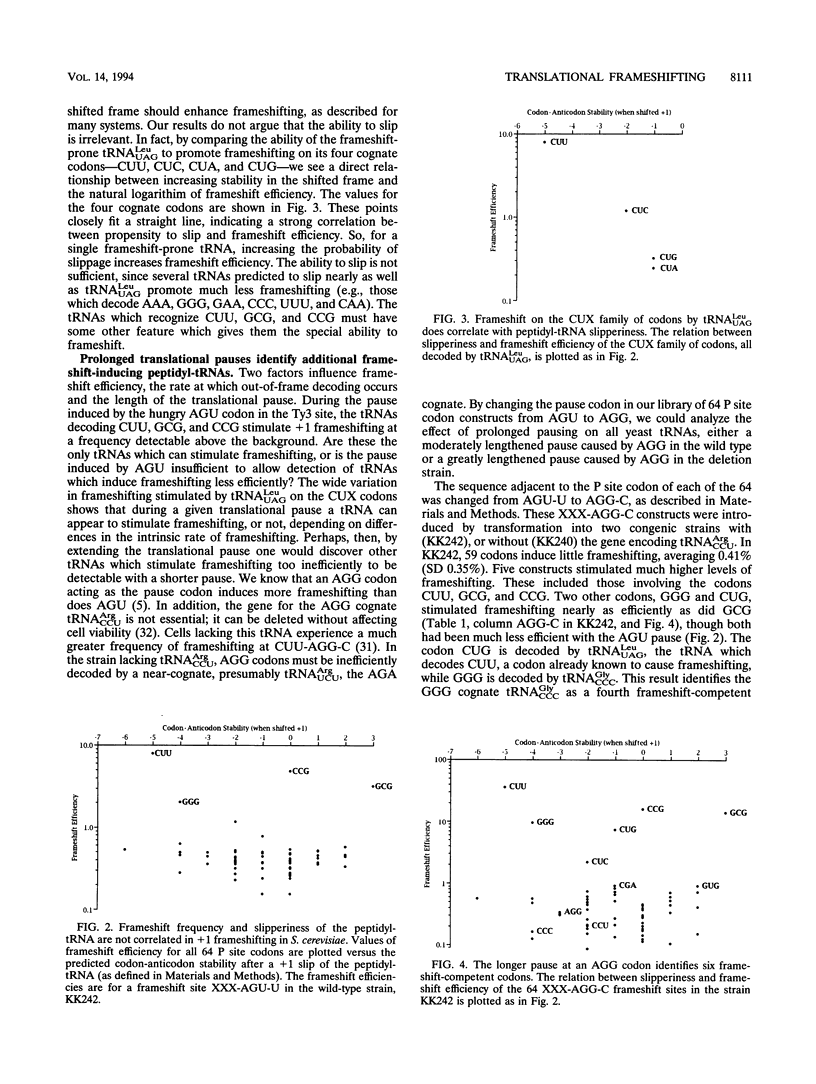
Recently we described an unusual programmed +1 frameshift event in yeast retrotransposon Ty3. Frameshifting depends on the presence of peptidyl-tRNA(AlaCGC) on the GCG codon in the ribosomal P site and on a translational pause stimulated by the slowly decoded AGU codon. Frameshifting occurs on the sequence GCG-AGU-U by out-of-frame binding of a valyl-tRNA to GUU without slippage of peptidyl-tRNA(AlaCGC). This mechanism challenges the conventional understanding that frameshift efficiency must correlate with the ability of mRNA-bound tRNA to slip between cognate or near-cognate codons. Though frameshifting does not require slippery tRNAs, it does require special peptidyl-tRNAs. We show that overproducing a second isoacceptor whose anticodon had been changed to CGC eliminated frameshifting; peptidyl-tRNA(AlaCGC) must have a special capacity to induce +1 frameshifting in the adjacent ribosomal A site. In order to identify other special peptidyl-tRNAs, we tested the ability of each of the other 63 codons to replace GCG in the P site. We found no correlation between the ability to stimulate +1 frameshifting and the ability of the cognate tRNA to slip on the mRNA--several codons predicted to slip efficiently do not stimulate frameshifting, while several predicted not to slip do stimulate frameshifting. By inducing a severe translational pause, we identified eight tRNAs capable of inducing measurable +1 frameshifting, only four of which are predicted to slip on the mRNA. We conclude that in Saccharomyces cerevisiae, special peptidyl-tRNAs can induce frameshifting dependent on some characteristic(s) other than the ability to slip on the mRNA.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Gesteland R. F., Reid B. R., Anderson C. W. Normal tRNAs promote ribosomal frameshifting. Cell. 1979 Dec;18(4):1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Thompson S., Gesteland R. F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Anderson S., Bankier A. T., de Bruijn M. H., Chen E., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3164–3166. doi: 10.1073/pnas.77.6.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A. J., Inglis S. C. Mutational analysis of the "slippery-sequence" component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992 Sep 20;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Atkins J. F., Gesteland R. F. tRNA anticodon replacement experiments show that ribosomal frameshifting can be caused by doublet decoding. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5062–5066. doi: 10.1073/pnas.83.14.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Curran J. F. Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site. Nucleic Acids Res. 1993 Apr 25;21(8):1837–1843. doi: 10.1093/nar/21.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987 Dec 11;238(4833):1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- Curran J. F., Yarus M. Use of tRNA suppressors to probe regulation of Escherichia coli release factor 2. J Mol Biol. 1988 Sep 5;203(1):75–83. doi: 10.1016/0022-2836(88)90092-7. [DOI] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donly B. C., Edgar C. D., Adamski F. M., Tate W. P. Frameshift autoregulation in the gene for Escherichia coli release factor 2: partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 1990 Nov 25;18(22):6517–6522. doi: 10.1093/nar/18.22.