Abstract
Mouse monocytes exposed to macrophage colony-stimulating factor (M-CSF) and interferon-γ (IFN-γ) were driven to a novel suppressor phenotype. These regulatory macrophages (M regs) expressed markers distinguishing them from M0-, M1-, and M2-polarized macrophages and monocyte-derived dendritic cells (DCs). M regs completely suppressed polyclonal T cell proliferation through an inducible nitric oxide synthase (iNOS)-dependent mechanism. Additionally, M regs eliminated cocultured T cells in an allospecific fashion. In a heterotopic heart transplant model, a single intravenous administration of 5 × 106 donor-strain M regs before transplantation significantly prolonged allograft survival in fully immunocompetent recipients using both the stringent C3H-to-BALB/c (32.6 ± 4.5 versus 8.7 ± 0.2 days) and B6-to-BALB/c (31.1 ± 12 versus 9.7 ± 0.4 days) strain combinations. Nos2-deficient M regs did not prolong allograft survival, proving that M reg function in vivo is iNOS-dependent and mediated by living cells. M regs were detectable for at least 2 weeks postinfusion in allogeneic recipients. In their origin, development, phenotypic relationship with other in vitro-derived macrophages and functions, there are solid grounds to assert a near-equivalence of mouse and human M regs. It is concluded that mouse M regs represent a novel, phenotypically distinct subset of suppressor macrophages. Clinical applications of M reg therapy as an adjunct immunosuppressive therapy are currently being investigated within The ONE Study.
Introduction
Although very effective in preventing acute transplant rejection, conventional maintenance immunosuppression has many unwanted consequences, including renal toxicity, metabolic disturbances, cardiovascular disease, and increased susceptibility to infection and malignancy.1 For this reason, and because improvements in long-term transplant survival have not mirrored recent short-term improvements, the reliable induction of donor-specific immunological nonreactivity remains an active topic of transplantation research.2 Sadly, attempts to translate tolerance-promoting protocols from animal models into clinical practice have, so far, not resulted in a clinically acceptable, safe and reproducible treatment.3 Nevertheless, the identification and immunological characterization of spontaneously tolerant kidney and liver transplant recipients encourages the belief that clinical tolerance induction remains a tenable objective.4,5
Adoptive transfer of allograft tolerance with immunoregulatory cells is a common method in experimental immunology, but its translation to clinical transplantation has only recently received serious attention.6,7 At the present time, several alternative cell types are approaching the point of preclinical development that might allow them to be properly investigated as adjunct immunosuppressive therapies in early stage clinical trials.8 One particularly promising candidate cell type, the regulatory macrophage, has been a focus of research in our laboratory.9 From a technological perspective, regulatory macrophages (M reg) exhibit many properties which suit them for clinical application in tolerance-promoting protocols, most importantly, the cells are simply and reliably obtained, terminally differentiated and potently T cell-suppressive.10,11 Human M regs are a phenotypically homogenous population of suppressor macrophages, which are relatively refractory to activation by lipopolysaccharides (J.A. Hutchinson, unpublished results). A Good Manufacturing Practice (GMP)-compliant procedure for generating human M regs from peripheral blood monocytes has been established. This process typically yields 2–8 × 108 M regs, which are sufficient to treat most recipients with 2.5–7.5 × 106 cells/ kg. To date, two living-donor renal transplant recipients have been treated with M regs in this dose range and, at three years post-transplant, both are maintained with only low-dose tacrolimus monotherapy and have stable renal function.9 Intriguingly, marker gene expression in the peripheral blood of both patients converged over time upon the tolerance-associated profile defined by the IOT-RISET study.4 Therefore, although M reg therapy, and cell-based immunosuppressive treatments in general, are still in their infancy, our first-in-man studies in solid organ transplantation point toward the potential clinical impact of this revolutionary approach.
