Abstract
It has long been recognized that a suite of proteins exists in coral skeletons that is critical for the oriented precipitation of calcium carbonate crystals, yet these proteins remain poorly characterized. Using liquid chromatography-tandem mass spectrometry analysis of proteins extracted from the cell-free skeleton of the hermatypic coral, Stylophora pistillata, combined with a draft genome assembly from the cnidarian host cells of the same species, we identified 36 coral skeletal organic matrix proteins. The proteome of the coral skeleton contains an assemblage of adhesion and structural proteins as well as two highly acidic proteins that may constitute a unique coral skeletal organic matrix protein subfamily. We compared the 36 skeletal organic matrix protein sequences to genome and transcriptome data from three other corals, three additional invertebrates, one vertebrate, and three single-celled organisms. This work represents a unique extensive proteomic analysis of biomineralization-related proteins in corals from which we identify a biomineralization “toolkit,” an organic scaffold upon which aragonite crystals can be deposited in specific orientations to form a phenotypically identifiable structure.
Keywords: acid-rich proteins, collagen, cadherin, carbonic anhdyrase
Biomineralizing organisms are found in all biological kingdoms and incorporate a variety of metals, from sodium to lead, as major components of minerals whose nucleation and growth are under a range of biological control (1–5). The skeletal organic matrix (SOM), occluded in the mineral, has been implicated as a source of the increased strength of biominerals over comparable geominerals and has long been hypothesized to aid in the stabilization, nucleation, growth, and spatial orientation of biominerals (6–10). However, the mechanisms for the role of the SOM in coral biomineral formation remain to be elucidated, primarily because the organic molecules have yet to be characterized.
At present, the best-characterized SOM is that contained in mammalian bones and teeth and is divided into structural and highly acidic protein categories. Collagen, a well-characterized fibrillar (i.e., framework) protein, plays a structural role in mammal SOM (reviewed in ref. 10), and has been investigated in the structural components of several coelenterates (11, 12), of which Order Scleractinia (i.e., stony corals) is a member. However, the greatest emphasis for nucleation proteins rests on the highly acidic proteins, such as bone sialoproteins and dentin matrix proteins in teeth in vertebrates (13, 14). Highly acidic proteins also have been identified or hypothesized in mineralizing invertebrates, and several have recently been described, including the Asprich family in the pen shell, Atrina rigida (15), Pif and Aspein in the pearl oyster, Pinctada fucata (16, 17), and the Adi-SAPs (highly acidic proteins proposed to be soluble or secreted) in the stony coral Acropora digitifera (18). Sequence-based homologs of each have been found in a variety of other invertebrates, yet these proteins, and their consequent biomineralization reactions, appear to have originated several times independently (19, 20), and their sequence similarity is most likely explained by convergent evolution. Proteins containing acidic amino acids (Asp and Glu) or phosphorylation sites (on Ser residues) are thought to be used at various stages of aragonite and calcite mineralization to temporarily stabilize amorphous calcium carbonate or nucleate the mineral under appropriate conditions (reviewed in ref. 21).
Stony corals (Class Anthozoa) are early-branching metazoans composed of four cell layers (22). Whereas the oral endodermal cells host endosymbiotic photosynthetic algae of the genus Symbiodinium, the aboral ectodermal cells, or calicoblastic cells, are the sites of biomineralization. Calicoblastic cells are thought to secrete SOM, which adheres the cells to recently formed extracellular skeleton, and are also considered to be intimately involved in nucleation and growth of aragonite crystals (23, 24). SOM is retained in the coral skeleton and the amino acid composition of the protein fraction indicates that the SOM is distinct from cellular or mucus protein (25, 26). To date, only one coral SOM protein, galaxin, has been fully sequenced and its role in the biomineralization process is not well understood because it does not bind calcium (27). Other proteins hypothesized to play a role in the mineralizing space between the calicoblastic cells and the skeleton include carbonic anhydrases (28), collagen (12), ion transporters (29), cysteine-rich proteins (30), von Willebrand factor type A domain-containing proteins and zona pellucidas (31), and secreted acidic proteins (18, 32). Together, these proteins may represent a “biomineralization toolkit” of calcifying proteins in corals. However, most of these proteins remain to be independently confirmed in coral skeleton, described as complete genes, or characterized with respect to function.
Here we use a proteomics approach to describe the SOM proteins in the widely distributed, Pocilloporid coral, Stylophora pistillata. Using liquid chromatography-tandem mass spectrometry (LC-MS/MS) protein sequencing, and a draft genome from S. pistillata, we identify partial and complete sequences of proteins in the S. pistillata aragonite skeleton. Comparison of our results with genome and transcriptome data from other mineralizers suggests that a coral skeleton contains a complex group of proteins that guide the biomineralization process to form specific, genetically determined structures. This, a unique proteome from a coral skeleton, identifies a biomineralization toolkit in these key, ecologically critical organisms.
Results and Discussion
Thirty-six proteins, predicted from the draft S. pistillata genome assembly, were detected by three LC-MS/MS analyses of SOM proteins (Table 1). This amount is similar in number, but not sequence identity, to the 33 shell matrix proteins recently identified by LC-MS/MS from mollusk nacre (aragonite) (33). Twenty-five of the coral SOM protein candidates were only observed after deglycosylation (Table S1). Thirty-one of these candidates could be observed with tryptic digestion and the remaining five were observed only after proteinase K digestion (Table S1).
Table 1.
