Abstract
Previous studies showed that the cytoplasmic transport of nanoparticles to the nucleus is driven by a vesicular sorting system. Artificial approaches for targeting a microtubule-associating motor complex is also a challenge. We describe herein the development of a liposomal nanoparticle, the surface of which is modified with stearylated octa-arginine (STR-R8), and a dynein light chain (LC8)-associated peptide derived from an African swine fever virus protein p54 (p54149-161) with polyethyleneglycol (PEG) as a spacer (p54149-161-PEG/R8-liposomal nanoparticles (LNPs)). The p54149-161-PEG/R8-LNPs preferentially gain access to the nucleus, resulting in a one- to two-order of magnitude higher transfection activity in comparison with p54149-161-free nanoparticles (PEG/R8-LNPs). Further studies of particle tracking in HeLa cells stably expressing green fluorescent protein (GFP)-tagged tubulin (GFP/Tub-HeLa) indicate that p54149-161 stimulated the transport of nanoparticles along fibrous tubulin structures. Moreover, a part of the p54149-161-PEG/R8-LNPs appeared to undergo quasi-straight transport without sharing the tracks corresponding to PKH67, the plasma membrane of which had been prestained with a marker just before transfection, while corresponding movement was never observed in the case of PEG/R8-LNPs. These findings suggest that a portion of the p54149-161-modified nanoparticles can use microtubule-dependent transport without the need for an assist by a vesicular sorting system.
Introduction
For the successful delivery of therapeutic genes, nuclear targeting is an important issue that needs to be overcome.1,2 In addition to biomembranes (i.e., plasma/endosomal membranes and nuclear membranes), the cytoplasm is also a crucial obstacle for the nuclear delivery of DNA. The diffusion of DNA in the cytoplasm is severely limited; the diffusion coefficient of naked DNA >2,000 bp in the cytoplasm is <1% of that in water,3 which is most likely due to restricted movement by actin cytoskeletal filament.4 Moreover, DNA is easily degraded by nucleases in the cytoplasm with a half-life of dozens of minutes.5 The condensation and/or encapsulation of DNA to form nano-sized particles provide protection against the action of cellular nucleases. Furthermore, the design of a nanoparticle to exploit the cellular factors involved in nuclear targeting is of great importance for efficient nuclear transport.
Real-time particle tracking is a powerful technology that has the capability to provide new insights into the mechanism of the cytoplasmic transport of viruses6 and artificial nanoparticles (i.e., polyethyleneimine-based nanoparticles7,8,9 and liposomal nanoparticles10,11). It is generally thought that the directional transport of polyplexes7 and lipoplexes11 occurs via vesicular transport, since the major fraction of the particles (>90%) were co-localized with a fluid-phase marker. We also provided support for these conclusions by using octa-arginine (R8)-modified liposomal nanoparticles (R8-liposomal nanoparticles (LNPs)).10 In this case, microtubule-dependent transport was observed only in particles that were co-localized with fluid-phase markers (endosomes). More importantly, in relation to this study, particles free from co-localization with endosomes never exhibited directional motion. These data prompted us to target a motor protein, in an attempt to artificially control the cytoplasmic transport of nanoparticles after endosomal escape.
Recent studies with live cells have clarified that the nuclear transport of endogenous proteins such as p53,12 parathyroid hormone-related protein13 and nuclear factor kappa B14 is assisted by microtubule-dependent transport. In addition, various types of incoming viruses15,16,17 including adenovirus6,18,19,20 and herpes simplex virus19,21 also use this machinery to deliver their genomes to the nucleus. Cytoplasmic dyneins comprise a superfamily of molecular motors that deliver the cargos to the minus-end of microtubules (retrograde transport). The dynein motor is a multi-subunit protein that contains two heavy chains, two intermediate chains (ICs), four light intermediate chains and a variable number of homodimeric light chains (LCs).15,22 To date, a yeast-hybrid system and a proteomics approach revealed that a large number of endogenous23,24,25,26 and virus-derived proteins15,21,27,28,29 interact with the components of LCs; LC8 or t-complex testis-expressed-1 (Tctex-1).30 Therefore, the findings reported in these earlier investigations suggest that LC8 functions as an adapter of these cargos. However, later structural and thermodynamic studies contradicted these models; the binding sites of the LC8 homodimer to the proposed cargos were sequestered by the multivalent binding with ICs in dynein complex formation.31 Therefore, LC8 is now thought to function as a dimerization stabilizer of their binding partners (scaffold proteins of various complexes), as well as the ICs in the dynein motor complex,32 but not as a mediator of the cargos to the dynein complex.
Nevertheless, we unexpectedly found that modification of the 13-amino acid peptide derived from an African swine fever virus protein p54 (YTTVTTQNTASQT; p54149-161), a peptide reported to be associated with LC8 using yeast-hybrid system27 stimulated the gene expression of pDNA-encapsulating nanoparticle. In this study, we report on an analysis of the intracellular trafficking by means of a particle tracking.
Results
Design of p54149-161-modified liposomal nanoparticle
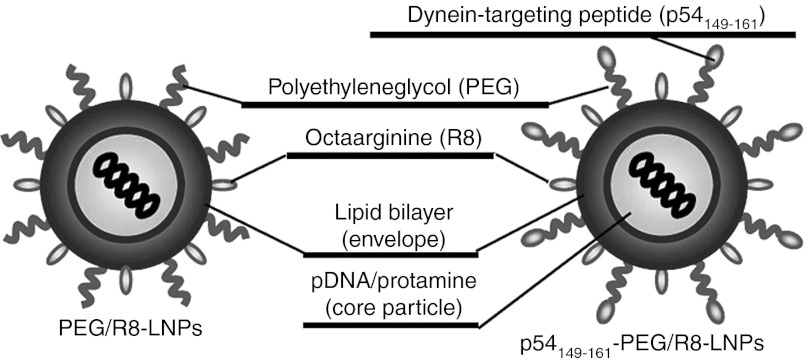
In this study, we used a pDNA-encapsulating LNPs, in which positively charged pDNA/polycation condensed particles were encapsulated in negatively charged lipid bilayers.33,34 A combination of 1.2-dioleoyl sn-glycero-3-phosphatidylethanolamine and phosphatidic acid was used as an endosome-fusogenic lipid envelope.35 The surface of the LNPs was modified with a high density (5% of total lipid) of octa-arginine (R8-LNPs), which marks the LNPs for efficient internalization into cells.36
Suh et al.37 demonstrated that the diffusivity of liposomes coated with polyethyleneglycol (PEG) was enhanced, as compared with PEG-unmodified liposomes when they were microinjected into the cytoplasm, presumably because the hydrophilic PEG layer prevented the aggregation of particles. Furthermore, we recently reported that liposomes modified with transferrin with a PEG spacer (transferrin-PEG-Lips) weretaken up by cells much more efficiently and faster than particles that had been directly modified with transferrin on the head group of the lipid (transferrin-Lips),38 presumably because modification of the ligand on the surface of the flexible PEG moiety facilitates its binding to the receptor. These results prompted us to modify the p54149-161 on the R8-modified nanoparticle with PEG as a spacer (p54149-161-PEG/R8-LNPs) (Figure 1). A cysteine-introduced and C-terminally amidated peptide (NH2-CYTTVTTQNTASQT-CONH2) was conjugated to the distearoyl-sn-Glycero-3-phoshoethanolamine-N-[(3-maleimide-1-oxopropyl)aminopropyl (polyethylene glycol)-2000] (Mal-PEG2000-DSPE) via a Michael addition reaction to prepare the p54149-161-conjugated PEG lipid (p54149-161-PEG2000-DSPE). Conjugation of the peptide to Mal-PEG2000-DSPE was confirmed by determining the molecular weight using MALDI-TOF MS (Supplementary Figure S1 online). The systematic introduction of a mutation in p54149-161 revealed that the 3 C-terminal amino acids (SQT) are essential for the binding of LC8.27 Therefore, we conjugated the peptide via its N-terminally inserted cysteine to the edge of the PEG so that the SQT domains were oriented outward from the particle surface. The synthesized p54149-161-PEG2000-DSPE was added to the lipid component (1–5% of total lipid) to prepare the p54149-161-PEG/R8-LNPs.