Macrophages are extremely versatile effector cells which engage in diverse, often antagonistic activities: They participate in both tissue-destructive and reparative processes, and can alternatively heighten or diminish immunological reactions.12 This functional adaptability is reflected by a remarkable phenotypic plasticity, which itself reflects the potential of macrophages to inducibly express a very broad range of genes.13 Macrophages are exquisitely sensitive to stimuli from their environment and other immunological effector cells, responding in the most appropriate manner by coordinately varying their expression of soluble mediators and surface phenotype.14,15 Thus, activated macrophages are often classified within a spectrum of polarization states: At one extreme, interferon-γ (IFN-γ) and lipopolysaccharides direct macrophages toward the M1-polarized phenotype, when they preferentially drive Th1 responses; at the other extreme, interleukin (IL)-4 or IL-13 pushes macrophages toward M2-polarization, when they promote Th2 responses.16 Macrophages exposed to other stimuli can, in general, be categorized by their phenotypic similarity to either M1-polarized or M2-polarized macrophages.17
In this article, it is demonstrated that the mouse M reg represents a novel state of macrophage polarization, distinct from previously described M1- and M2-polarized macrophage subsets and monocyte-derived dendritic cells (DCs). Mouse M regs are profound inhibitors of mitogen-driven T cell proliferation, primarily through the action of inducible nitric oxide synthase (iNOS), and delete activated T cells in an alloantigen-specific manner. A single administration of donor-derived M regs before transplantation significantly prolongs heterotopic heart allograft survival in an iNOS-dependent manner. Mouse and human M regs are near-equivalent cell types in their origin and development, morphology, marker phenotype and in vitro functions. Therefore, insights drawn from studying the therapeutic benefits of mouse M regs in mice, such as the combinatorial action of rapamycin and M reg treatment, are of immediate relevance to our on-going clinical investigations with human M reg therapy.
Results
Phenotypic characterization of mouse M regs
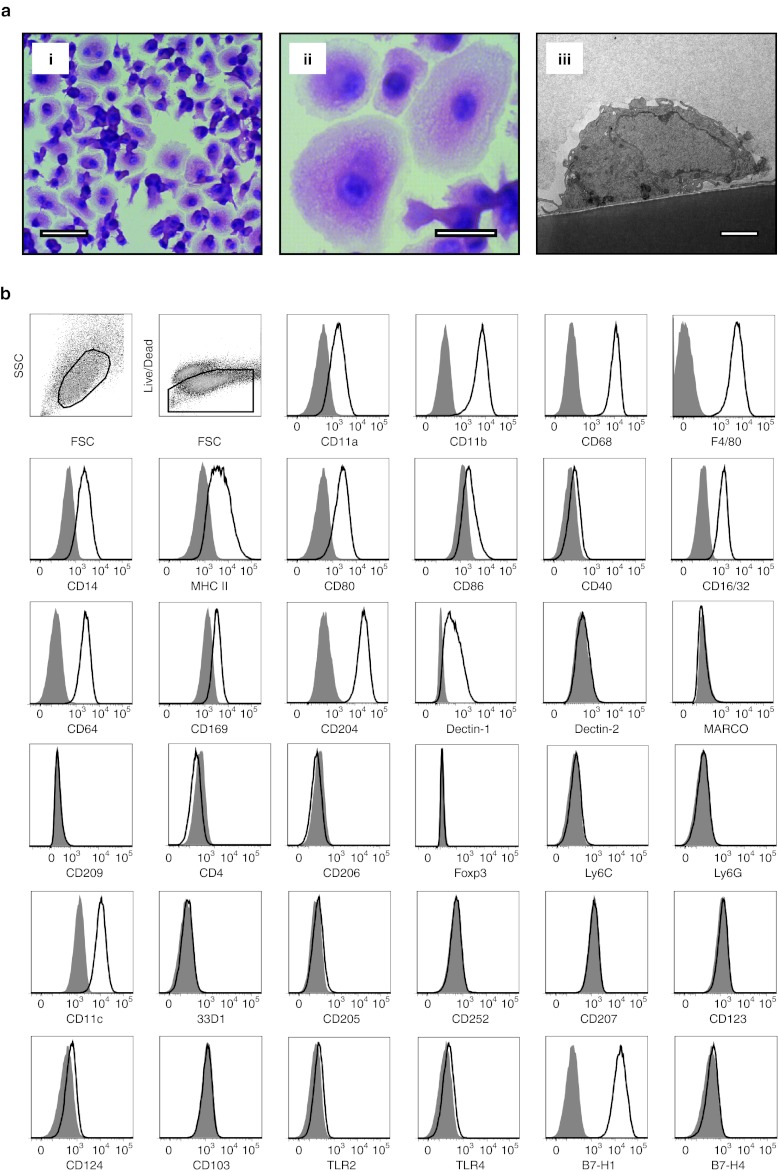
M regs arose from isolated CD11b+ Ly6C+ bone marrow monocytes over a 7-day culture period during which the cells were propagated in medium containing 10% fetal calf serum (FCS) and 10% human AB serum supplemented with 5 ng/ml macrophage colony-stimulating factor (M-CSF); on the final day of culture, the cells were pulsed with 25 ng/ml IFN-γ. Plated at a density of 105 monocytes/cm2, most of the resulting cells