Thirty-six predicted proteins in S. pistillata SOM samples detected by LC-MS/MS and their bioinformatics analysis
| Protein | Gene | Accession no. | Name | P. damicornis | A. digitifera | Favia sp. | N. vectensis | P. maxima | S. purpuratus | E. huxleyii | R. filosa | H. sapiens | T. pseudonana |
| P1 | g11108 | KC509948 | Protocadherin fat-like | – | + | +*†‡ | + | + | – | – | – | – | – |
| P2 | g11187 | KC493647 | CARP8 | + | +‡ | +†‡ | – | – | – | – | – | – | – |
| P3 | g12510 | KC342189 | Thrombospondin | – | + | +† | + | – | – | – | – | – | – |
| P4 | g9861 | KC342190 | Viral inclusion protein | +* | + | +*† | + | – | – | – | – | – | – |
| P5 | g11674 | KC150884 | Hemicentin | + | +† | +‡ | +‡ | + | + | – | – | +‡ | – |
| P6 | g11666 | KC149520 | Actin | +*† | +* | +* | +* | +* | +* | +* | +* | +* | +* |
| P7 | g4601 | KC342191 | Actin | +* | +*‡ | +* | +*† | +* | +* | +* | +* | +* | +* |
| P8 | g9654 | KC342192 | Major yolk protein | +‡ | + | +†‡ | – | + | – | – | – | – | – |
| P9 | g10811 | KC000002 | Protocadherin fat-like | – | +‡ | +†‡ | +‡ | – | – | – | – | – | – |
| P10 | g11107 | KC509947 | Cadherin | +* | + | +†‡ | + | – | – | – | – | – | – |
| P11 | g13727 | KC342193 | Actin | +* | +*† | +* | +* | +* | +* | +* | +* | +* | +* |
| P123 | g2385 | JX891654 | — | – | – | +†‡ | – | – | – | – | – | – | – |
| P13 | g6918 | KC342194 | Sushi domain-containing | + | +† | + | – | – | – | – | – | – | – |
| P14 | g9951 | KC342195 | Collagen - alpha | – | + | +† | – | – | – | – | – | – | – |
| P15 | g1532 | KC493648 | CARP9 | – | +‡ | +†‡ | – | – | – | – | – | – | – |
| P16 | g11702 | KC342196 | — | – | +† | +* | + | – | – | – | – | – | – |
| P17 | g12472 | KC149521 | Glyceraldehyde 3-phosphatase dehydrogenase | +* | +*† | +* | +* | + | + | + | + | +* | + |
| P18 | g810 | KC342197 | Collagen - alpha | – | + | +† | + | – | – | – | – | – | – |
| P19 | g20041 | KC342198 | Contactin-associated protein | – | + | +†‡ | + | – | – | – | – | – | – |
| P20 | g6066 | KC342199 | MAM domain anchor protein | + | +†‡ | +* | + | – | – | + | – | +‡ | – |
| P21 | g18277 | KC479163 | Zona pellucida | +*†‡ | + | +*‡ | + | – | – | – | – | – | – |
| P22 | g19762 | KC493649 | — | – | – | – | – | – | – | – | – | – | – |
| P23 | g1057 | KC000004 | Protocadherin | + | + | +* | +† | + | + | – | – | – | – |
| P24 | g15888 | KC479164 | Vitellogenin | – | + | +*†‡ | – | – | – | – | – | – | – |
| P25 | g11220 | KC479165 | Ubiquitin | +* | +*‡ | +* | +*† | +* | +* | +* | +* | +* | +* |
| P26 | g1441 | KC479166 | Vitellogenin | + | – | +†‡ | – | – | – | – | – | – | – |
| P27 | g18472 | KC479167 | Integrin - alpha | +* | + | +†‡ | – | – | – | – | – | – | – |
| P28 | g11651 | KC149519 | Late embryogenesis protein | +† | – | – | – | – | – | – | – | – | – |
| P29 | g13377 | KC479168 | Tubulin - beta | +* | +* | +*† | +* | +* | +* | +* | +* | +* | + |
| P30 | g11056 | KC000003 | Myosin regulatory light chain | +* | + | +†‡ | – | + | – | – | – | – | – |
| P31 | g20420 | KC479169 | Neurexin | – | + | +†‡ | – | – | – | – | – | – | – |
| P32 | g5540 | KC479170 | Kielin/chordin llke | +*† | +‡ | +‡ | – | – | – | – | – | +‡ | – |
| P33 | g8985 | KC479171 | Flagellar associated protein | +*† | +* | +* | + | – | – | – | – | – | – |
| P34 | g1714 | KC479172 | MAM/LDL receptor domain containing protein | + | +‡ | +*† | + | – | + | + | – | + | – |
| P35 | g7349 | EU532164.1 | Carbonic anhydrase (STPCA2) | +*† | +‡ | + | – | + | – | – | – | + | – |
| P36 | g13890 | KC479173 | Zonadhesion-like precursor | + | +*†‡ | + | +* | – | + | – | – | – | – |
Returned sequences with e-values ≤10−10 are presented in order of decreasing e-value. “Protein name” is the best BLAST hit in NCBI. “Gene” is the code number in our S. pistillata gene prediction model. The “+” and “–” represent presence and absence, respectively, of similar sequences in comparison species.
*Sequence similarity is greater than 70%.
†Most similar sequence by bit score.
‡Indicates export signal.