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M., Rojas A. M., Weiser J., Kurland C. G. How many EF-Tu molecules participate in aminoacyl-tRNA binding and peptide bond formation in Escherichia coli translation? J Mol Biol. 1990 Feb 20;211(4):739–749. doi: 10.1016/0022-2836(90)90074-V. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J. Alternative readings of the genetic code. Cell. 1993 Aug 27;74(4):591–596. doi: 10.1016/0092-8674(93)90507-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Vimaladithan A., Türkel S., Johnson R., Zhao H. Three downstream sites repress transcription of a Ty2 retrotransposon in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2081–2090. doi: 10.1128/mcb.13.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Zhao H., Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993 Jul 16;74(1):93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P., Liao X. B., Belcourt M., Zhao H., Kapakos J., Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Nov;9(11):4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Hansen L. J., Chalker D. L., Orlinsky K. J., Sandmeyer S. B. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol. 1992 Mar;66(3):1414–1424. doi: 10.1128/jvi.66.3.1414-1424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman J. E., Sarnoff J., Alzner-DeWeerd B., Yin S., RajBhandary U. L. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Jørgensen F., Adamski F. M., Tate W. P., Kurland C. G. Release factor-dependent false stops are infrequent in Escherichia coli. J Mol Biol. 1993 Mar 5;230(1):41–50. doi: 10.1006/jmbi.1993.1124. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Pande S., Faiola B., Moore D. P., Boeke J. D., Farabaugh P. J., Strathern J. N., Nakamura Y., Garfinkel D. J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993 Oct;135(2):309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Shafer B. K., Garfinkel D. J., Strathern J. N., Nakamura Y. Ty element-induced temperature-sensitive mutations of Saccharomyces cerevisiae. Genetics. 1992 Aug;131(4):821–832. doi: 10.1093/genetics/131.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J., Sandmeyer S. B., Forrest D. B. Transposition of a Ty3 GAG3-POL3 fusion mutant is limited by availability of capsid protein. J Virol. 1992 Oct;66(10):6081–6092. doi: 10.1128/jvi.66.10.6081-6092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. G. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. Unorthodox codon reading and the evolution of the genetic code. Cell. 1981 Feb;23(2):305–306. doi: 10.1016/0092-8674(81)90124-0. [DOI] [PubMed] [Google Scholar]
- Menninger J. R. Ribosome editing and the error catastrophe hypothesis of cellular aging. Mech Ageing Dev. 1977 Mar-Apr;6(2):131–142. doi: 10.1016/0047-6374(77)90014-8. [DOI] [PubMed] [Google Scholar]
- Ninio J. A semi-quantitative treatment of missense and nonsense suppression in the strA and ram ribosomal mutants of Escherichia coli. Evaluation of some molecular parameters of translation in vivo. J Mol Biol. 1974 Apr 5;84(2):297–313. doi: 10.1016/0022-2836(74)90586-5. [DOI] [PubMed] [Google Scholar]