Figure 1.
Design of nanoparticles. pDNA was condensed with protamine, and thereafter encapsulated in the lipid envelope. A cysteine-introduced and C-terminally amidated peptides was conjugated to the Mal-PEG-DSPE via a Michael addition reaction to prepare the peptide-conjugated PEG lipid (peptide-PEG-DSPE). The surface of the nanoparticles was modified with octa-arginine (R8) and PEG by incorporating the stearylated-R8 (STR-R8) and synthesized peptide-PEG-DSPEs. PEG, polyethylene glycol.
As a comparison, we used dynein intermediate chain (DIC)-derived peptides (DICLC8; NH2-CVSYSKETQTPL-CONH2 and DICTctex-1; NH2-LGRRLHKLGVSKVTQVDFLC-CONH2), which includes peptide motifs of (K/R)XTQT and (R/K)(R/K)XX(R/K) to associate with LC823 and Tctex-1,39 respectively (Supplementary Figure S2 online). As a nontargeting control, distearoyl-sn-Glycero-3-phosphoethanolamine-N-[methoxy (polyethylene glycol)-2000] (PEG2000-DSPE), a peptide-unmodified PEG-lipid was alternatively added to prepare R8-LNPs (PEG/R8-LNPs).
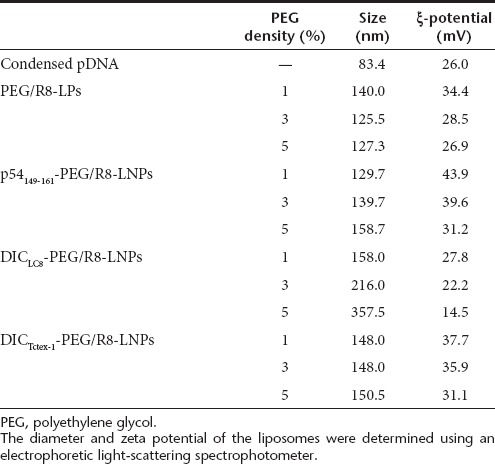
The physicochemical characteristics of PEG/R8-LNPs, p54149-161- PEG/R8-LNPs, DICLC8-PEG/R8-LNPs and DICTctex-1-PEG/R8-LNP are summarized in Table 1. The average sizes of all of the particles are quite comparable (120–150 nm). The ξ-potentials tend to decrease with the density of p54149-161-PEG2000-DSPE or PEG2000-DSPE, most likely because these incorporated PEG-lipids possess one anionic charge in their structures. However, the parameters were mutually comparable between p54149-161-PEG/R8-LNPs and PEG/R8-LNPs, when compared at the same density of PEG-lipids. Therefore, the following differences in the function and intracellular trafficking of these particles cannot be explained by the physicochemical characteristics of the preparations.
Table 1. Physicochemical characters of LNPs.
Functional evaluation of PEG/R8-LNPs and peptide-PEG/R8-LNPs
We first investigated the effect of p54149-161 on the transfection activities of encapsulated pDNA (Figure 2a). PEG/R8-LNPs and p54149-161- PEG/R8-LNPs were incubated with HeLa cells in serum-free Dulbecco's modified Eagle medium at a concentration of 1.6 µg pDNA/ml for 3 hours. The medium was then replaced with fresh culture medium, and cultured for additional 21 hours. It is noteworthy that the gene expression of PEG/R8-LNPs was gradually decreased when the density of PEG-modification increased. This is most likely because the cellular uptake and/or endosomal escape triggered by the R8-driven membrane association would be hampered by the hydrophilic PEG layer.40 Nevertheless, p54149-161-PEG/R8-LNPs exhibited a one-order (1 % of peptide modification) or approximately two-orders (3 and 5% of peptide modification) of magnitude higher transfection activity as compared with PEG/R8-LNPs. In contrast, the transfection activity was only marginally enhanced by the modification of DICLC8 and DICTctex-1. It thus appears that p54149-161 possesses a unique function that permits the intracellular trafficking of nanoparticles to be modulated. In the following studies, we focused on PEG/R8-LNPs and p54149-161-PEG/R8-LNPs that were modified with 5% of PEG2000-DSPE and p54149-161-PEG2000-DSPE, respectively. Real-time PCR was used to determine the intracellular copy numbers of pDNA at 6 hours post-transfection, which are a hybrid parameter of the cellular uptake efficacy and intracellular degradation. Although the copy number of pDNA delivered by the p54149-161-PEG/R8-LNPs was significantly greater than PEG/R8-LNP, the extent (at most twofold) does not explain a two-orders of magnitude difference in transfection activity (Figure 2b). Therefore, it is most plausible to assume that p54149-161 improved the intracellular trafficking processes.
Figure 2.
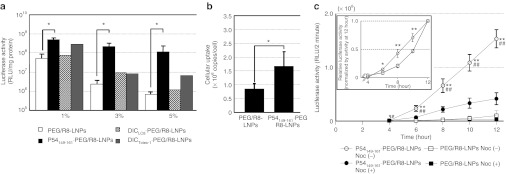
Transfection activity and intracellular copy numbers of pDNA delivered with LNPs. (a) PEG/R8-LNPs and peptide-conjugated PEG/R8- LNPs were transfected to HeLa cells for 24 hours. Each bar represents the mean gene expression of the reporter gene (luciferase) ± SD. Indicated molar percentage (to the total lipid amount) of peptide-PEG-DSPE or PEG-DSPE was incorporated into the lipid envelope. Asterisks represent a significant difference, as determined by the Mann–Whitney U-test (*P < 0.05). (b) PEG/R8-LNPs and p54149-161-PEG/R8-LNPs (2 µg pDNA) at 37 oC for 6 hours. After purification of the cellular DNA, intracellular copy numbers were quantified by Real-time PCR. Data are represented as the mean ± SD (N = 3). Asterisks represent a significant difference determined by one-way ANOVA, followed by Student's t-test. (c) The cells were incubated with PEG/R8- LNPs or p54149-161-PEG/R8-LNPs in serum-free medium in the presence or absence of 10 µmol/l nocodazole for 3 hours, followed by incubation in Dulbecco's modified Eagle medium containing 10% serum and 100 µmol/l D-luciferin. The time shown in the x-axis started from the addition of LNP solutions and the measurement started from 4 hours. The insert represents the relative gene expressions of LNPs from 4 to 12 hours in the absence of nocodazole, normalized by those at 12 hours. Statistical analyses were performed by one-way ANOVA followed by Bonferroni's multiple comparison test (**P < 0.01 against PEG/R8-LNPs, and ##P < 0.01 against nocodazole-treated condition) or one-way ANOVA followed by Student's t-test (inset; *P < 0.05 and **P < 0.01 against PEG/R8-LNPs). LNP, liposomal nanoparticles; PEG, polyethylene glycol.