A major problem in isolating and identifying specific proteins from corals is their posttranslational modification, primarily by glycosylation (32). Although we consistently observed five distinct protein bands in size-fractionated SOM proteins, there was always a strong background smear on silver-stained polyacrylamide gels (Fig. S1). Periodic acid-Schiff staining confirmed the abundance of glycans in these samples (Fig. S1). Hence, the results presented here are almost certainly a conservative estimate of the total number of proteins in the skeletal matrix.
Nearly all proteins detected by LC-MS/MS show similarities to proteins known to play roles in the structure and adhesion of cells (Table 1). We detected multiple proteins with hits (e-value ≤ 10−5) to precursor cadherins (P1, P9, P10, and P23) and von Willebrand factor (P3, P5, P13, P14, P18, and P32) domains. Blast hits for other proteins include two collagens (P14 and P18), three actins (P6, P7, and P11), and a carbonic anhydrase (P35), among others. Five candidate proteins exhibit poor (e-value > 10−5) or no blast hits in National Center for Biotechnology Information (NCBI) (P2, P12, P15, P16, and P22). Although amino acid analyses of SOM from a variety of corals suggest that highly acidic proteins are compositionally important (25, 34), we only identified two such predicted proteins by LC-MS/MS.
Structural Proteins.
The highest scoring predicted protein, P1, is an incomplete protocadherin fat 1-like protein that contains a von Willebrand factor type A domain. We identified three separate PCR amplicons that map to this gene in S. pistillata cDNA, validating its expression. These transcripts encompass 11 of the 15 MS-sequenced peptides for this candidate protein (Fig. S2). Comparison with a very similar predicted protein from A. digitifera and Favia sp. (Table 1) suggests that the S. pistillata protein is incomplete and can be extended ∼750 residues toward the N terminus, likely containing a secretory signal, and over 1,000 residues toward the C terminus (Table S1) (18, 35). Two additional proteins among the top 10 detected also show cadherin precursor-like qualities (P9, P10) (Table 1).
Cadherins are calcium-dependent cell-cell adhesion molecules with specificity for certain cell types, such as neurons or osteoblasts (36). Apoptotic osteoblasts and intact retinal cells can cleave the extracellular portion of their respective N-cadherins to produce a 90-kDa soluble protein that may have roles in cell survival and protein expression. In addition, these cadherins have been implicated in the aggregation of cells into which they have been cloned (37), and could help explain the proto-polyps recently observed in S. pistillata cell cultures (34).
Of the six detected proteins that contain von Willebrand factor type A (an adhesion glycoprotein), four (P3, P5, P14, and P18) show strong sequence similarity between the predicted sequences from S. pistillata and those from A. digitifera, Favia sp., and Pocillopora damicornis (Table 1 and Table S1) (18, 35, 38). We compared these four predicted proteins from S. pistillata with pre- and postsettlement genes in Acropora millepora larvae as described by Hayward et al. (31). A. millepora genes A9, A90, and A102 are ≥40% similar to the P5 protein found in this study, with blastp e-values ranging from 10−15 to 10−23. All three proteins in the A. millepora expression study were up-regulated in presettlement planulae relative to the postsettlement stage and two, A9 and A90, were localized to the aboral region postsettlement (31). This finding suggests that A9, A90, and A102—and therefore, possibly P5—are involved in adhesion of calicoblastic cells to the skeleton. The highly acidic mollusk nacre protein, Pif, also contains a von Willebrand factor type A, although there is no sequence similarity between Pif and the coral proteins described here (16).
Collagen and chitin share similar roles in biomineralizing organisms (39), although chitin would not be detectable by the procedures we used here. Two α-collagen–like proteins were detected both before and after deglycosylation (P14 and P18) (Table S1). We have confirmed the transcription of a portion of P14 by PCR amplification of S. pistillata cDNA (Fig. S2). P14 shows strong similarities to two predicted sequences from A. digitifera (e-value 10−16) and Favia sp. (e-value 10−41) (Table 1) (18, 35). Collagen, a fibrillar protein common in extracellular matrix, is generally considered as a place for noncollagenous proteins to bind and nucleate the mineral (reviewed in ref. 10), but may also form sites of nucleation in “holes” of packed collagen molecules (40). Although collagen has been confirmed in sea pen axial stalks (11) and gorgonian skeletons (12), little work has been conducted on its presence in scleractinian skeletons. The extracellular matrix portion of coral cell cultures has been shown to positively stain for collagen (41), and so it seems highly likely that collagen-like molecules are a component of S. pistillata SOM.
Strong similarities were observed for a carbonic anhydrase (P35), which was found only in deglycosylated samples, and gene sequences from P. damicornis, Favia sp., A. digitifera, Pinctada maxima, and Emiliania huxleyi (Table 1 and Table S1) (18, 35, 38, 42). The complete sequence of P35 has previously been determined, and was named S. pistillata carbonic anhydrase 2 (STPCA2; accession number ACE95141.1) by Bertucci et al. (43); it has been immunolocalized to the cytosol of endo- and ectodermal cells of S. pistillata tissue slides and has a very high enzyme efficiency for interconverting HCO3− and metabolic CO2 (43). Although SPTCA1 and STPCA2 show 35% sequence identity, peptides we detected by LC-MS/MS were unique to STPCA2. Whereas only STPCA1 has previously been localized in S. pistillata skeleton (44), our results strongly suggest that STPCA2 is also present in the calicoblastic space and is retained in the skeleton after mineralization. Presence of both STPCA1 and -2 in the calcifying space likely appears to be an enzymatic “bet-hedging,” a strategy that permits integral pH and bicarbonate control for both coral skeleton (45) and mollusk shell (46) formation.