displayed a characteristic morphology with a prominent central cell body surrounded by a thin skirt of cytoplasm (Figure 1a). M regs were predominantly mononuclear, although occasional binucleate forms could be observed, and their cytoplasm appeared densely granular. Transmission electron microscopy revealed M regs adhere tightly to underlying cell culture surfaces, extend numerous cell-surface processes and harbor lipid inclusions (Figure 1a).
Figure 1.
Morphology and cell-surface phenotype of B6 M regs. (a) M regs grow as semiconfluent cell layers (i: bar = 50 µm) with individual cells adopting a spreading morphology (ii: bar = 25 µm). Transmission electron microscopy shows M regs adhere tightly to the underlying tissue culture surface. M regs exhibit a single nucleus, numerous cell-surface projections and harbor lipid inclusions (iii: bar = 2 µm). (b) B6 M regs displayed a cell-surface phenotype consistent with their classification as partially matured macrophages. Open traces represent specific signals and shaded traces represent fluorescence-minus-one (FMO) isotype controls. Data are representative of three independent experiments.
M regs expressed a broad selection of typical macrophage markers, including CD11a, CD11b, CD68, F4/80, and CD14 (Figure 1b). Intermediate levels of major histocompatibility complex (MHC) class II and CD80 expression, and low or absent expression of CD86 and CD40, were indicative of a partially matured antigen-presenting cell.18 M regs expressed the Fc-receptors CD16/32 and CD64. Sialoadhesin (CD169), macrophage scavenger receptor (CD204) and Dectin-1 were expressed by M regs, but other markers of notable tissue macrophage subsets were absent, including Dectin-2, MARCO, CD209, CD4, CD206, and Foxp3.19 M regs expressed neither Ly6C nor Ly6G (which together constitute the Gr1 antigen) that define populations of myeloid-derived suppressor cells.20 CD11c was homogeneously expressed by M regs, but they lacked expression of several other DC-defining markers, including 33D1 and OX40L (CD252), as well as CD205, CD207, and CD103.21 Importantly, M regs did not express TLR2 or TLR4. Consistent with having been exposed to IFN-γ, M regs expressed PD-L1 (B7-H1) whereas B7-H4 expression was not detected. M regs derived from C3H and BALB/c mice shared the same phenotypic profile as C57BL/6 (B6) M regs (data not shown). Considering their origin, mode of derivation, form and cell-surface phenotype, it seems most appropriate to classify mouse M regs as a type of macrophage; however, no correspondence between M regs and previously described in vitro-derived or physiologically occurring macrophage types was immediately obvious (Supplementary Figure S1).