To gain more insights into the mechanism of the intracellular trafficking of these particles, the time-dependent profile for transfection efficiency was monitored with AB-2550 KronosDio (ATTO, Tokyo, Japan), which allows continuous measurement of the bioluminescence by luciferase gene-transfected cells. After transfection, a medium, modified to include luciferin, was used in culturing the cells, and bioluminescence was monitored at 20-minute intervals. As shown in Figure 2c, the higher gene expressionin p54149-161-PEG/R8- LNPs as compared with PEG/R8-LNPs (>30-fold) was confirmed by this analysis. Of note, a significantly higher gene expression in p54149-161-PEG/R8-LNPs was initially observed immediately after the transfection (at 4–6 hours). The inset shows a plot of the relative gene expressions of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs, normalized by their respective ones at 12 hours. Gene expression starts to increase at 4 hours in proportion to the time of exposure to the p54149-161-PEG/R8-LNPs, while a quadratic dependence ontime was observed in the case of PEG/R8-LNPs. These results indicate that p54149-161-PEG/R8-LNPs are associated with an earlier onset of gene expression. Moreover, treatment with nocodazole, a microtubule-disruption agent, significantly impaired the gene expression of p54149-161-PEG/R8-LNPs. These collective data indicate that microtubule-dependent transport plays a key role in the nuclear delivery of pDNA. In the following studies, we focused on the impact of p54149-161-modification on the microtubule-dependent transport of the nanoparticles.
Time-dependent accumulation of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs toward the nucleus
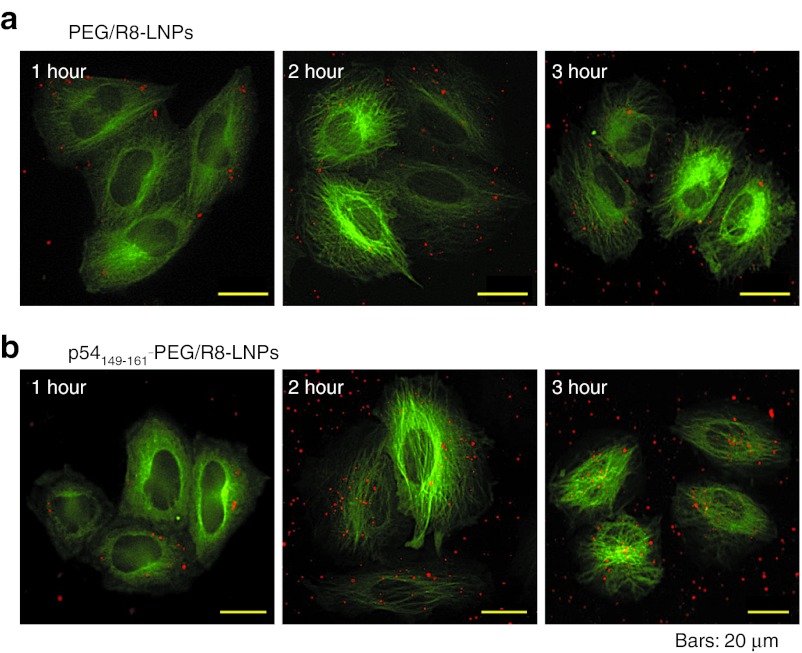
To visualize the intracellular trafficking of nanoparticles, the pDNA was labeled with rhodamine.26 HeLa cells stably expressing green fluorescent protein (GFP)-tagged tubulin (GFP/Tub-HeLa) were used to visualize microtubules. In the following imaging studies, cells were incubated with LNPs in 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (HEPES) solution at a concentration of 0.25 µg pDNA/ml for 1 hour. Under normal transfection conditions, intracellular particles are too active to permit their tracking across a time span. Therefore, the concentration of LNPs was reduced to permit the particles to be tracked in sequential frames. The images were acquired by multi-color wide-field fluorescence microscopy. Following a 1-hour pulse incubation of GFP/Tub-HeLa with PEG/R8-LNPs and p54149-161-PEG/R8-LNPs, the cells were washed to remove unbound particles, and immediately observed by fluorescence microscopy in HEPES buffer, or additionally incubated for various durations to examine the further distribution of these carriers (Figure 3). The pictures were captured at nearly the bottom of the focal plane. In fact, the settling of particles in the dish bottom could also be detected in regions that were free of GFP/Tub-HeLa cells. Of note, the particles observed around the nucleus at the bottom focal plane were not detected at the top (beneath the top of the plasma membrane) focal planes. On the contrary, particles detected at the top focal plane were not observed in the bottom focal plane (data not shown). Therefore, the nuclear accumulation observed at the bottom plane cannot be an artifact due to the leakage of the fluorescence signals derived from the particles simply just bound on the cellular surface. Collectively, it is plausible to assume that the particles detected here are located inside of the cells.
Figure 3.
Imaging of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs for the analysis of intracellular trafficking. (a) PEG/R8-LNPs and (b) p54149-161- PEG/R8-LNPs encapsulating rhodamine-labeled pDNA were transfected to GFP/Tub-HeLa cells for 1, 2, and 3 hours (represented in left, middle, and right panels, respectively). GFP, green fluorescent protein; PEG, polyethylene glycol.
At 1 hour after transfection, both particles were located beneath the plasma membrane. However, the p54149-161-PEG/R8- LNPs gradually gained access around the nucleus, especially regions that were rich in GFP/Tub (nucleus-neighboring MTOC) as shown in Figure 3b, while nuclear accumulation was poorly observed in PEG/R8-LNPs (Figure 3a).
