Abstract
Quantitative measurement of the levels of mRNA expression using real-time reverse transcription polymerase chain reaction (RT-PCR) has long been used for analyzing expression differences in tissue or cell lines of interest. This method has been used somewhat less frequently to measure the changes in gene expression due to perturbagens such as small molecules or siRNA. The availability of new instrumentation for liquid handling and real-time PCR analysis as well as the commercial availability of start-to-finish kits for RT-PCR has enabled the use of this method for high-throughput small-molecule screening on a scale comparable to traditional high-throughput screening (HTS) assays. This protocol focuses on the special considerations necessary for using quantitative RT-PCR as a primary small-molecule screening assay, including the different methods available for mRNA isolation and analysis.
Keywords: Real-time PCR, High Throughput Screening, phenotypic screening, qRT-PCR, gene expression
INTRODUCTION
Phenotypic small-molecule screening, in which a biological system is probed for a change of interest independent of a known target, has been shown to be valuable in the generation of hits for successful lead and drug development (Swinney and Anthony, 2011). When a pathway of interest is to be targeted, a common technique is to use an engineered system such as a reporter gene assay. In such a system, any molecule that modulates a component of the pathway will change the expression level of an easily measured protein such as luciferase or beta-galactosidase. Some disadvantages of this method are the time required to create the modified cell line and the potential for the reporter construct to lead to artifacts. By contrast, a method such as quantitative RT-PCR (qRT-PCR) that directly measures levels of the native mRNA expression is faster to develop and more biologically relevant. Until recently, cost and throughput limited the use of real-time qRT-PCR as a method for screening the tens or hundreds of thousands of compounds typical of primary HTS. As described in this protocol, there are now standard reagents and instruments available that allow the miniaturization and rapid execution of real-time PCR in a robust, cost-effective manner, compatible with high-throughput small-molecule screening. The choice of commercial kits depends in part on the type of cells being analyzed. Protocols for isolating and analyzing cDNA from adherent and suspension cells are described, as well as a third option that bypasses a separate cDNA isolation step and uses cell lysates directly in the qRT-PCR analysis.
STRATEGIC PLANNING
The essential piece of equipment for qRT-PCR is the real-time thermal cycler, which is a thermally controlled multi-well block mounted below a detection system that can simultaneously measure the fluorescence signal in all wells of a multiwell assay plate. Instruments are available in 96, 384, and 1536 format; for large-scale HTS, miniaturization to 384 or 1536 is advisable for throughput and cost reasons. Some of the 96 and 384 well instruments available are sold by Applied Biosystems/Life Technologies/Invitrogen (7900, Viia 7), Bio-Rad (CFX line), and Roche (LightCycler 480) and are common in many labs, core facilities, and academic screening centers. In addition, Roche offers the 1536 LightCycler, which enables sub-uL reactions on a scale compatible with ultra-high throughput screening (uHTS (Schlesinger et al., 2010)). Depending on the throughput desired, integration with an automated plate handler or screening system may also be an advantage.
The degree of miniaturization and number of compounds to be screened also dictate whether some form of automated liquid handling may be necessary (Rudnicki and Johnston, 2009). At a minimum, a multichannel pipette is required. Ideally, a bulk reagent dispenser such as the Combi Multidrop (Thermo) and a multichannel pipetting station (96 or 384 tip head) such as the Beckman Multimek or CyBio Vario are used for reagent and cDNA transfer. Unlike qPCR analysis of many different genes from a small number of cDNA preparations (Arany 2008), HTS of small molecules requires repeated analysis of the same gene or small set of genes, typically 2 or 3 genes. For this reason, the detection reagents for the qPCR step can be distributed from a master mix to the plate using a bulk reagent dispenser, but the individual wells with different cDNA samples must be transferred using separate tips with a multichannel pipette or a 96- or 384-tip pipetting robot. For further miniaturization, especially to 1536 format, more specialized instruments are required, including sub-uL bulk dispensers (e.g. Thermo Combi-nL or Biotek BioRaptr) for use with mastermixes and an acoustic dispenser (e.g. Labcyte Echo or EDC ATS) to transfer cDNA samples.
As with all highly parallel experiments such as HTS, sample management is essential. Up to three whole-plate transfers are required for qRT-PCR analysis, so it is essential to track the relationships between the different source and destination plates very carefully. Ideally a laboratory informatics management system (LIMS) database is used to record the transfers; at a minimum all plates should be uniquely labeled, such as with a barcode, and all plate identities should be tracked in a spreadsheet.
BASIC PROTOCOL TWO-STEP cDNA GENERATION AND qPCR ANALYSIS
The goal of high-throughput compound screening using qRT-PCR as a detection format is to obtain an accurate measurement of the effect of small molecules on the mRNA expression levels of a gene or genes of interest. To detect variation due to nonspecific effects, the expression level of the gene or genes of interest are normalized to the expression level of a control gene, typically a general metabolic or “housekeeping” gene such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or TATA-binding protein (TBP) (Schmittgen 2008). The choice of the control gene depends on the expression level of the gene(s) of interest. Ideally, a housekeeping gene with expression levels similar to the target gene(s) is used. The calculations for normalization and data analysis will be discussed below.