Mouse M regs represent a novel and unique state of macrophage activation
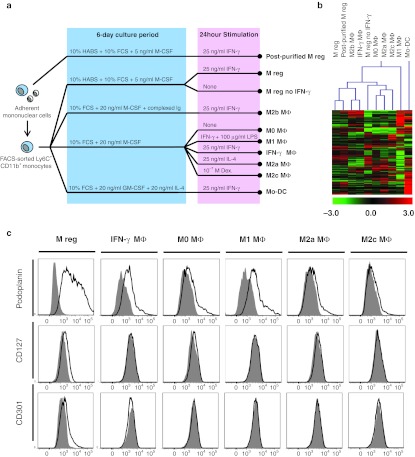
To better understand the interrelationship of mouse M regs and macrophages in previously defined states of polarization, a panel of macrophage types was generated for comparison to M regs in whole genome transcriptional profiling studies by microarray (Figure 2a). This approach enabled an unbiased assessment of the degree of overall phenotypic similarity between M regs and the comparator macrophages. Additionally, this approach identified differentially expressed genes which most accurately discriminated M regs from the comparator macrophage types.
Figure 2.
Defining the phenotypic proximity of M regs to macrophages in other states of activation. (a) M regs and a panel of nine comparator cell types were generated from fluorescence-activated cell sorting (FACS)-sorted CD11b+ Ly6C+ Ly6G– B6 bone marrow monocytes. Three series of comparator cell types were generated in fully independent experiments. (b) Hierarchical clustering (Manhattan; average linked) of the reporter sets returned by one-way ANOVA which were significantly and highly differentially regulated in any two of the comparator macrophage populations. Red shading indicates a higher expression of a certain reporter in the respective sample compared to the median of all samples, a green shading indicates downregulation. (c) Confirmation by flow cytometry of the relative upregulation of podoplanin, CD127, and CD301 (MGL1) cell-surface expression in B6 M regs, but not other macrophage types. Open traces represent specific signals and shaded traces represent fluorescence-minus-one (FMO) isotype controls. Data are representative of three independent experiments.
CD11b+ Ly6C+ monocytes were isolated by fluorescence-activated cell sorting (FACS)-sorting from pooled bone marrow of B6 mice before being differentially stimulated in vitro to generate M regs and a panel of nine comparator cell types. In addition to monocytes, classically activated (M1) macrophages and alternatively activated (M2a) macrophages, comparisons of M regs were made to resting (M0) macrophages, immune complex-stimulated (M2b) and dexamethasone-treated (M2c) macrophages, as well as IFN-γ-stimulated macrophages (IFN-γ MΦ) and monocyte-derived DCs (mo-DC). To assess the relative contribution of IFN-γ stimulation to the M reg phenotype, M regs were generated omitting stimulation with IFN-γ. “Post-purified” M regs (generated from plastic-adherent monocytes and FACS-sorted at the end of the culture period) were included in the comparison to evaluate the possible influence of leucocytes other than monocytes on M reg development.
Three separate series of M regs and comparator cell types were generated in fully independent experiments, giving rise to a microarray dataset comprising triplicate samples of ten cell types produced by single-colour RNA hybridizations to Agilent Whole Mouse Genome Oligo Microarrays. To gain an impression of inter-sample variability, samples were compared to one another in an interexperiment correlation analysis based on the unfiltered, normalized, log2-transformed intensity data (Supplementary Figure S2). Triplicate samples of each cell type tightly co-clustered (with the exception of M regs and post-purified M regs, which are highly similar to one another) and five distinct clusters of cells types emerged. Unsurprisingly, monocytes proved to be the most distinct cell type. M0, M2a, and M2c macrophages constituted a distinct cluster, as did M2b and IFN-γ MΦ, and the M reg samples clustered separately (Supplementary Figure S2).