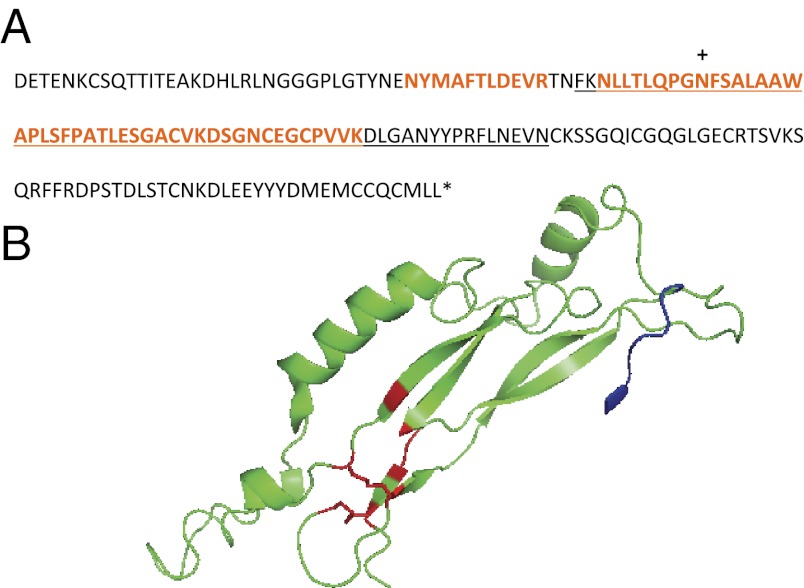
The final structural protein of interest is a 184-aa 21-kDa protein with a theoretical isoelectric point of 5.06 (P12) (Table 1). This gene contains a secretory signal at the N terminus, ends with a stop codon, shows significant sequence identity to a predicted protein from Favia sp. (35), contains no known domains, and is predicted to have an N-linked glycosylation site at Asn-79 (Fig. 1). Finally, it does not contain disproportionate amounts of acidic, basic, or sulfur-bearing residues. We used PCR amplification of S. pistillata cDNA to confirm the correct transcription of an internal portion of P12 that contains one of the LC-MS/MS sequences (Fig. S2).
Fig. 1.
Predicted structure of P12, a potential BMP inhibitor. The amino acid sequence (A) and predicted secondary structure (B) of a unique SOM protein after cleavage of the export signal peptide. For the sequence, peptides sequenced by LC-MS/MS are colored orange; translated sequence confirmed from PCR amplification of S. pistillata cDNA is underlined; stop codon is marked with an asterisk; the predicted glycosylation site, Asn79 is indicated by a “+.” In the structure, the N-terminal region corresponding to a potential binding site with a bone morphogenic protein is shown in blue, and Cys potentially involved in a cystine knot fold disulfide bonds between Cys107 and Cys144, and Cys-137 and Cys181 are colored red.
Consensus structure predictions of P12 in I-TASSER and Phyre2 (Fig. 1), suggest that it may be related to the secreted protein noggin that binds to and inhibits the function of some bone morphogenic proteins (BMPs), of which one, a BMP2/4 ortholog, is known to be present in coral calicoblastic cells (47). The predicted structure (I-TASSER: Tm-score of 0.74, rmsd of 3.5 Å, and query sequence coverage of 94.6%) exhibits a β-sheet portion and cystine-knot cytokine fold found in the noggin protein family (98.9% confidence by Phyre2). In the complete structure, disulfide bonds could occur between Cys107 and Cys144, and Cys-137 and Cys181 (Fig. 1). The P12 sequence contains CXGC and CXC and the last Cys in the knot is immediately followed by a stop codon, which is standard for this type of fold (48). These structure predictions suggest that P12 may have a role in inhibiting skeleton formation by blocking the coral BMP2/4 ortholog’s receptor binding regions.
CARP Subfamily.
Two coral acid-rich proteins (CARPs), CARP4 and CARP5, were sequenced by LC-MS/MS and we propose that they belong to a highly acidic subfamily of proteins that is well conserved across Order Scleractinia but appears to be absent from other known biomineralizers. Six internal peptides of CARP4 (P2) and four internal peptides of CARP5 (P15) were sequenced by LC-MS/MS with best sequencing following proteinase K digestion (Fig. S2). CARP4 and CARP5 have predicted isoelectric points of 4.02 and 4.04, respectively. Two sites of glycosylation, Asn-98 and Asn-126, are predicted for CARP4, whereas only one, Asn-133, is predicted for CARP5 (Fig. S3). Detection of CARP5 and the C-terminal end of CARP4 only in deglycosylated samples supports this prediction. Glycosylation of CARP4 is also suggested by the difference between its predicted size and that of a 55-kDa protein that was partially analyzed by Puverel et al. (49), and which contains two internal peptides matching CARP4 (Fig. S3). Additionally, anomalous migration of highly charged proteins in SDS/PAGE systems has previously been observed for Aspein, a highly acidic molluscan protein (17).