qPCR relies on the generation of a fluorescent signal correlated with the amount of PCR product present after each round of thermal cycling. The fluorescent signal can be generated by a general intercalator (Sybr green) that measures all DNA present. Alternatively, a fluorophore-quencher pair attached to a DNA sequence specific for the gene of interest is digested by the DNA polymerase during amplification, releasing the fluorophore and generating a signal. Thus, there are two methods for fluorescent quantification of both a control gene and one or more target genes. Using Sybr green, it is necessary to run a separate qPCR reaction for each gene to be quantified, as the intercalator does not distinguish between the PCR products generated either from each template sequence or from non-specific amplification of other sequences. The readings from the separate plates are then used for normalization. Using hydrolysable probes with different fluorophores for each gene sequence provides additional specificity to the target sequences and enables both genes to be analyzed in the same reaction, as nearly all real-time PCR instruments have the capability to monitor multiple excitation/emission wavelength channels. Multiplexing using multicolor hydrolysis probes is easier and faster in execution and provides more consistent normalization to the control gene. Design of primer-probe sets that perform well together is more difficult than simply identifying PCR primer pairs for use with Sybr green detection, but the up-front investment in assay design is well worth the effort. In addition, although probes are more expensive than unmodified PCR primers, the same probe set is used for the entire screen. Therefore, unlike genotyping experiments where many probes are required at a high cost, the reagents for small-molecule HTS can be purchased or synthesized in bulk at considerable savings.
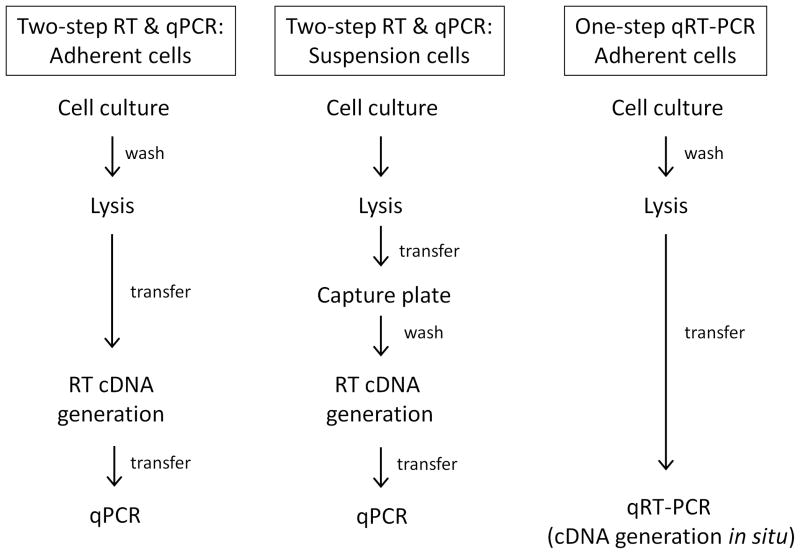
The difference between the three protocols presented herein (Figure 1) involves the method used to isolate the mRNA and generate cDNA for analysis. The appropriate protocol depends primarily on the cell type being analyzed (adherent or suspension) and by the desired workflow timing. The first method requires a washing step prior to cell lysis to remove cell media that can interfere with cDNA generation, and is therefore only appropriate for adherent cells. While it is possible to remove cell culture components from suspension cells using filter plates or by centrifuging cells, attempting to use such methods in multiwell formats introduces unacceptable variation in the experiment; therefore an alternate protocol using mRNA capture plates that is more ideal for suspension cells is described. Kits also exist that generate cDNA in situ prior to real-time analysis rather than isolating cDNA in a separate step. Overall, the two-step adherent protocol is currently the most cost effective and has the fewest time constraints, as the cDNA is isolated as an intermediate product and can be stored prior to transfer to multiple analysis plates.
Figure 1.
Three alternate protocols for qRT-PCR depending on cell type
Materials
Consumables
Cells and cell culture components (optimized for cell line to be analyzed) and related tissue-culture materials (CO2 incubator, serological pipettes, cell culture flasks)
Multiwell sterile cell-culture treated plates
Multiwell RNAse-free PCR plates
Sealing film for PCR plates
Multiwell qPCR plates
Optical sealing film for qPCR plates
RNAse free tips, either for multichannel pipettor or robotic pipetting station
Small molecule compounds to be screened
Phosphate-buffered saline pH 7.4
RNA isolation & cDNA preparation kit (e.g., Applied Biosystems Cells-to-Ct, Qiagen Fastlane, Roche RealTime ready cell lysis & transcriptor cDNA kit)
RNAse/DNAse free water
Oligonucleotide primers for each gene of interest (for Sybr green detection) or primer-probe sets for each gene (for multicolor probe detection)
Real-time qPCR mastermix containing dNTPs, buffer, polymerase (with or without Sybr green depending on detection method; e.g., Roche Sybr green master or Roche probes master)
Instrumentation
Bulk reagent dispenser (e.g., Thermo Combi Multidrop) and accessories
Multichannel pipette or robotic pipetting station (e.g., CyBio Vario)
Real-time qPCR instrument (see Strategic Planning)
Standard multiwell PCR block (optional)
Plate washer (e.g., Biotek EX-405) (optional)
Centrifuge for multiwell plates
Optional: Acoustic dispenser (Labcyte Echo or EDC ATS-100) and acoustic-certified source plates
Treat cells with compounds and lyse
-
1
Culture & expand cells using established methods (Phelan 2007).
-
2
Plate cells in multiwell tissue culture (TC) treated plates. Incubate overnight in TC incubator (37 °C, 5% CO2, 95% humidity).
Optimal plating density is dependent on cell type and growth rate. Cells should not be overgrown at the end of the compound treatment period. For 96 well plates,5–10k cells in 200 uL is typical; for 384 well plates, 1000–2000 cells in 50 uL.
-
3
Treat cells with compounds to desired screening concentration.