A one-way ANOVA returned a list of genes which were highly (at least 20-fold difference between any two cell types) and significantly (P < 0.01) differentially regulated in any two of the cell types. (Monocytes were excluded from the analysis to avoid the list being dominated by genes they expressed.) The reporter-wise median-centered log2 intensity data were averaged for each cell type and subjected to hierarchical clustering (Figure 2b). The salient finding of this analysis is that M regs form a unique cluster with post-purified M regs, IFN-γ MΦ, and M2b macrophages, which is entirely distinct from monocyte-derived DCs and M0-, M1-, M2a-, and M2c-polarized macrophages. In this analysis, the phenotypic similarity of M regs to either IFN-γ MΦ or M2b was less than the similarity between M0, M2a, and M2c macrophages. Therefore, it is concluded the mouse M reg represents a novel and unique state of macrophage polarization.
From the microarray dataset, differentially up- and downregulated genes which distinguish M regs from the comparator macrophages were identified and the discriminatory value of an arbitrary selection of these genes as markers of M regs was assessed by flow cytometry (Figure 2c). Higher cell-surface expression of podoplanin was detected in M regs than in M0-, M1-, M2a-, M2c-, or IFN-γ Mф. CD127 (IL-7R) and CD301 (MGL1) were also specifically expressed by M regs.
IFN-γ and factors present in human serum drive M reg development
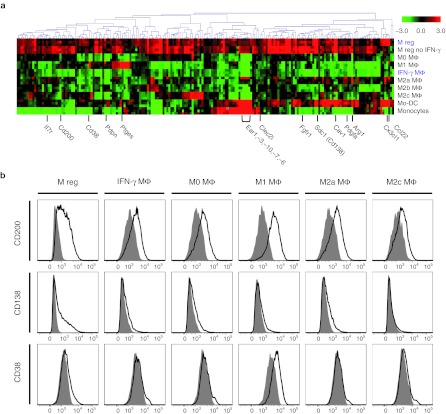
Having established that M regs represent a novel state of macrophage polarization, the question naturally arises, which factors drive monocytes to this unique fate? Insights into this problem can be drawn from reconsidering the microarray dataset: M regs show greatest similarity to IFN-γ MΦ and M2b-polarized macrophages, indicating that IFN-γ exposure is a major determinant of the M reg phenotype. Accordingly, macrophages generated under the same conditions as M regs, but omitting IFN-γ treatment, were found not to cluster with M regs (Figure 2b). Genes significantly differentially expressed between IFN-γ treated and untreated M regs were identified by significance analysis of microarrays (SAM) (Supplementary Figure S3). Among the large number of genes induced by IFN-γ stimulation were MHC class II, components of the IFN-γ signalling pathway, known IFN-γ-inducible genes22 and many others. In contrast, very few genes were found to be downregulated by IFN-γ in M regs.
Our work with human M regs has conclusively demonstrated the need for several specific components of human serum in driving their differentiation to a stably unactivated state (ref. 23 and J.A. Hutchinson, unpublished results). To simulate human M reg development as accurately as possible, mouse M regs were also cultivated in the presence of human serum; accordingly, any differences between M regs and IFN-γ-stimulated macrophages (IFN-γ MΦ) can be attributed to the exposure of M regs to human serum. SAM returned 156 genes significantly upregulated in M regs compared to IFN-γ MΦ, including genes with known immunoregulatory functions, such as Cd200 and Arg1 (arginase 1). Sdc1 (syndecan-1, CD138), Cav1 (caveolin 1), Cx3cl1, and Ccl22 were among the genes most highly upregulated in M regs with respect to IFN-γ MΦ (Figure 3a). To validate these results, expression of CD200, CD138, and CD38 was assessed by flow cytometry: CD200 and CD138 were most highly expressed in M regs, whereas CD38 was expressed by M regs and M1 macrophages (Figure 3b). Of the genes differentially regulated in M regs and IFN-γ MΦ, 94% were common to the list of genes returned by SAM as differentially regulated in M reg versus M2b macrophages. Strikingly, most of the genes differentially expressed in M regs compared to IFN-γ-stimulated or M2b-polarized macrophages showed similar expression profiles in M regs not exposed to IFN-γ. From these observations, we conclude that exposure to unknown factors present in human serum (but not in FCS) is a second major determinant of the mouse M reg phenotype.