Real-time image acquisition of microtubule-dependent intracellular transport
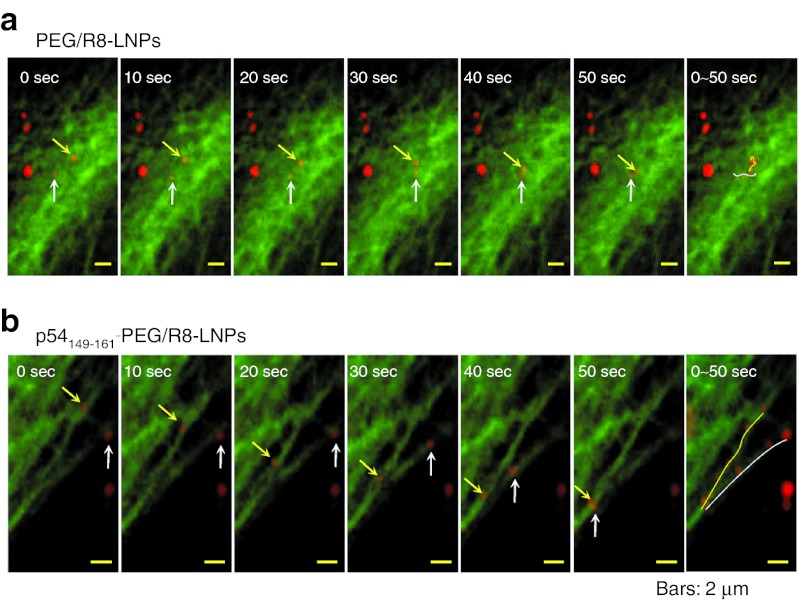
To examine the directional transport properties along with microtubules, the movement of PEG/R8-LNPs and p54149-161-PEG/R8- LNPs was tracked by multi-color real-time imaging. At an earlier time (<2 hour), the major fraction of particles were located on the upper focal plane (beneath the top of the plasma membrane) where tubulin structures were rarely observed. Therefore, fluorescence images were recorded after 2.5-hour incubation with PEG/R8-LNPs and p54149-161-PEG/R8-LNPs, when a significant number of these particles had accumulated in the focal plane that was rich in GFP/Tub signals. In the case of PEG/R8-LNPs, the major fraction of the particles showed fluctuating motion (Figure 4a and Supplementary Video S1 online). In contrast to the PEG/R8-LNPs, the cytoplasmic movement of p54149-161-PEG/R8-LNPs was active, and moreover, directional transport was well observed while the association with the microtubule filamentous structure was maintained (Figure 4b and Supplementary Video S2 online). Furthermore, when the samples were treated with nocodazole, the directional transport of p54149-161-PEG/R8-LNPs was completely lost (Supplementary Video S3 online). Together with the above data (Figure 2c) concerning the nocodazole-mediated inhibition ingene expression, these data collectively indicate that p54149-161 stimulates the microtubule-dependent transport of the nanoparticles.
Figure 4.
A series of time lapse images for PEG/R8-LNPs and p54149-161- PEG/R8-LNPs. Typical particle tracks of (a) PEG/R8-LNPs and (b) p54149-161-PEG/R8-LNPs obtained for 10-second frame intervals are represented. The original videos are represented in Supplementary Video S1 and S2 online. In the far right column, overlay images are represented to show the trajectories. PEG, polyethylene glycol.
Involvement of vesicular transport to the microtubule-dependent transport of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs
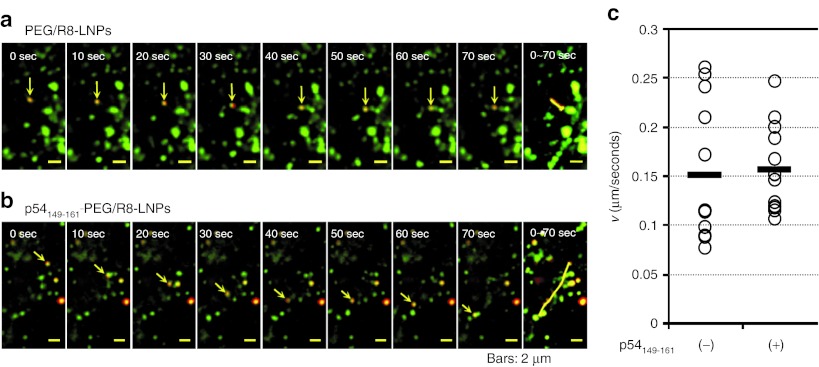
To investigate the mechanism of microtubule-dependent transport, we examined the issue of whether the directional transport of p54149-161-PEG/R8-LNPs occurred before or after endosomal release. To address this issue, the plasma membranes of HeLa cells were preliminarily labeled with PKH67 GREEN FLUORESCENT CELL LINKER (Sigma-Aldrich, St Louis, MO). This probe is incorporated into the lipid region of the plasma membrane.41 Therefore, most of the internalized vesicles should be labeled regardless of the internalization pathway. LNPs were incubated with the cells immediately after staining with PKH67 for a short duration (1 hour) to be sure that LNPs were taken up by the cells before all of the labeled PKH67 on the plasma membrane had been internalized. The punctate signals (red), irrespective of whether they were co-localized with PKH67 (green), were defined as particles that were in the process of vesicular transport or particles that had possibly escaped from endosomes (particles in cytoplasm). The particles that were subject to directional transport were observed both in PEG/R8-LNPs and p54149-161-PEG/R8-LNPs (Figures 5a,b). The movements of the particles were categorized into active transport or diffusion based on the plots of two-dimensional mean-square displacements (MSD) over time, as described previously.7,8,9,10,42 The average velocity (v) was determined by the fitting of a MSD-Δt plot (see Experimental Procedures). As shown in Figure 5c, the mean velocities and the variations of the p54149-161-PEG/R8-LNPs were comparable with those of PEG/R8-LNPs (0.16 ± 0.04 µm/second and 0.15 ± 0.07 µm/seconds, respectively), and PEG-unmodified R8-LNPs (0.21 ± 0.19 µm/second) as reported previously.10
Figure 5.
Dual imaging of vesicular transport and LNPs. (a and b) Typical images for the directional transport of PEG/R8-LNPs (a) and p54149-161-PEG/R8-LNPs (b) encapsulating rhodamine-labeled pDNA. Rhodamine-pDNA was represented as red. The transport vesicles stained with PKH67 are represented as green. In the rightmost column, overlay images were represented to show the trajectories. (c) The average velocities (v) obtained by the curve fitting of MSD-Δt curves for quasi-straight trajectories of PEG/R8-LNPs (n = 12) and p54149-161-PEG/R8-LNPs (n = 12) was plotted. Black bars represent the mean values. PEG, polyethylene glycol.
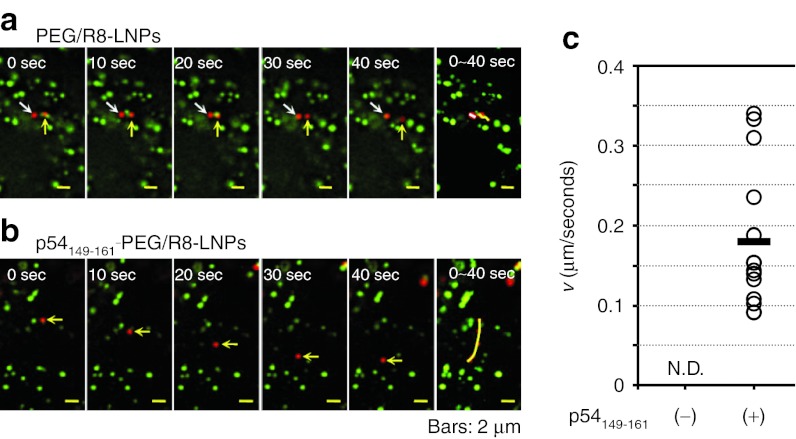
The most striking difference between PEG/R8-LNPs and p54149-161-PEG/R8-LNPs was observed when we focused on the particles that had not co-localized with the PKH67. As shown in typical images shown in Figure 6a, PEG/R8-LNPs exhibited only fluctuating movement. The plots of MSD with time could be fitted to a linear regression, indicating that these can be categorized as diffusive motion. In contrast, a part of the p54149-161-PEG/R8- LNPs showed quasi-straight motion without co-localization with PKH67 signals (Figure 6b). The average velocity (v) obtained by the fitting of a MSD-Δt plot was determined to be 0.18 µm/second, and highly variable, ranging from 0.08 µm/second to 0.35 µm/second (SD= 0.09) (Figure 6c).