Typical compound screening concentration is 10 uM. Compounds can be transferred directly from concentrated DMSO stocks (e.g with a pin transfer device) or transferred with a multichannel robot or manual pipettor to an intermediate dilution plate as necessary for volume transfers compatible with the precision of available instrumentation. Mock treatment control wells (e.g. DMSO vehicle only) must also be included, representing 6–8% of the wells on the plate, typically equivalent to one column.
-
4
Incubate cells in TC incubator (37 °C, 5% CO2, 95% humidity).
Length of compound treatment depends on desired biological effect, typically 24–48 hr. This timing can be optimized in assay development if a positive control compound is available, which can be used to determine assay parameters such as Z′ factor (Zhang et al., 1999). The positive control can also be included in each screening plate for monitoring of HTS performance.
After treatment, follow recommended lysis and cDNA procedure from kit. The following steps are adapted from the ABI Cells-to-Ct kit for 384 well format plates, volumes should be adjusted accordingly for 96 well format:
-
5
Wash plates twice with 100 uL PBS using plate washer. If a plate washer is not available, media can be removed with multichannel pipettor and PBS added with bulk dispenser.
Ensure that appropriate settings are used to minimize cell disruption. If removing media is problematic and residual media causes poor lysis or poor cDNA synthesis, plates can be briefly centrifuged upside down on paper towel to remove residual media.
-
6
Add 10 uL of the kit lysis solution containing DNAse I. Incubate 5 minutes at 25 °C.
For most adherent cell lines, shaking is not necessary. If lysis is incomplete, as determined by poor cDNA yield, vortex the plated cells for one minute, incubate 5 minutes at 25 °C, then centrifuge for 1 minute. Do not over-incubate.
-
7
Add 1 uL of the stop solution. Incubate 2 minutes at 25 °C.
Following stop, lysates can be stored frozen at −20 °C until processed for cDNA generation.
Generate cDNA
-
8
Prepare cDNA RT mastermix. Per 10 uL reaction, mix:
5 uL 2× RT buffer
0.5 uL 20× RT enzyme mix
2.5 uL RNAse/DNAse free water
Include additional mastermix to account for dead volume in the bulk dispenser lines.
-
9
Dispense RT mastermix to a 384-well PCR plate using bulk dispenser, 8 uL per well.
-
10
Transfer 2 uL of cell lysate to the RT reaction plate using a multichannel or robotic pipettor.
Be sure to record source and destination plate pairings.
-
11
Seal plates and incubate in a PCR block with a heated lid for 1 hour at 37 °C, followed by 5 minutes at 95 °C.
If a standard PCR block is not available in addition to the real-time PCR instrument, the real time instrument can be used for the cDNA step; however, this will reduce instrument time available for the qPCR step.
A 37 °C incubator can be used for the 1 hour incubation, but it is advisable to use a PCR block that has a heated lid or “hot bonnet” to avoid evaporation and improve well-to-well and plate-to-plate consistency.
-
12
Remove cDNA plates and cool to room temperature prior to opening. If condensation is present on the inside of the seal, centrifuge for 1 minute. If qPCR is not to be performed immediately, store cDNA at −20 °C for up to 6 months.
One advantage of the two-step method is that a 10 uL cDNA reaction is sufficient for 10–20 qPCR reactions, depending on the volume of qPCR reaction used. This enables analysis of multiple genes by Sybr green or even reanalysis several months later should other genes of interest in the particular cell line be identified.
If an acoustic dispenser is to be used to transfer cDNA to the qPCR plate, first transfer the cDNA to a certified acoustic source plate. All subsequent transfers would then use the acoustic dispensing system, which is advisable for transfers to the 1536 qPCR plates used by the Roche 1536 Lightcycler. These transfers require an acoustic dispenser that is calibrated for aqueous source solutions.
Perform qPCR analysis
-
13
Prepare qPCR mastermix. Per 5 uL reaction, mix:
2.5 uL qPCR mastermix, Sybr green or probes as appropriate
0.125 uL 20× primer set (for Sybr green) or probes/primer set, for each gene to be analyzed
Nuclease free water to 4 uL
Include additional mastermix to account for dead volume in the bulk dispenser lines.
-
14
Dispense qPCR mastermix to a 384-well qPCR plate using bulk dispenser, 4 uL per well.
-
15
Transfer 1 uL of cDNA to the qPCR reaction plate using a multichannel or robotic pipettor.
Be sure to record source and destination plate pairings.
Include 1–2 wells of no-template control on the plate to test for contamination. This can be done by removing tips from the desired no-template wells prior to transferring the cDNA.
-
16
Seal plates firmly with optical film, avoiding any wrinkles that will interfere with top reading.
Sealed plates should be protected from direct light but are stable at room temperature for at least 24 hours. Stability is dependent on the qPCR mastermix kit used; “Fast” chemistry qPCR reagents tend to be less stable and may need refrigeration prior to reading but can be thermal cycled at a higher rate.
-
17
Load plates into real-time PCR instrument and thermal cycle:
95 °C 10 minutes
95 °C 10 seconds
60 °C 30 seconds, reading necessary fluorescent channels during this step.
Repeat steps 2 & 3 for 40 to 55 cycles, depending on abundance of genes being analyzed.
Optional last step: For quality control (QC) with Sybr reactions only, generate product melting curve by ramping from 40 to 90 degrees and reading fluorescence every 0.1 degrees.
Incubation temperature and time for the low temperature anneal/extension step may need to be adjusted depending on PCR primer characteristics. For poorly annealing primers, the annealing and extension steps may need to be separated into two incubation temperatures, for example an annealing temperature of 50 to 60 °C for 15–30 seconds followed by an extension temperature of 72 °C for 15–30 seconds.
The melting curve for Sybr products confirms that a PCR product of consistent length has been generated and can be used to QC across multiple plates to ensure that the desired product has been measured in all wells. This QC step is recommended during assay development and validation but is not essential during HTS, as it reduces throughput.