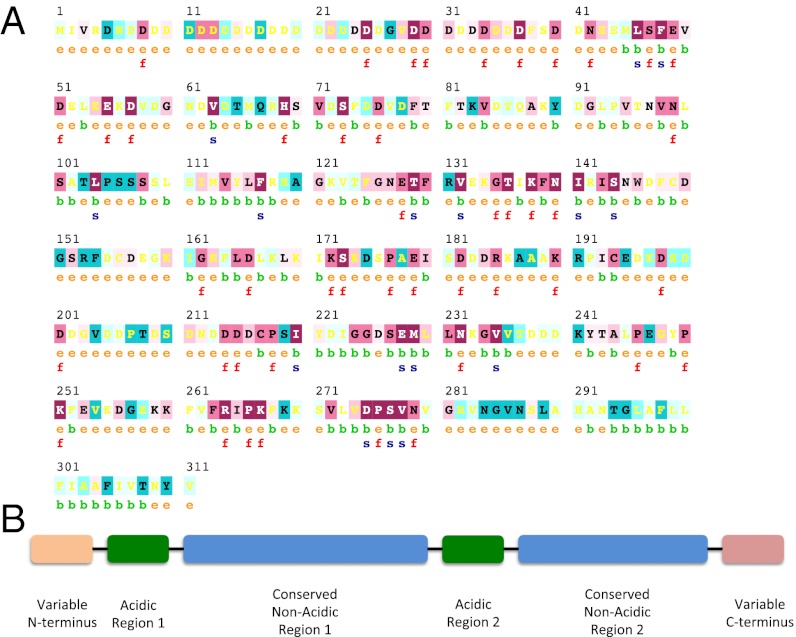
CARPs 4 and 5 exhibit significant sequence identity (31–85%) with predicted genes from three other stony corals (Fig. S3). However, similar sequences are absent outside of Order Scleractinia (Table 1). Multiple sequence alignment of CARP4- and CARP5-like proteins from P. damicornis, A. digitifera, and Favia sp. reveals several highly conserved regions of this unique coral protein subfamily (18, 35, 38). The general pattern appears to be a variable N terminus followed by one highly acidic region plus a highly conserved nonacidic region; this combination of acidic-plus-nonacidic regions is then repeated before ending in a variable C terminus (Fig. 2). The duplication and then variation of the general gene pattern within each species examined could allow for redundancy in supporting the activity of the protein subfamily. A similar pattern of redundancy, but not sequence similarity, in a highly acidic protein subfamily is observed in the Asprich proteins of A. rigida (15).
Fig. 2.
CARP subfamily general pattern. (A) Conservation of residues in the CARP subfamily as predicted by ConSurf, with CARP4 as the query. Warmer (more red) and cooler (more blue) colors represent conserved and variable amino acid positions, respectively. Residues are predicted to be exposed (e), buried (b), functional (i.e., highly conserved and exposed; f), or structural (i.e., highly conserved and buried, s). Numbers indicate residue number of CARP4. (B) Schematic of the CARP subfamily of SOM proteins. An N-terminal variable region is followed by a repeat of a highly acidic region plus a highly conserved nonacidic region; the C terminus is variable.
Like the Asprich protein family described by Gotliv et al. (15) for the mollusk, A. rigida, we propose that the two acidic regions of the CARP subfamily in corals are templates on which calcium carbonate nucleation or growth could occur (7). Unlike the Asprich subfamily, these CARPs contain two nonacidic, yet highly conserved regions that we propose to represent potential protein–protein interaction sites based on their degree of conservation (Fig. 2). Binding to structural proteins described above would allow these highly acidic proteins to be arrayed in an ordered fashion in the calcifying space for a tighter control by corals over biomineralization.
Additional, highly acidic proteins will almost certainly be found in coral skeleton by other methods. The lack of peptide sequence variability in these proteins makes them poor candidates for identification by LC-MS/MS. Hence, CARPs 4 and 5 should be considered the first, but likely not the only, acidic proteins involved in coral biomineralization.
In summary, the proteomics approach used here identified 36 proteins in S. pistillata SOM. Our results suggest that the in vivo coral SOM protein complex is laid out as follows: cadherins, integrins, contactin, and similar adhesion proteins play a dual role to (i) constitute an extracellular matrix that adheres to newly formed skeleton and (ii) attach calicoblastic cells to this skeleton-blanketing matrix. Actins and tubulins allow for flexibility of the calicoblastic space’s size and shape during aragonite crystal growth. Collagens provide a structural support within the calicoblastic space to which CARP subfamily and analogous proteins can bind as sites of mineral nucleation and growth, and at least two carbonic anhydrases mediate the subsequent carbonate chemistry effects. Mineral reworking proteins, although not found in this study, are also likely present and P12 may modulate their activity. We suggest that together, these 36 proteins constitute part of the biomineralization toolkit of Order Scleractinia; some proteins may be part of the general toolkit of all calcium carbonate mineralizers (e.g., recent proteomic analyses by refs. 50 and 33), and others, particularly the CARP subfamily described here, are clearly limited to the aragonite precipitating corals. Whereas almost certainly more proteins will be discovered in the SOM, this initial set provides a basis for understanding the spatial relationships between the major components within the skeleton and how their relative expression influences rates of calcification. Future expression studies of the effects of disturbance on coral biomineralization will be guided by pinpointing the spatial and temporal arrangement and function of these 36 SOM proteins in the calicoblastic space.
Methods
Model Organism.
S. pistillata, a common hermatypic coral found throughout the Pacific and Indian oceans (51), has been well studied both in situ and in laboratory settings. We grew coral nubbins at 28 °C in an 800-L flow through system, as previously described (34).
SOM Extraction.