Figure 6.
A series of time lapse images for PEG/R8-LNPs and p54149-161- PEG/R8-LNPs free from the vesicular transport. (a) PEG/R8-LNPs and (b) p54149-161-PEG/R8-LNPs encapsulating rhodamine-pDNA was transfected to the HeLa cells prestained by PKH67. Typical examples of the particles free from co-localization with endosomal compartment are shown. In the rightmost column, overlay images were represented to show the trajectories. (c) The average velocity (v) obtained by the curve fitting of MSD-Δt curves for quasi-straight trajectories of p54149-161-PEG/R8- LNPs (n = 14) was plotted. Black bar represent the mean values. Quasi-straight trajectory was not observed in PEG/R8-LNPs. N.D., not detected; PEG, polyethylene glycol.
Discussion
Several studies reported that naked pDNAs possessing nuclear transcription factors (i.e., nuclear factor kappa B43 and cyclic adenosine monophosphate responsive-element binding protein)44 are recognized by these responsible transcription factors, and are transported to the nucleus in a microtubule-dependent manner. However, naked pDNA is susceptible to digestion by DNase and thus has a short half-life (<1–2 hours).5 Therefore, the targeting of the pDNA-loaded nanoparticles to the motor complex would also be a desirable strategy for the nuclear delivery of pDNA with protection from enzymatic degradation. However, to the best of our knowledge, a consensus sequence that is capable of targeting the dynein complex has not been reported. Since DIC-originated peptides targeting LC8 and Tctex-1; (K/R)XTQT and (R/K)(R/K)XX(R/K), respectively are conserved in the potential protein cargos of dynein,23,39 it was assumed that LC8 and Tctex-1 served to connect these motifs in potential cargos tothe dynein complex.24 However, recent studies indicate that LC8 is not a cargo adaptor of the dynein complex, but rather functions to promote the formation of dimeric IC.32 This hypothesis is consistent with the fact that surface modification of these two peptides (DICLC8 and DICTctex) showed marginal effects on gene expression (Figure 2).
In contrast, the most significant findings of this study is that the transfection activity of pDNA-encapsulating nanoparticles was drastically enhanced by one- or two-orders of magnitude as the result of the modification of the functional peptide; p54149-161, which was identified as a LC8-associating domain in African swine fever virus-derived protein p54 (Figure 2).27 Of note, the enhanced transfection activity was consistent with a stimulated access of p54149-161-PEG/R8-LNPs around the nucleus (Figure 3). Real-time particle tracking clearly showed that pDNA encapsulated in the p54149-161-PEG/R8-LNPs underwent directional motion associated with a fibroustubulin structure (Figure 4b and Supplementary Video S2 online). A detailed particle tracking analysis revealed that the direction of the transport of p54149-161-PEG/R8-LNPs was not limited to nucleus-directed transport (Figure 4b and Supplementary Video S2 online). The apparent nuclear accumulation of p54149-161-PEG/R8-LNPs (Figure 3b) might be the result of a dynamic equilibrium involving the more frequent minus end-directed transport toward the nucleus and the less frequent plus end-directed transport as observed in adenovirus.6
To gain further insights into the mechanism of microtubule-dependent transport, we attempted to distinguish between p54149-161- PEG/R8-LNPs in the cytosol and those in endocytic vesicles. One of the major strategies is to label the vesicles with specific markers (i.e., Lysotracker for lysosomes, fluorescent protein-fused Rab5/Rab7 for early/late endosomes and Alexa Fluor Dextran for macropinocytosis). However, a certain type of nonviral vector can be taken up by the cells not only by the classical endocytosis pathway,45 but also by other pathways such as macropinocytosis46 depending on the cell line and the composition of the LNP.47 It is likely that multiple pathways are also involved in the transport of the PEG/R8-LNPs and/or p54149-161-PEG/R8-LNP. In fact, our preliminary study showed that only a small portion of the LNPs was co-localized with an early endosome marker (Venus-fused Rab5), which were genetically overexpressed in HeLa cells at 15, 40, and even 120 minutes post-transfection (data not shown). Under these circumstances, we cannot be absolutely assured that the lack of co-localization with specific endosome markers means the cytoplasmic localization of LNPs, since these markers can label only a part of the intracellular vesicles. Therefore, in this study, we used a nonspecific marker; PKH67, which is incorporated into the lipid bilayer of the plasma membrane, and would then be expected to share its intracellular fate with endocytic vesicles via membrane invagination regardless of the internalization pathway.48,49
p54149-161-PEG/R8-LNPs and PEG/R8-LNPs as well are subject to vesicular transport (Figure 5). In addition, the most unique characteristic of p54149-161-PEG/R8-LNPs is that a part of the particles are subject to quasi-straight transport without co-localization with the signals of PKH67, whereas p54149-161- unmodified PEG/R8-LNPs free from this probe did not show any directional transport (Figure 6). While this assay system might have a risk in terms of over estimating the efficiency of endosomal escape due to the release of PHK67 from endocytic vesicles, and/or quenching concomitant with intracellular degradation, this result can be explained by assuming that p54149-161-PEG/R8-LNPs are recognized by motor proteins. As shown in a prevalent model considered in earlier studies, LC8 may mediate the binding of p54149-161-PEG/R8-LNPs to the dynein complex; one binding site of the homodimeric LC8 captures the p54149-161-modified particle, and the other binds to the ICs. Alternatively, p54149-161 might bind to LC8 at a different position from where the DIC-derived (K/R)XTQT motif binds.
Meanwhile, quantitative particle tracking revealed that the directional transport of p54149-161-PEG/R8-LNPs (0.18 ± 0.09 µm/second; Figure 6) and endosomes with them (0.16 ± 0.04 µm/second; Figure 5) were slower than particle-free endosomes (0.47 ± 0.14 µm/second) as reported previously.10 One possible explanation is that the speed of motor-driven transport may be slowed down by the highly dense cargos. Alternatively, particle transport would be slowed down by intermissive association with RNAs or negatively charged organelles such as mitochondria, since the p54149-161-PEG/R8- LNP is positively charged (ζ-potential of +30 mV).
Finally, some comments are in order concerning the future design of a nanoparticle to maximize the function of this peptide. Although modification with p54149-161 stimulated transgene expression, the activity is still less than that for a commercially available transfection reagent (Lipofectamine Plus; Life Technologies, Carlsbad, CA). Also, the gene expression of EGFP was still observed to be heterogeneous (~30%) as shown in Supplementary Figure S3 online. Therefore, further attempts to maximize the function of p54149-161 are needed in the future. One of the key factors is the topology of the peptide. In this study, we modified the surface of the LNPs with p54149-161 using PEG as a spacer (Figure 1). However, surface PEGylation presents a dilemma; while PEG is useful for controlling the size and dispersibility of a carrier, it is undesirable for cellular association and endosomal escape.40 In fact, fewer numbers of intracellular particles were detectable in the case of PEG/R8-LNPs (Figure 3a) in comparison with PEG-unmodified R8-LNPs.10 Therefore, PEG/R8-LNPs that were trapped in endosomes were readily degraded in lysosomes, while a small fraction ofthe PEG/R8-LNPs underwent vesicular transport (Figure 5). To maximize the function of the p54149-161 peptide, it would be desirable to combine other technologies for the release of the peptide-conjugated particle to the cytosol. One such strategy involves coating the p54149-161-modified particle with an additional lipid bilayer that is optimized to fuse with the endosomal membrane. We previously developed a multilayered lipid nanoparticle, in which condensed DNA cores are coated with two types of lipid bilayers, both of which are designed to overcome the endosomal membrane and nuclear membranes, respectively.35 Modification of the inner membrane with p54149-161 is currently in progress.