Analyze data
-
18
Using the RT-PCR instrument software, generate a cycle call for when each well enters log phase amplification.
Depending on the calculation method used by the instrument, this is called variously Ct (for when the background-normalized fluorescence trace crosses a common threshold) or Cp (for when the trace is maximally curved, determined by the maximum of the second derivative.) The general term is Cq for Quantification Cycle.
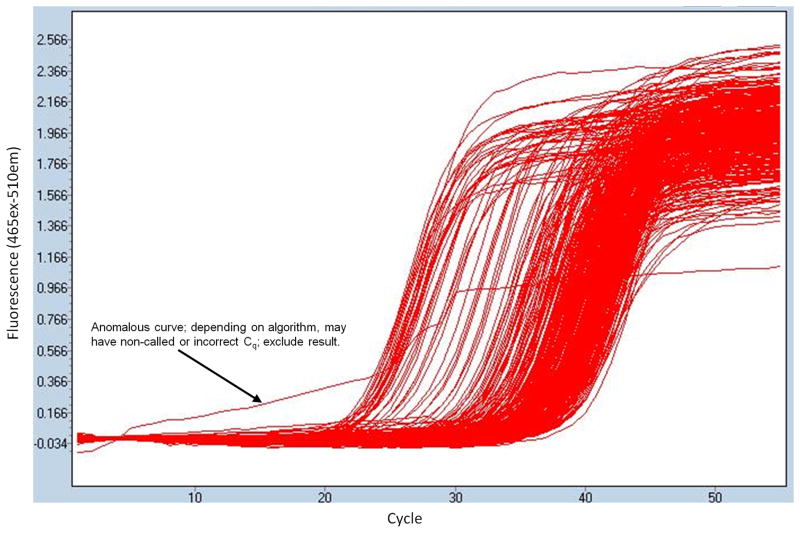
Wells that do not have a value called by the machine for a given color channel did not amplify. The no-template controls should exhibit this behavior. To simplify downstream calculations, these wells can either be omitted or arbitrarily assigned a Cq value equal to the total number of cycles run (Figure 2).
-
19
Determine the ΔCq by subtracting the Cq value of the control gene from the Cq value of the target gene in each well.
-
20
Determine the ΔΔCq value of each compound treatment by averaging the ΔCq values of the mock control wells on each plate and subtracting that average from the ΔCq value of each compound well.
Depending on the desired effect of the target gene (increased or decreased expression) the sign of ΔΔCq of “hits” may be positive or negative (Table 1):
If the goal is to decrease expression of the target gene, ΔCq of the treated wells will be greater than ΔCq of mock wells (i.e., Cq of the test gene increased due to compounds, representing fewer mRNA copies) and a positive ΔΔCq is desired (Table 1, Cmpd #2.)
Conversely, if the goal is to increase target expression, ΔCq of treated wells will be less than ΔCq of mock wells (i.e., Cq of the test gene decreased due to compounds, representing more mRNA copies) and a negative ΔΔCq is desired (Table 1, Cmpd #3.)
It is important to distinguish compounds that specifically affect the target gene(s) versus those that affect both target and control genes. A significant ΔΔCq may be observed if control and target Cq both change in response to compound and one changes more than the other. Superficially the ΔΔCq suggests this well is a hit, but such compounds are likely acting promiscuously and are less likely to be of interest. Examples of compounds with such effects are toxic compounds, compounds that elicit a stress response, or translational inhibitors (e.g., cyclohexamide) (Table 1, Cmpd #4.)
-
21
Determine the outliers in ΔΔCq to select hit compounds for follow-up, using standard HTS hit calling methods for normally distributed data such as Z-score (number of standard deviations away from the mock well treatment average) or MAD score (Zhang 2011).
Using the above normalizations, the average ΔΔCq of the DMSO wells on each plate should be approximately 0 and hits can be selected from the compound population distribution. It is appropriate to apply standard screening statistics as the noise in ΔΔCq of the DMSO wells is generated by instrument and experimental error and should be normally distributed. As minor pipetting errors such as failing to transfer cDNA due to bubbles can result in extreme outliers, it is best to use robust statistical metrics such as MAD score.
Figure 2.
Amplification curves from one channel of a two-color 384-well PCR experiment. Anomalously shaped curves (one shown) or non-amplifying curves (none shown) can be discarded or assigned arbitrarily high Cq values for downstream calculations.
Table 1.
Example Cq data for controls and test compounds, derived calculations and interpretations.
| Well type | Cq target | Cq ctrl | ΔCq | ΔΔCq | Result interpretation |
|---|---|---|---|---|---|
| DMSO | 27.12 | 18.66 | 8.46 | −0.03 | no treatment (average ΔCq of mock treatment wells = 8.49) |
| DMSO | 26.94 | 18.55 | 8.39 | −0.1 | no treatment |
| DMSO | 26.88 | 18.27 | 8.61 | 0.12 | no treatment |
| Cmpd #1 | 27.05 | 18.48 | 8.57 | 0.08 | No cmpd effect |
| Cmpd #2 | 31.33 | 18.61 | 12.72 | 4.23 | Selective repression of target |
| Cmpd #3 | 24.88 | 18.47 | 6.41 | −2.08 | Selective induction of target |
| Cmpd #4 | 34.25 | 22.34 | 11.91 | 3.42 | Superficially appears to be selective target repression based on ΔΔCq, but probably cytotoxic artifact(note increased Cq ctrl) |
Alternate protocol: Generation of cDNA using oligo-dT capture plates
As suspension cells cannot be reliably washed in multiwell plates, an alternative protocol is necessary for suspension cells such as those derived from blood lineages. For this protocol, the turbocapture kit from Qiagen can be used as directed for suspension cells to generate cDNA.