S. pistillata skeletons were soaked for 4 h in 3% (wt/vol) sodium hypochlorite, copiously rinsed in deionized water, and dried overnight at 60 °C. Dried skeletons were ground to a fine powder with an agate mortar and pestle and again bleached, rinsed, and dried. The skeletal powder was decalcified in 1 N HCl at room temperature while shaking. HCl was added gradually so that the solution reached neutral pH within 30 min of acid addition; more HCl was only added if skeleton powder remained after 30 min. pH of the decalcification solution was brought to neutral with 1 M NaOH. Water-soluble and -insoluble organic fractions were separated by centrifugation and analyzed separately. Trichloroacetic acid (TCA)-acetone precipitations were used to clean and precipitate proteins from the decalcification solution (52). Briefly, one volume of 60% (wt/vol) TCA was added to five volumes soluble SOM samples and 1 mL 60% (wt/vol) TCA was added to insoluble SOM pellets. Both fractions were incubated at 4 °C overnight, centrifuged at 10,000 × g at 4 °C for 30 min, washed twice with ice-cold 90% (vol/vol) acetone at 4 °C for 15 min, and centrifuged at 10,000 × g at 4 °C for 30 min. Additionally, SOM proteins were enzymatically deglycosylated with O-glycosidase, N-glycosidase F, sialidase, B1-4 galactosidase, and B-N-acetylglucosaminidase in a deglycosylation mix per manufacturer instructions (New England BioLabs).
Protein Separation and Characterization.
SOM proteins were separated by SDS/PAGE and bands were visualized by silver staining (Pierce silver stain for mass spectrometry) and Periodic acid-Schiff staining (Pierce glycoprotein staining kit). Smearing of proteins in gels precluded extraction of individual bands for sequencing.
Proteomics.
SOM complexes were digested either by trypsin or proteinase K, and masses and charges of the digested peptides were analyzed on a Thermo LTQ-Orbitrap-Velos ETD mass spectrometer with Dionex U-3000 Rapid Separation nano LC system. The LC-MS/MS data were searched using predicted gene models from S. pistillata by X! Tandem using an in-house version of the Global Proteome Machine (GPM USB; Beavis Informatics) with carbamidoethyl on cysteine as a fixed modification and oxidation of methionine and tryptophan as a variable modification (53). Spectra were also analyzed against a suite of potential microbial genomes to exclude possible microbial contamination of the dry skeleton. Data for LC-MS/MS sequenced proteins have been deposited in GenBank (Table 1).
Gene Confirmation.
Internal sequences of predicted genes were confirmed in DNA and cDNA by PCR using gene-specific primers (Table S2). Holobiont DNA and cDNA were prepared as previously described from S. pistillata colonies maintained in in-house aquaria (34). All PCR tubes contained 0.25-μg template, 0.2 mM dNTPs, 1× High Fidelity reaction buffer, 0.5 μM of each primer, and 0.04 units μL−1 of Phusion polymerase (New England BioLabs) in a 25-μL reaction volume. Amplifications were performed in a Veriti Thermal Cycler (Applied Biosystems) at 35 cycles of 98 °C for 10 s, primer-specific annealing temperature for 30 s, and 72 °C for 30–180 s. PCR products were sequenced by GENEWIZ.
Bioinformatics.
LC-MS/MS results were filtered to remove hits from standard contamination (common Repository of Adventitious Proteins, or cRAP, database). A nonredundant list of all proteins detected with e-values ≤10−10 was used for blast analysis against NCBI and to query a database we created that contains translated sequences from Homo sapiens (54), Thalassiosira pseudonana [diatom (55)], Nematostella vectensis [anemone (56)], Strongylocentrotus purpuratus [urchin (57)], E. huxleyi CCMP1516 (coccolithophore; draft genome), and A. digitifera [hard coral (18)] genomes; a transcriptome from P. damicornis [hard coral (38)]; and expressed sequence tag (EST) libraries from Favia sp. [hard coral (35)], Reticulomyxa filosa [foraminiferan (58)], and P. maxima [oyster (42)]. N. vectensis and R. filosa do not biomineralize; all other comparison species produce calcium- or silica-based minerals. Predicted proteins from the comparison species with similarities greater than 35% and e-values ≤10−10 were retained for further analysis.
For CARP subfamily homologs, predicted proteins in comparison species were combined if they closely mimicked matched S. pistillata CARPs. These combinations are noted in protein names when they are presented in the multiple sequence alignment. Residues whose conservation suggests a functional role were predicted in ConSurf (59) using CARP4 as the query sequence.
Structures of selected proteins were predicted using both I-TASSER (60) and Phyre2 (61). We used these two programs to obtain a consensus in structure matching, particularly in the case of one S. pistillata protein that showed no similarity to proteins in the NCBI and contained no known domains. Glycosylation sites were predicted using the EnsembleGly server at the AIRL at Iowa State University. Images of predicted structures were generated in MacPyMOL v1.3r1 (Schrödinger).
Supplementary Material
Acknowledgments
We thank Joseph Yaiullo of the Long Island Aquarium and Frank Natale of the Institute of Marine and Coastal Sciences for providing and maintaining corals used in this study; N. Kröger for helpful comments on an earlier draft of this manuscript; and Rutgers Center for Advanced Biotechnology and Medicine Biological Mass Spectrometry Facility for generating and helping analyze LC-MS/MS data. This research was supported by the National Science Foundation Grant EF1041143.
Footnotes
The authors declare no conflict of interest.
Data deposition: GenBank accession numbers for genes identified in the paper can be found in Table 1 in the text.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301419110/-/DCSupplemental.