- Peter K., Lindsley D., Peng L., Gallant J. A. Context rules of rightward overlapping reading. New Biol. 1992 May;4(5):520–526. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D., Yarus M. tRNA-tRNA interactions within cellular ribosomes. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4397–4401. doi: 10.1073/pnas.86.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard R. A., Chen K., Walker J. R., van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg). Nucleic Acids Res. 1990 Sep 11;18(17):5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanjaard R. A., van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7967–7971. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W. P., Brown C. M. Translational termination: "stop" for protein synthesis or "pause" for regulation of gene expression. Biochemistry. 1992 Mar 10;31(9):2443–2450. doi: 10.1021/bi00124a001. [DOI] [PubMed] [Google Scholar]
- Thompson R. C. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem Sci. 1988 Mar;13(3):91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Tu C., Tzeng T. H., Bruenn J. A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L. A second tRNA binding site on elongation factor Tu is induced while the factor is bound to the ribosome. Proc Natl Acad Sci U S A. 1985 May;82(10):3212–3216. doi: 10.1073/pnas.82.10.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L. GTPase center of elongation factor Tu is activated by occupation of the second tRNA binding site. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4617–4621. doi: 10.1073/pnas.83.13.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Noort J. M., Kraal B., Bosch L., La Cour T. F., Nyborg J., Clark B. F. Cross-linking of tRNA at two different sites of the elongation factor Tu. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3969–3972. doi: 10.1073/pnas.81.13.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Lazdunski C. Effect of distribution of unfavourable codons on the maximum rate of gene expression by an heterologous organism. J Theor Biol. 1986 May 7;120(1):99–110. doi: 10.1016/s0022-5193(86)80020-0. [DOI] [PubMed] [Google Scholar]
- Weijland A., Parmeggiani A. Toward a model for the interaction between elongation factor Tu and the ribosome. Science. 1993 Feb 26;259(5099):1311–1314. doi: 10.1126/science.8446899. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Ribosomal frameshifting from -2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R., Lindsley D., Falahee B., Gallant J. On the mechanism of ribosomal frameshifting at hungry codons. J Mol Biol. 1988 Sep 20;203(2):403–410. doi: 10.1016/0022-2836(88)90008-3. [DOI] [PubMed] [Google Scholar]
- Weissenbach J., Dirheimer G. Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim Biophys Acta. 1978 May 23;518(3):530–534. doi: 10.1016/0005-2787(78)90171-5. [DOI] [PubMed] [Google Scholar]
- Williams J. M., Donly B. C., Brown C. M., Adamski F. M., Trotman C. N., Tate W. P. Frameshifting in the synthesis of Escherichia coli polypeptide chain release factor two on eukaryotic ribosomes. Eur J Biochem. 1989 Dec 22;186(3):515–521. doi: 10.1111/j.1432-1033.1989.tb15237.x. [DOI] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]