Materials
Turbocapture cDNA kit (Qiagen, P/N 72271 for 384 well, 72251 for 96 well. Buffer TCL, P/N 1031586 also required for suspension cells.)
Following cDNA generation as described in the Qiagen kit, qPCR analysis can be performed as above (Step 13 and onwards).
Alternate protocol: One-step qRT-PCR
For more streamlined HTS using qRT-PCR, kits are available that allow cell lysates to be directly analyzed in the qPCR instrument. This requires that the RT step occur in the real-time PCR instrument. As with the two-step method above, this is only appropriate for adherent cells as wash steps are required.
Materials
One-step qPCR kit (e.g, Roche Real-time Ready cell lysis kit, P/N 05943523001)
RNA RT-qPCR mastermix (e.g., Roche RNA probes master P/N 04991885001)
Optional: acoustic liquid dispenser (Labcyte Echo, EDC ATS) and TC-treated acoustic source plates
This protocol is adapted from the Roche Real-time Ready kit for 384 well format plates, volumes should be adjusted accordingly for 96 well format:
-
1
Culture, treat, and wash cells with compound as in steps 1–5 above.
Optional: For maximal throughput, cells can be cultured and treated in TC-treated acoustic source plates, available from Labcyte for the Echo dispenser (1536 well former part number 1536LDV-TC, 384 well format 384LDV-TC.)
-
2
Add 10 uL of cell lysis reagent containing RNAse inhibitor to each well.
-
3
Incubate 5 minutes at room temperature.
Lysates are stable 24–48 hours at 4 °C or several months at −20 °C, but may only be thawed once due to degradation of the RNAse inhibitor. For this reason, screens requiring more flexibility in running the qPCR analysis step, for example due to limited instrument access, should use the two-step protocol above.
-
4
Prepare RNA RT-qPCR mastermix. Per 5 uL reaction, mix:
1.85 uL 2.7× RNA probes master mix or RNA Sybr master mix
0.125 uL 20× primer set (for Sybr green) or probes/primer set, for each gene to be analyzed
Nuclease free water to 4 uL
-
5
Transfer 1 uL of cell lysate to the qPCR reaction plate.
Be sure to record source and destination plate pairings.
Include 1–2 wells of no-template control on the plate to test for contamination. This can be done by removing tips from the desired no-template wells prior to transferring the cDNA.
-
6
Seal plates firmly with optical film, avoiding any wrinkles that will interfere with top reading.
-
7
Load plates into real-time PCR instrument and thermal cycle:
63 °C 5 minutes (RT step)
95 °C 10 minutes
95 °C 10 seconds
60 °C 30 seconds, reading necessary fluorescent channels during this step.
Repeat steps 2 & 3 for 40 to 55 cycles, depending on abundance of genes being analyzed.
-
8
Perform data analysis as above in steps 18–21.
Reagents and solutions
All reagents described are specific to the cell culture requirements of the cells to be used or are included in the commercial kits described. The manufacturer’s recommendations with respect to storage and expiration should be followed.
Commentary
Background information
Shortly after the introduction of PCR in 1983, it was recognized that the initial amount of template could be quantitated by stopping the reaction at regular cycle intervals and measuring double-stranded DNA levels by methods such as absorbance or gel quantitation (VanGuilder et al. 2008). Over a decade later, methods were described for generating a signal in situ during amplification that could be read concurrent with thermal cycling (Heid et al. 1996).
While initial quantitative PCR methods were used for determining viral loads, the majority of the applications of the method have been for measuring expression levels of a number of genes in different biological samples such as diseased vs. non-diseased cells or various tissue types, generating transcriptional profiles characteristic of the underlying biology (Derveaux et al., 2010). It serves as a complement to expression microarrays and is often used to validate the latter, offering lower throughput but greater sensitivity and reliability (Ding and Cantor 2004, Arikawa et al. 2008). More recently it has been used to detect other cellular nucleic acids, including non-coding RNAs such as miRNAs (Benes and Castoldi 2010).
Due to the cost of designing detection assays for each separate gene, real time qRT-PCR is often not used in high-throughput format for genotyping. However, applications in which it is desirable to measure the response of a small number of genes to many different treatments are well suited for high-throughput analysis using this technique. In addition to changes in gene expression caused by compound treatment, described here and elsewhere (Hakamatsuka and Tanaka 1997, Maley et al. 2004, Wagner and Arany 2009), any system in which a nucleic acid can be generated as a biomarker or a surrogate for another analyte (such as a protein) can also be analyzed by qPCR (Swartzman et al., 2010). Thus, in addition to describing the biology of a natural or perturbed biological system, it can also be used to detect natural or artificial constructs for diagnostic or screening uses.
Critical Parameters
Assay probe design
The most important factor for a successful high-throughput gene expression assay is the design of the primers and probes used to detect the gene(s) of interest through the qRT-PCR reaction. Fortunately, bioinformatics resources and commercial products released in the last 5 years have made this a much less daunting task. In particular, many of the same companies that sell the instrumentation and consumables for real-time PCR are happy to assist with the design of assays to maximize the use of their products. Bio-Rad, Applied Biosystems/Life
Technologies/Invitrogen, and Roche all offer pre-designed packages of primers and/or probes. These sets are often referred to as assays and are sold as a complete package for real-time analysis. In addition, working with public genome databases, these same sites will design an assay based on any known sequence or gene, optimizing primer sequence and amplicon length. Most assays are available in the two formats described in this protocol, primers alone for use with Sybr green or primer-probe sets for use as Taqman assays. Additional formats, such as molecular beacons or FRET probes, are also available and may have an advantage for specific applications, such as when a loss of signal rather than a gain of signal is desired during amplification.