References
- 1.Lowenstam HA, Weiner S. On Biomineralization. New York, NY: Oxford Univ Press; 1989. [Google Scholar]
- 2.Dove PM, Yoreo JJD, Weiner S, editors. Reviews in Mineralogy and Geochemistry: Biomineralizaiton. Washington, DC: Mineralogical Society of America; 2003. Vol 54, p 381. [Google Scholar]
- 3.Dobbs FC, Zimmerman RC, Drake LA. Occurrence of intracellular crystals in leaves of Thalassia testudinum. Aquat Bot. 2004;80(1):23–28. [Google Scholar]
- 4.Drescher B, Dillaman RM, Taylor AR. Coccolithogenesis In Scyphosphaera apsteinii (Prymnesiophyceae) J Phycol. 2012;48(6):1343–1361. doi: 10.1111/j.1529-8817.2012.01227.x. [DOI] [PubMed] [Google Scholar]
- 5.Boonrungsiman S, et al. The role of intracellular calcium phosphate in osteoblast-mediated bone apatite formation. Proc Natl Acad Sci USA. 2012;109(35):14170–14175. doi: 10.1073/pnas.1208916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wainwright SA. Skeletal organization in the coral, Pocillopora damicornis. Q J Microsc Sci. 1963;s3-104(66):169–183. [Google Scholar]
- 7.Weiner S, Hood L. Soluble protein of the organic matrix of mollusk shells: a potential template for shell formation. Science. 1975;190(4218):987–989. doi: 10.1126/science.1188379. [DOI] [PubMed] [Google Scholar]
- 8.Addadi L, Joester D, Nudelman F, Weiner S. Mollusk shell formation: A source of new concepts for understanding biomineralization processes. Chemistry. 2006;12(4):980–987. doi: 10.1002/chem.200500980. [DOI] [PubMed] [Google Scholar]
- 9.Asenath-Smith E, Li H, Keene EC, Seh ZW, Estroff LA. Crystal growth of calcium carbonate in hydrogels as a model of biomineralization. Adv Funct Mater. 2012;22(14):2891–2914. [Google Scholar]
- 10.Beniash E. Biominerals—Hierarchical nanocomposites: The example of bone. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2011;3(1):47–69. doi: 10.1002/wnan.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks MH, Bear RS, Blake CH. X-ray diffraction evidence of collagen-type protein fibers in the Echinodermata, Coelenterata and Porifera. J Exp Zool. 1949;111(1):55–78. doi: 10.1002/jez.1401110104. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg WM. Evidence of a sclerotized collagen from the skeleton of a gorgonian coral. Comp Biochem Physiol B. 1974;49(3):525–529. doi: 10.1016/0305-0491(74)90188-6. [DOI] [PubMed] [Google Scholar]
- 13.Hunter GK, Goldberg HA. Nucleation of hydroxyapatite by bone sialoprotein. Proc Natl Acad Sci USA. 1993;90(18):8562–8565. doi: 10.1073/pnas.90.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He G, Dahl T, Veis A, George A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein 1. Nat Mater. 2003;2(8):552–558. doi: 10.1038/nmat945. [DOI] [PubMed] [Google Scholar]
- 15.Gotliv B-A, et al. Asprich: A novel aspartic acid-rich protein family from the prismatic shell matrix of the bivalve Atrina rigida. ChemBioChem. 2005;6(2):304–314. doi: 10.1002/cbic.200400221. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki M, et al. An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science. 2009;325(5946):1388–1390. doi: 10.1126/science.1173793. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi T, Sarashina I, Iijima M, Endo K. In vitro regulation of CaCO(3) crystal polymorphism by the highly acidic molluscan shell protein Aspein. FEBS Lett. 2008;582(5):591–596. doi: 10.1016/j.febslet.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Shinzato C, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476(7360):320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- 19.Murdock DJE, Donoghue PCJ. Evolutionary origins of animal skeletal biomineralization. Cells Tissues Organs. 2011;194(2–4):98–102. doi: 10.1159/000324245. [DOI] [PubMed] [Google Scholar]
- 20.Knoll AH. Biomineralization and evolutionary history. Rev Mineral Geochem. 2003;54(1):329–356. [Google Scholar]
- 21.Marin F. Unusually acidic proteins in biomineralization. In: Baurlein E, Picket-Heaps J, editors. Handbook of Biomineralization. Max Planck Institute for Biochemistry; 2007. pp. 273–286. [Google Scholar]
- 22.Veron J. Corals of Australia and the Indo-Pacific. Honolulu, HI: Univ of Hawaii Press; 1986. p. 644. [Google Scholar]
- 23.Clode PL, Marshall AT. Low temperature FESEM of the calcifying interface of a scleractinian coral. Tissue Cell. 2002;34(3):187–198. doi: 10.1016/s0040-8166(02)00031-9. [DOI] [PubMed] [Google Scholar]
- 24.Goffredo S, et al. The skeletal organic matrix from Mediterranean coral Balanophyllia europaea influences calcium carbonate precipitation. PLoS ONE. 2011;6(7):e22338. doi: 10.1371/journal.pone.0022338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young SD. Organic material from scleractinian coral skeletons - I. Variation in composition between several species. Comp Biochem Physiol. 1971;40B(1):113–120. [Google Scholar]
- 26.Ducklow HW, Mitchell R. Composition of mucus released by coral reef coelenterates. Limnol Oceanogr. 1979;24(4):706–714. [Google Scholar]
- 27.Watanabe T, Fukuda I, China K, Isa Y. Molecular analyses of protein components of the organic matrix in the exoskeleton of two scleractinian coral species. Comp Biochem Physiol B Biochem Mol Biol. 2003;136(4):767–774. doi: 10.1016/s1096-4959(03)00177-5. [DOI] [PubMed] [Google Scholar]
- 28.Moya A, et al. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol Ecol. 2012;21(10):2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x. [DOI] [PubMed] [Google Scholar]
- 29.Kaniewska P, et al. Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS ONE. 2012;7(4):e34659. doi: 10.1371/journal.pone.0034659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunagawa S, DeSalvo MK, Voolstra CR, Reyes-Bermudez A, Medina M. Identification and gene expression analysis of a taxonomically restricted cysteine-rich protein family in reef-building corals. PLoS ONE. 2009;4(3):e4865. doi: 10.1371/journal.pone.0004865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayward DC, et al. Differential gene expression at coral settlement and metamorphosis—A subtractive hybridization study. PLoS ONE. 2011;6(10):e26411. doi: 10.1371/journal.pone.0026411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarashina I, Endo K. Skeletal matrix proteins of invertebrate animals: Comparative analysis of their amino acid sequences. Paleontological Research. 2006;10(4):311–336. [Google Scholar]
- 33.Marie B, et al. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci USA. 2012;109(51):20986–20991. doi: 10.1073/pnas.1210552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mass T, et al. Aragonite precipitation by “proto-polyps” in coral cell cultures. PLoS ONE. 2012;7(4):e35049. doi: 10.1371/journal.pone.0035049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehr SFP, et al. De novo RNA-seq assembly and clustering of expressed proteins from uncharacterized corals. BMC Genomics.