Collectively, we here report on the utility of an African swine fever virus-derived p54149-161 peptide to improve the cytoplasmic transport of its encapsulated pDNA toward the nucleus and subsequent gene expression. Particle tracking revealed that a part of the particles move by microtubule-dependent transport and the movement is independent of vesicular trafficking. Therefore, the peptide will be useful for developing a motor complex-targeting nanoparticle for realizing a rational strategy for satisfying the protection of DNA from DNase in the cytoplasm and achieving efficient nuclear targeting.
Materials and Methods
Materials. The pDNA was purified using a Qiagen Endofree plasmid Mega Kit (Qiagen GmbH, Hilden, Germany). 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine was purchased from Avanti Polar Lipids (Alabaster, AL). Phosphatidic acid was purchased from Sigma-Aldrich. Stearylated octa-arginine (STR-R8) was synthesized as described previously.50 All other chemicals used were commercially available and reagent grade products. HeLa cell cultures and the establishment of its transfectant stably expressed with pEGFP/Tub (GFP/Tub-HeLa) were carried out described previously.10 To prepare the reporter gene vector (pcDNA3.1-GL3), an insert fragment encoding the luciferase (GL3) was obtained by the Hind III/Xba I digestion of the pGL3-basic vector (Promega, Madison, WI), and ligated to the HindIII/Xba I digested site of pcDNA3.1 (Invitrogen, Carlsbad, CA).
Synthesis of peptide-PEG-DSPE. Equimolar quantities of cystein-introduced peptides (p54149-161; NH2-CYTTVTTQNTASQT-CONH2, DICLC8; NH2-CVSYSKETQTPL-CONH2 and DICTctex-1; NH2-LGRRLHKLGVSKV TQVDFLC-CONH2) were mixed in DMSO at 30 oC for 24 hours. Conjugation of the peptide to PEG2000-DSPE was confirmed by determining the molecular weight of the resulting products by MALDI-TOF MS (Supplementary Figure S1 online).
Preparation of the R8-liposome encapsulating pDNA. R8-Lip encapsulating pDNA particles were prepared by the lipid hydration method as reported previously.10 In a typical run, pDNA (0.1 mg/ml) was condensed with protamine (0.1 mg/ml) in 10 mmol/l HEPES (pH 7.4), at N/P ratio of 2.0. A lipid film was prepared in a glass test tube by evaporating a chloroform solution of the lipids, containing 1.2-dioleoyl sn-glycero-3-phosphatidylethanolamine and phosphatidic acid at a molar ratio of 7:2 (total lipid amount: 82.5 nmol) plus indicated mol% of PEG2000-DSPE or p54149-161- PEG2000-DSPE. The prepared lipid film was then hydrated with the condensed DNA solution (150 µl) for 10 minutes at room temperature; the final lipid concentration was 0.55 mmol/l. After hydration, the tube was sonicated for 1 minute in a bath-type sonicator to complete the lipid coating of the condensed DNA (AU-25C; Aiwa, Tokyo, Japan). The diameter and zeta potential of the liposomes were determined using an electrophoretic light-scattering spectrophotometer (Zetasizer; Malvern Instruments, Malvern, WR, UK).
Transfection studies. 5 × 104 cells were seeded on a 24-well plate (Corning incorporated, Corning, NY) in 0.5 ml of culture medium 1 day before transfection. For the transfection, a 0.25 ml aliquot of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs in serum- and antibiotics-free Dulbecco's modified Eagle medium (including 0.4 µg DNA) was incubated with the cells for 3 hours. The medium was then replaced with fresh medium containing 10% serum and the cells were incubated for a further 3 hours. The cells were then washed with 0.25 ml of phosphate-buffered saline two times and lysed with 75 µl of reporter lysis buffer (Promega, Madison, WI). Luciferase activity was initiated by the addition of 50 µl of luciferase assay reagent (Promega) into 20 µl of cell lysate, and was measured by means of a luminometer (Luminescencer-PSN; ATTO, Tokyo, Japan). The amount of protein in the cell lysate was determined using a bicinchoninic acid protein assay kit (PIERCE, Rockford, IL).
Time-dependent monitoring of transfection. 8 × 104 cells were seeded on 35-mm culture dishes in 2 ml of culture medium 1 day before transfection. For the transfection, the cells were incubated with a 2 ml aliquot of PEG/R8-LNPs and p54149-161-PEG/R8-LNPs in serum- and antibiotics-free Dulbecco's modified Eagle medium (including 1.6 µg DNA) for 3 hours in the presence or absence of 10 µmol/l nocodazole. The medium was then replaced with fresh phenolred-free medium containing 10% serum and 100 µmol/l D-luciferin. The dishes were set in a luminometer incorporated in a small CO2 incubator (ATTO), and the bio luminescence was monitored at 20-minute intervals (2-minute collection time).
Comparison of the intracellular copy numbers of p54149-161-PEG/R8- LNPs and PEG/R8-LNPs. HeLa cells (2 × 105 cells) were incubated with PEG/R8-LNPs and p54149-161-PEG/R8-LNPs (2 µg pDNA) in 6-well platesat 37 oC for 6 hours. After purification of the cellular DNA by means of GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich), intracellular copy numbers were quantified by Real-time PCR. The values were normalized by cell numbers, which was quantified by the number of copies of the genomic β-actin gene. The primers used in this analysis are listed in Table 2.
Table 2. List of primers used in the quantification of intracellular copy numbers.

Fluorescence image acquisition and real-time particle tracking. For visualization of pDNA, the molecule was labeled with rhodamine by means of a Mirus Label IT CX-rhodamine nucleic acid labeling kit (Mirus, Madison, WI). The pDNA was labeled in the optimized buffer supplied with the kit, but the Label IT solution was mixed at a 1/4 concentration of the recommended protocol for labeling with rhodamine by diluting the Label IT solution with distilled water. pDNA was incubated for 120 minutes, and the rhodamine-labeled pDNA was purified by ethanol precipitation. To demonstrate particle tracking along microtubule filamentous structures, 0.5 × 105 of GFP/Tub-HeLa cells were seeded on a 3.5 cm glass base dish (IWAKI, Osaka, Japan) in 2 ml of culture medium 2 days before transfection.