Internal controls
Another key part of assay design is selection of a proper control gene for normalizing against effects such as cytotoxicity or variation in cell plating. Ideally a gene unrelated to any pathway affected by the target gene should be used, such as the housekeeping genes TBP, GAPDH, or various rRNA sequences. In addition, it is best to use a control gene that amplifies in roughly the same number of cycles as the target gene, as this allows more robust normalization. In extreme cases where target genes are very highly expressed or underexpressed and the target and control signals are not both in the linear detection range of the assay, one of the assays can be adjusted by limiting one of the reagents such as a primer (known as primer-limiting) to artificially change the Cq value by reducing the efficiency of each round of PCR.
It is also ideal to measure the control gene and the target gene in the same reaction plate by using multiplexed probes with differentially emitting fluorophores. This reduces any variability between the control and target genes that is due to pipetting error or unequal treatment (light or temperature exposure) when separate plates are needed for each gene, as is the case when using Sybr green.
Assay development
Prior to any small molecule high-throughput screen, extensive assay development is required, and is especially critical in an assay as sensitive as qRT-PCR. All steps of a protocol, such as cell culturing, seeding, washing, and reagent transfers should be validated to minimize variation due to technical limitations. Untreated control plates should be run to determine the variability of the measurements and the corresponding sensitivity of the assay. When properly designed, these controls can also serve to test other technical aspects such as consistency of plate positioning (avoiding rotations) and recording transfers among the multiple plates that are generated in the course of an experiment.
In addition to technical sensitivity as measured by standards such as the percent coefficient of variation, in phenotypic assays it is important to establish a threshold for a biologically relevant response. Ideally a small molecule positive control can be used to both establish this biological response during assay development and to monitor the biological consistency during HTS. In the absence of a small molecule control, a genetic control, such as an overexpression vector or siRNA, can be used to establish assay sensitivity. However, genetic controls suffer from the limitation that the effect seen is often orders of magnitude greater than the response that can be expected from a small molecule, making comparison to screening results difficult. Finally, in the absence of either of these controls, a technical control such as a known stress inducer or cytotoxic agent can be used as a technical control simply to track and confirm the execution of reagent transfers, even though there is no relationship of this type of control to the desired biology.
Overall, the design and validation of the assay is key for successful small-molecule screening by real-time qRT-PCR. This type of assay is among the most expensive used for small-molecule screening, and while the biological relevance of the data can provide a good return on this investment, it is easy to waste a large amount of expensive reagents if experiments are not properly planned and tested prior to screening.
Data analysis and concentration-response curves
The ΔΔCq method described above is used to relate two differently treated samples, in this case compound and mock treatment, where each sample has a target gene compared to a control gene (Schmittgen and Livak, 2008). The data for the mock treatments is typically normally distributed due to experimental error, and single point values can be compared to this distribution for determining likelihood of a significant effect due to compound treatment.
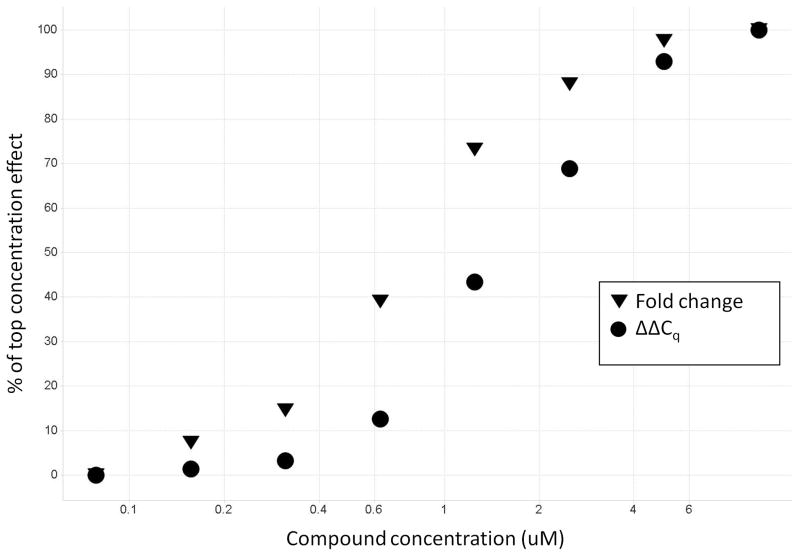
The next step following hit calling is typically retesting of the compounds in the assay at a range of dilutions to construct a concentration response curve. There are two additional considerations in concentration-response tests compared to the primary HTS. First, the goal is not to measure the significance of an outlier, but to observe the biological effect of the compounds. This requires converting an exponential measurement given by Cq to a linear scale that represents the actual relative change in copy number. This conversion is done by the calculation: Fold change = 2ΔΔCq (Table 2.) These values should then be plotted against the tested concentration values for curve fitting (Figure 3.)
Table 2.