- 36.Derycke L, et al. Soluble N-cadherin fragment promotes angiogenesis. Clin Exp Metastasis. 2006;23(3–4):187–201. doi: 10.1007/s10585-006-9029-7. [DOI] [PubMed] [Google Scholar]
- 37.Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54(7):993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- 38.Traylor-Knowles N, et al. Production of a reference transcriptome and transcriptomic database (PocilloporaBase) for the cauliflower coral, Pocillopora damicornis. BMC Genomics. 2011;12(1):585. doi: 10.1186/1471-2164-12-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehrlich H. Chitin and collagen as universal and alternative templates in biomineralization. Int Geol Rev. 2010;52(7–8):661–699. [Google Scholar]
- 40.Silver FH, Landis WJ. Deposition of apatite in mineralizing vertebrate extracellular matrices: A model of possible nucleation sites on type I collagen. Connect Tissue Res. 2011;52(3):242–254. doi: 10.3109/03008207.2010.551567. [DOI] [PubMed] [Google Scholar]
- 41.Helman Y, et al. Extracellular matrix production and calcium carbonate precipitation by coral cells in vitro. Proc Natl Acad Sci USA. 2008;105(1):54–58. doi: 10.1073/pnas.0710604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson DJ, et al. Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 2010;27(3):591–608. doi: 10.1093/molbev/msp278. [DOI] [PubMed] [Google Scholar]
- 43.Bertucci A, Tambutté S, Supuran CT, Allemand D, Zoccola D. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol (NY) 2011;13(5):992–1002. doi: 10.1007/s10126-011-9363-x. [DOI] [PubMed] [Google Scholar]
- 44.Moya A, et al. Carbonic anhydrase in the scleractinian coral Stylophora pistillata: Characterization, localization, and role in biomineralization. J Biol Chem. 2008;283(37):25475–25484. doi: 10.1074/jbc.M804726200. [DOI] [PubMed] [Google Scholar]
- 45.Venn AA, et al. Impact of seawater acidification on pH at the tissue–skeleton interface and calcification in reef corals. Proc Natl Acad Sci USA. 2012;110(5):1634–1639. doi: 10.1073/pnas.1216153110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyamoto H, et al. A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci USA. 1996;93(18):9657–9660. doi: 10.1073/pnas.93.18.9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoccola D, et al. Specific expression of BMP2/4 ortholog in biomineralizing tissues of corals and action on mouse BMP receptor. Mar Biotechnol (NY) 2009;11(2):260–269. doi: 10.1007/s10126-008-9141-6. [DOI] [PubMed] [Google Scholar]
- 48.Vitt UA, Hsu SY, Hsueh AJW. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 2001;15(5):681–694. doi: 10.1210/mend.15.5.0639. [DOI] [PubMed] [Google Scholar]
- 49.Puverel S, et al. Soluble organic matrix of two Scleractinian corals: Partial and comparative analysis. Comp Biochem Physiol B Biochem Mol Biol. 2005;141(4):480–487. doi: 10.1016/j.cbpc.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Mann K, Wilt FH, Poustka AJ. Proteomic analysis of sea urchin (Strongylocentrotus purpuratus) spicule matrix. Proteome Sci. 2010;8:33. doi: 10.1186/1477-5956-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veron JEN. Corals of the World. Monterey, CA: Sea Challengers; 2000. [Google Scholar]
- 52.Jiang L, He L, Fountoulakis M. Comparison of protein precipitation methods for sample preparation prior to proteomic analysis. J Chromatogr A. 2004;1023(2):317–320. doi: 10.1016/j.chroma.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 53.Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 54.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306(5693):79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan JC, et al. StellaBase: The Nematostella vectensis genomics database. Nucleic Acids Res. 2006;34(Database issue) suppl 1:D495–D499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sodergren E, et al. Sea Urchin Genome Sequencing Consortium The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314(5801):941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burki F, Nikolaev SI, Bolivar I, Guiard J, Pawlowski J. Analysis of expressed sequence tags from a naked foraminiferan Reticulomyxa filosa. Genome. 2006;49(8):882–887. doi: 10.1139/g06-048. [DOI] [PubMed] [Google Scholar]
- 59.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38(Web Server issue) suppl 2:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.