To evaluate the co-localization of carriers with transport vesicles, plasma membrane were preliminarily stained by PKH67 GREEN FLUORESCENT CELL LINKER (Sigma-Aldrich) following to the protocol with some arrangements. 0.5 × 105 cells seeded on a 35 mm glass base dish for 2 days were washed with serum-free medium. After a 100-fold dilution of PKH67 with diluent C supplied in the kit (10 µmol/l), a 100 µl aliquot of the solution was applied on the center of the glass bottom region, and then incubated for 3 minutes at room temperature. After the removal of PKH67 solution, the labeling reaction was completely blocked by adding 100 µl of 100% serum. The cells were incubated for 1 minute under gentle pipetting.
After washing with HEPES solution (135 mmol/l NaCl, 5.4 mmol/l KCl, 1 mmol/l MgCl2, 1.8 mmol/l CaCl2, 5 mmol/l HEPES, and 10 mmol/l glucose), GFP/Tub-HeLa cells or PHK67-stained HeLa cells were incubated with 1 ml of HEPES solution including PEG/R8-LNPs or p54149-161-PEG/R8-LNPs prepared with rhodamine-labeled pDNA (corresponding to 0.25 µg pDNA). At 1 hour post-transfection, the cells were washed to remove external LNPs, and then further incubated in HEPES solution until being observed.
Images were acquired by Nikon ECLIPSE TE-2000-U wild field fluorescence microscopy equipped with a Nikon Plan Apo 60×/1.4 oil immersion objective (Nikon, Tokyo, Japan). Control of the microscopy and acquisition of digital images were performed with NIS -Elements software (Nikon). A mercury lamp was used for illumination. Green fluorophores (i.e., GFP/Tub and PKH67) and red fluorophores (i.e., rhodamine-labeled pDNA) were excited with light filtered through 492/18 and 580/20 excitation filters, respectively. Fluorescence was collected in the epi direction. The fluorescence was passed through a dichromatic mirror, reflections at the exciting wavelength (82100v2bs; Chroma Technology, Rockingham, VT) were further filtered from residual excitation light by bandpass filters 535/30 and 630/60, respectively). Image sequences were captured with an electron multiplier charge coupled device camera (ImagEM; Hamamatsu Photonics, Hamamatsu, Japan). The automatic particle-finding and quantitative analysis of trajectories were demonstrated by the G-track software ver1.2 (G-Angstrom K.K., Sendai, Japan), in which the center of the fluorescent spot is located by two-dimensional Gaussian fitting.
Data analysis. For each trajectory of a particle, the MSD for every time interval was calculated using the formula below:
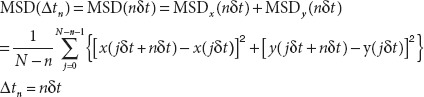 |
where (x(jΔt+nΔt), y(jΔt+nΔt)) describes the particle position following a time interval Δtn = nΔt after starting at position (x(jΔt), y(jΔt)), N is the total number of frames in the video recording sequence, Δt is the frame interval for image acquisitionand n and j are positive integers, with n determining the time increment.
Particles that exhibited quasi-straight motion through more than five successive frames allowing them to move at least 5 µm were initially selected, and thereafter, the MSD values were plotted against Δt.
The MSD-Δt plot for particles undergoing active transport is described by a quadratic curve and can be expressed as
where v is the mean velocity.7,8,9,10
The values of D and v can be obtained by fitting the MSD-Δt plot by means of an iterative nonlinear least-squares method using the MULTI program (downloaded from http://www.kobegakuin.ac.jp/~pharm/asc/excel/index.html). The input data were weighted as the reciprocal of the square of the observed values, and the Damping–Gauss–Newton method was used as the algorithm for the fitting.
SUPPLEMENTARY MATERIAL Figure S1. Conjugation of P54149-161 with PEG-lipid. Figure S2. Schematic diagram illustrating a dynein complex and its targeting peptide. Figure S3. Transfection activities of PEG/R8-NLPs and p54149-161-PEG/R8-NLPs. Video S1. Videos to Figure 4a. PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells. Video S2. Videos to Figure 4b. p54149-161-PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells. Video S3. p54149-161-PEG/R8-LNPs (5mol% modification) encapsulating the rhodamine-labeled pDNA were transfected to the HeLa cells in the presence of 10 µM nocodazole.
Acknowledgments
This work was supported in part by Funding Program for Next Generation World-Leading Researchers (NEXT Program), and Grant-in-Aid for Young Scientists (A) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. H.A. is also supported by the Asahi Glass Foundation, and by Photographic Research Fund of Konica Minolta Imaging Science Foundation. The authors would also like to thank Dr. M. S. Feather for his helpful advice in writing the English manuscript. The authors declared no conflict of interest.
Supplementary Material
Conjugation of P54149-161 with PEG-lipid.
Schematic diagram illustrating a dynein complex and its targeting peptide.
Transfection activities of PEG/R8-NLPs and p54149-161-PEG/R8-NLPs.
Videos to Figure 4a. PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells.
Videos to Figure 4b. p54149-161-PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells.
p54149-161-PEG/R8-LNPs (5mol% modification) encapsulating the rhodamine-labeled pDNA were transfected to the HeLa cells in the presence of 10 µM nocodazole.
REFERENCES
- Kamiya H, Akita H., and, Harashima H. Pharmacokinetic and pharmacodynamic considerations in gene therapy. Drug Discov Today. 2003;8:990–996. doi: 10.1016/s1359-6446(03)02889-7. [DOI] [PubMed] [Google Scholar]
- Mastrobattista E, van der Aa MA, Hennink WE., and, Crommelin DJ. Artificial viruses: a nanotechnological approach to gene delivery. Nat Rev Drug Discov. 2006;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N., and, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Dauty E., and, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J Biol Chem. 2005;280:7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW.et al. (1999Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer Gene Ther 6482–497. [DOI] [PubMed] [Google Scholar]
- Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP., and, Greber UF. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausinger R, von Gersdorff K, Braeckmans K, Ogris M, Wagner E, Bräuchle C.et al. (2006The transport of nanosized gene carriers unraveled by live-cell imaging Angew Chem Int Ed Engl 451568–1572. [DOI] [PubMed] [Google Scholar]
- de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M.et al. (2007Cellular dynamics of EGF receptor-targeted synthetic viruses Mol Ther 151297–1305. [DOI] [PubMed] [Google Scholar]
- Suh J, Wirtz D., and, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci USA. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita H, Enoto K, Masuda T, Mizuguchi H, Tani T., and, Harashima H. Particle tracking of intracellular trafficking of octaarginine-modified liposomes: a comparative study with adenovirus. Mol Ther. 2010;18:955–964. doi: 10.1038/mt.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer AM, de Bruin KG, Ruthardt N, Mykhaylyk O, Plank C., and, Bräuchle C. Dynamics of magnetic lipoplexes studied by single particle tracking in living cells. J Control Release. 2009;137:136–145. doi: 10.1016/j.jconrel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Giannakakou P, Sackett DL, Ward Y, Webster KR, Blagosklonny MV., and, Fojo T. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat Cell Biol. 2000;2:709–717. doi: 10.1038/35036335. [DOI] [PubMed] [Google Scholar]
- Lam MH, Thomas RJ, Loveland KL, Schilders S, Gu M, Martin TJ.et al. (2002Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules Mol Endocrinol 16390–401. [DOI] [PubMed] [Google Scholar]