Hypothetical concentration-response data converted from ΔΔCq (calculated from raw data as described in protocol steps 19 & 20) to fold change in expression and normalized to % of maximal effect. Fold repression is equal to 2ΔΔCq. Each calculated value (ΔΔCq or fold repression) is then separately normalized to a percentage of effect, with DMSO treatment (ΔΔCq = 0) set to 0% effect and the top concentration treatment (10 uM) set to 100%.
| Compound concentration (uM) | ΔΔCq | Fold repression | % of top concentration effect, ΔΔCq | % of top concentration effect, Fold repression |
|---|---|---|---|---|
| 0.078125 | 0 | 1.0 | 0 | 0 |
| 0.15625 | 0.3 | 1.2 | 7.0 | 1.3 |
| 0.3125 | 0.6 | 1.5 | 15 | 3.1 |
| 0.625 | 1.6 | 3.0 | 39 | 13 |
| 1.25 | 3.0 | 8.0 | 73 | 43 |
| 2.5 | 3.6 | 12 | 88 | 69 |
| 5 | 4.0 | 16 | 98 | 93 |
| 10 | 4.1 | 17 | 100 | 100 |
Figure 3.
Plotting of concentration response curve of hit compounds for reduced expression of a target gene. By converting to the relevant biological measurement, fold change, the apparent EC50 has shifted approximately 2-fold. Hypothetical data shown in Table 2.
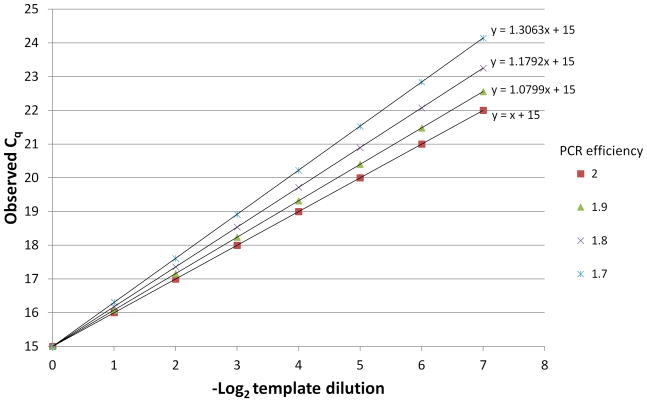
The conversion Fold change = 2ΔΔCq (or alternatively 2−ΔΔCq depending on whether a fold repression or fold induction is being described) assumes a perfect PCR efficiency; that is, it assumes that each round of PCR exactly doubles the amount of DNA product during the logarithmic PCR expansion. In reality, PCR efficiencies are usually near 1.9 but can be as low as 1.7 (Tuomi et al. 2010). If more precision is desired, a standard curve can be generated for the qPCR assay to be used by making a serial dilution of a sample PCR template and measuring the actual fold amplification observed in each round of PCR, which can be used in the above calculation (Figure 4.)
Figure 4.
Construction of a standard curve to measure PCR efficiency. An arbitrary starting amount of template (in this case, sufficient to give a Cq of 15) is 2-fold serially diluted and the dilution series is measured by real time qPCR. The slope of the resulting Cq values vs. −log2 of the dilution is related to the PCR efficiency by the equation Eff = 21/slope. Idealized curves are shown.
The second consideration when trying to fit concentration-response curves is deciding whether the maximal effect can be determined. In the hypothetical graph in Figure 3, the response plateaus at high compound concentration, enabling the determination of an EC50 (50% of maximal effect) for the curve. More typically, either due to complex biology or weakly active compounds, it is not possible to determine the upper asymptote for the fitted curve, and the maximal effect may be very different for various compounds. In these cases, it is more appropriate to determine a threshold of N-fold induction or repression that represents a statistically and biologically significant effect, and set this ECN as the concentration at which the compound achieves the desired threshold. This allows more robust comparison of compounds with a wide range of biological effects and enables better selection for compounds to carry forward for further development.
Troubleshooting
The greatest difficulty in small-molecule screening is high variability, which can be observed by high variation in the mock treatment wells. This reduces the sensitivity of the experiment and increases the rate of false positives. In qRT-PCR experiments, it has been shown that with careful technique, the qPCR and RT generation together represent less than 20% of the total variation between replicate analyses (Kitchen et al. 2010). The majority of variability comes from biological variability, including sample preparation. In the protocol presented here, this includes cell culture of the cell line to be used, plating and treatment of the cells, and washing and lysis prior to cDNA generation. It is important to have cultured cells as consistent as possible across all compound tests, including minimizing passage number variation and exposure to variable environmental conditions. Plating of cells should be done in manageable batches to reduce variability due to plating order. Washing parameters should be set to minimally disturb the cells, and lysis should be optimized for the cells used to ensure complete reaction. While replicates may help determine the extent of variability seen in treatment wells, it is costly to run biological replicates (treating two separate plates of cells with the same compounds) as the cDNA generation accounts for the majority of the cost of the assay. Real-time qPCR replicates from the same cDNA well can confirm the robustness of the analysis step, but as mentioned above this is not expected to reveal much variability and is time consuming to perform for an entire HTS campaign.
Very high Cq values are indicative of poor mRNA isolation or cDNA generation and are problematic because very small amounts of contamination or variability can have a large effect on the results. If values are too low, confirm the efficiency of the PCR reaction and consider re-optimizing the assay design or even selecting a different target gene. Adjustment of cell number may have a small effect but is unlikely to improve low signals, as a 2-fold increase in cell seeding will only have ~1 cycle effect on Cq.
Finally, it is essential to include controls for non-amplification to ensure that there is no contamination. If no-template controls are showing signals that are within several cycles of experimental wells, the data cannot be accepted as valid. Contamination can be controlled by the same methods used for standard PCR, including isolating and cleaning a workspace and never bringing post-amplification materials into the PCR setup area.
Anticipated Results
It is expected that in a compound library of appropriate size or design, some amount of biological effect will be seen, if only due to cytotoxic compounds. Typically a validation library containing a sample of bioactive compounds is run early in the screen to confirm the presence of this effect. Figure 2 illustrates a typical assay plate with a range of Cq values due to compound treatment, and similar small-molecule high-throughput screening results have been reported and analyzed previously (Arany et al., 2008). Overall, the active well rate for compounds showing the desired change in target gene expression is highly dependent on the biological system under investigation. It is entirely possible that a particular gene is intractable to specific modulation by small molecules, and that any putative hits will be removed as artifacts in subsequent follow up assays. As with all discovery projects, the researchers needs to make an informed decision about the return on investment of additional screening or follow up.
Time Considerations
The largest investment of time is in the assay development and validation but is essential to avoid wasting time and money in downstream failures. Several rounds of mock runs are usually required to get low-noise consistent results through the entire process of cell culture, lysis, cDNA generation, and qPCR analysis. This will take several weeks of iteration.
Depending on the choice of protocol and number of compounds tested, the small-molecule HTS can be staged in various ways. Using a two-step protocol to isolate cDNA has the advantage of allowing the separate execution of each phase of the assay rather than multitasking, which adds to the total project time but can make execution more consistent. The only time-sensitive step in the protocol is the lysis treatment, which can ruin the nucleic acid sample if run too long. For large numbers of multiwell plates it is advisable to have at least two scientists working together to execute the protocol, as subsequent steps can back up and interfere with the key timing step. It is important to recognize other potential bottlenecks as well including standard PCR blocks for the RT step, robotic pipetting throughput, and real-time instrument availability. Proper planning will enable optimal use of these resources
Even though the real-time qPCR analysis step takes 1–1.5 hours per plate, it is now possible to screen tens or hundreds of thousands of compounds using real-time qRT-PCR thanks to miniaturization to 384- or 1536-well format. If an assay is robust and appropriate instrumentation is available, thousands of compounds can be screened each day during a campaign, enhancing the potential for discovery of specific small-molecule modulators of the target gene of interest.
Footnotes
Internet Resources with Annotations
Bio-rad gene expression gateway: http://www.biorad.com/genomicsProvides useful tutorials on experimental design and detection formats
Roche UPL: http://www.roche-applied-science.com/sis/realtimeready/index.jsp
Search of existing qPCR detection reagents and bioinformatics tools for designing custom assays.
Life technologies/Applied Biosystems designer for custom Taqman probes: https://www5.appliedbiosystems.com/tools/cadt/
Life technologies Taqman assays and assay designer.
Literature Cited
- 1.Arany Z. High-Throughput Quantitative Real-Time PCR. Current Protocols in Human Genetics. 2008:11.10.1–11.10.11. doi: 10.1002/0471142905.hg1110s58. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Wagner BK, Ma Y, Chinsomboon J, Laznik D, Spiegelman BM. Gene expression-based screening identifies microtubule inhibitors as inducers of PGC-1 alpha and oxidative phosphorylation. Proc Natl Acad Sci USA. 2008;105:4721–4726. doi: 10.1073/pnas.0800979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arikawa E, Sun Y, Wang J, Zhou Q, Ning B, Dial SL, Guo L, Yang J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods. 2010;50:244–249. doi: 10.1016/j.ymeth.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Ding C, Cantor CR. Quantitative analysis of nucleic acids--the last few years of progress. J Biochem Mol Biol. 2004;37:1–10. doi: 10.5483/bmbrep.2004.37.1.001. [DOI] [PubMed] [Google Scholar]
- 7.Hakamatsuka T, Tanaka N. Screening for bioactive compounds targeting the cellular signal transduction pathway using an RT-PCR-based bioassay system. Biol Pharm Bull. 1997;20:464–466. doi: 10.1248/bpb.20.464. [DOI] [PubMed] [Google Scholar]
- 8.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 9.Kitchen RR, Kubista M, Tichopad A. Statistical aspects of quantitative real-time PCR experiment design. Methods. 2010;50:231–236. doi: 10.1016/j.ymeth.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Maley D, Mei J, Lu H, Johnson DL, Ilyin SE. Multiplexed RT-PCR for high throughput screening applications. Comb Chem High Throughput Screen. 2004;7:727–732. doi: 10.2174/1386207043328300. [DOI] [PubMed] [Google Scholar]
- 11.Phelan M. Basic Techniques in Mammalian Cell Tissue Culture. Curr Protoc Cell Biol. 2007;1:1–15. doi: 10.1002/0471143030.cb0101s36. [DOI] [PubMed] [Google Scholar]
- 12.Rudnicki S, Johnston S. Overview of liquid handling Instrumentation for high-throughput screening applications. Curr ProtocChem Biol. 2009;1:43–54. doi: 10.1002/9780470559277.ch090151. [DOI] [PubMed] [Google Scholar]
- 13.Schlesinger J, Tönjes M, Schueler M, Zhang Q, Dunkel I, Sperling SR. Evaluation of the LightCycler 1536 Instrument for high-throughput quantitative real-time PCR. Methods. 2010;50:S19–S22. doi: 10.1016/j.ymeth.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 15.Swartzman E, Shannon M, Lieu P, Chen SM, Mooney C, Wei E, Kuykendall J, Tan R, Settineri T, Egry L, Ruff D. Expanding applications of protein analysis using proximity ligation and qPCR. Methods. 2010;50:S23–S26. doi: 10.1016/j.ymeth.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 16.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 17.Tuomi JM, Voorbraak F, Jones DL, Ruijter JM. Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods. 2010;50:313–322. doi: 10.1016/j.ymeth.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 18.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 19.Wagner BK, Arany Z. High-throughput real-time PCR for detection of gene-expression levels. In: Clemons PA, Tolliday NJ, Wagner BK, editors. Cell-based assays for high-throughput screening. Humana Press; New York: 2009. pp. 167–175. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XD. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screening. 2011;16:775–785. doi: 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]