- Mikenberg I, Widera D, Kaus A, Kaltschmidt B., and, Kaltschmidt C. Transcription factor NF-kappaB is transported to the nucleus via cytoplasmic dynein/dynactin motor complex in hippocampal neurons. PLoS ONE. 2007;2:e589. doi: 10.1371/journal.pone.0000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF., and, Way M. A superhighway to virus infection. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Hernáez B, Tarragó T, Giralt E, Escribano JM., and, Alonso C. Small peptide inhibitors disrupt a high-affinity interaction between cytoplasmic dynein and a viral cargo protein. J Virol. 2010;84:10792–10801. doi: 10.1128/JVI.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider MA, Spoden GA, Florin L., and, Lambert C. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol. 2011;13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E.et al. (2000Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis Hum Gene Ther 11151–165. [DOI] [PubMed] [Google Scholar]
- Mabit H, Nakano MY, Prank U, Saam B, Döhner K, Sodeik B.et al. (2002Intact microtubules support adenovirus and herpes simplex virus infections J Virol 769962–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer J., and, Vallee RB. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to pH-primed hexon. Viruses. 2011;3:1417–1431. doi: 10.3390/v3081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas MW, Diefenbach RJ, Homa FL, Miranda-Saksena M, Rixon FJ, Vittone V.et al. (2004Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport J Biol Chem 27928522–28530. [DOI] [PubMed] [Google Scholar]
- Mallik R., and, Gross SP. Molecular motors: strategies to get along. Curr Biol. 2004;14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- Lo KW, Naisbitt S, Fan JS, Sheng M., and, Zhang M. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem. 2001;276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- Moseley GW, Roth DM, DeJesus MA, Leyton DL, Filmer RP, Pouton CW.et al. (2007Dynein light chain association sequences can facilitate nuclear protein import Mol Biol Cell 183204–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Crespo I, Yélamos B, Roncal F, Albar JP, Ortiz de Montellano PR., and, Gavilanes F. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 2001;503:135–141. doi: 10.1016/s0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U., and, Sung CH. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Alonso C, Miskin J, Hernáez B, Fernandez-Zapatero P, Soto L, Cantó C.et al. (2001African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein J Virol 759819–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Badrane H, Ceccaldi PE., and, Tordo N. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raux H, Flamand A., and, Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB, Williams JC, Varma D., and, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC., and, Hendrickson WA. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci USA. 2007;104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P, Szenes á, Radnai L, Bakos A, Pál G., and, Nyitray L. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 2011;278:2980–2996. doi: 10.1111/j.1742-4658.2011.08254.x. [DOI] [PubMed] [Google Scholar]
- Kogure K, Akita H., and, Harashima H. Multifunctional envelope-type nano device for non-viral gene delivery: concept and application of Programmed Packaging. J Control Release. 2007;122:246–251. doi: 10.1016/j.jconrel.2007.06.018. [DOI] [PubMed] [Google Scholar]
- Kogure K, Akita H, Yamada Y., and, Harashima H. Multifunctional envelope-type nano device (MEND) as a non-viral gene delivery system. Adv Drug Deliv Rev. 2008;60:559–571. doi: 10.1016/j.addr.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R.et al. (2009Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process Biomaterials 302940–2949. [DOI] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Futaki S., and, Harashima H. High density of octaarginine stimulates macropinocytosis leading to efficient intracellular trafficking for gene expression. J Biol Chem. 2006;281:3544–3551. doi: 10.1074/jbc.M503202200. [DOI] [PubMed] [Google Scholar]
- Suh J, Choy KL, Lai SK, Suk JS, Tang BC, Prabhu S.et al. (2007PEGylation of nanoparticles improves their cytoplasmic transport Int J Nanomedicine 2735–741. [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Kogure K, Chaki S, Nakamura Y, Moriguchi R, Hamada H.et al. (2008An artificial virus-like nano carrier system: enhanced endosomal escape of nanoparticles via synergistic action of pH-sensitive fusogenic peptide derivatives Anal Bioanal Chem 3912717–2727. [DOI] [PubMed] [Google Scholar]
- Mok YK, Lo KW., and, Zhang M. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem. 2001;276:14067–14074. doi: 10.1074/jbc.M011358200. [DOI] [PubMed] [Google Scholar]
- Hatakeyama H, Akita H., and, Harashima H. A multifunctional envelope type nano device (MEND) for gene delivery to tumours based on the EPR effect: a strategy for overcoming the PEG dilemma. Adv Drug Deliv Rev. 2011;63:152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Iwasa A, Akita H, Khalil I, Kogure K, Futaki S., and, Harashima H. Cellular uptake and subsequent intracellular trafficking of R8-liposomes introduced at low temperature. Biochim Biophys Acta. 2006;1758:713–720. doi: 10.1016/j.bbamem.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Suh J, Wirtz D., and, Hanes J. Real-time intracellular transport of gene nanocarriers studied by multiple particle tracking. Biotechnol Prog. 2004;20:598–602. doi: 10.1021/bp034251y. [DOI] [PubMed] [Google Scholar]
- Mesika A, Kiss V, Brumfeld V, Ghosh G., and, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–208. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- Badding MA, Vaughan EE., and, Dean DA. Transcription factor plasmid binding modulates microtubule interactions and intracellular trafficking during gene transfer. Gene Ther. 2012;19:338–346. doi: 10.1038/gt.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhorn IS, Kalicharan R., and, Hoekstra D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J Biol Chem. 2002;277:18021–18028. doi: 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- Gonçalves C, Mennesson E, Fuchs R, Gorvel JP, Midoux P., and, Pichon C. Macropinocytosis of polyplexes and recycling of plasmid via the clathrin-dependent pathway impair the transfection efficiency of human hepatocarcinoma cells. Mol Ther. 2004;10:373–385. doi: 10.1016/j.ymthe.2004.05.023. [DOI] [PubMed] [Google Scholar]
- dos Santos T, Varela J, Lynch I, Salvati A., and, Dawson KA. Quantitative assessment of the comparative nanoparticle-uptake efficiency of a range of cell lines. Small. 2011;7:3341–3349. doi: 10.1002/smll.201101076. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Khalil IA, Kogure K, Futaki S., and, Harashima H. Octaarginine- and octalysine-modified nanoparticles have different modes of endosomal escape. J Biol Chem. 2008;283:23450–23461. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Masuda T, Khalil I, Akita H., and, Harashima H. Enhanced gene expression by a novel stearylated INF7 peptide derivative through fusion independent endosomal escape. J Control Release. 2009;138:160–167. doi: 10.1016/j.jconrel.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K.et al. (2001Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery J Biol Chem 2765836–5840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conjugation of P54149-161 with PEG-lipid.
Schematic diagram illustrating a dynein complex and its targeting peptide.
Transfection activities of PEG/R8-NLPs and p54149-161-PEG/R8-NLPs.
Videos to Figure 4a. PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells.
Videos to Figure 4b. p54149-161-PEG/R8-LNPs (5 mol% modification) encapsulating the rhodamine-labeled pDNA was transfected to the HeLa cells.
p54149-161-PEG/R8-LNPs (5mol% modification) encapsulating the rhodamine-labeled pDNA were transfected to the HeLa cells in the presence of 10 µM nocodazole.