Figure 3.
Exposure to human serum influences the development of mouse M regs. Mouse M regs are generated in the presence of human serum because development of human M regs depends upon factors present in human serum and not in fetal calf serum (FCS). (a) Hierarchically clustered heatmap (Pearson uncentered, average linked) depicting reporters returned by significance analysis of microarrays (SAM) (estimated FDR = 0) as significantly upregulated in M regs in comparison to interferon-γ (IFN-γ) MΦ. Three series of comparator cell types were generated in fully independent experiments. (b) Confirmation by flow cytometry of the relative upregulation of CD200, CD138, and CD38 cell-surface expression in B6 M regs. Open traces represent specific signals and shaded traces represent fluorescence-minus-one (FMO) isotype controls. Data are representative of three independent experiments.
M regs suppress mitogen-driven T cell proliferation in vitro
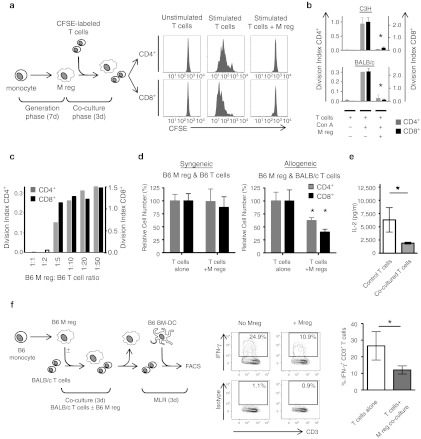
To assess the capacity of M regs to suppress T cell proliferation in vitro, M regs were set in direct 1:1 coculture with T cells for 3 days; subsequently, flow cytometry-based methods were used to quantify CD4+ and CD8+ T cell division (Figure 4a). T cells from C3H mice treated with ConA and T-Stim® proliferated strongly, but coculture with syngeneic M regs completely suppressed this proliferation (CD4+ P < 0.01; CD8+ P < 0.01; n = 3; Figure 4b). Likewise, M regs of BALB/c origin suppressed proliferation of syngeneic responder T cells (CD4+ P < 0.01; CD8+ P < 0.01). B6 M regs also suppressed mitogen-stimulated proliferation of syngeneic T cells, an effect that was evident at M reg: T cell ratios of up to 1:5 (Figure 4c). In the allogeneic setting, neither C3H nor B6 M regs caused alloantigen-driven proliferation of unstimulated BALB/c T cells; on the contrary, C3H and B6 M regs were capable of completely suppressing mitogen-stimulated proliferation of BALB/c T cells (Supplementary Figure S4a,b). B6 M regs also suppressed proliferation of BALB/c T cells in response to B6 tumor necrosis factor-α (TNF-α)-matured bone marrow-derived DCs (data not shown). M regs suppressed ConA-stimulated T cell proliferation through a transwell partition, implying that M regs act, at least in part, through diffusible mediators (Supplementary Figure S4c). Taken together, these results demonstrate that M regs are profoundly suppressive of T cell proliferation and that the mechanism of this suppression is not MHC-restricted. However, we qualify this statement by noting that, in our experimental system, T cells were nonspecifically activated with mitogen. Were T cell activation solely dependent on direct alloantigen presentation by M regs then only antigen-reactive T cells would be suppressed; hence, although M regs may suppress T cells through a nonspecific mechanism, their ultimate effect may be antigen-specific.
Figure 4.
M regs nonspecifically suppress T cell proliferation, but preferentially eliminate allogeneic T cells. (a) Polyclonally activated C3H CFSE-labeled T cells were cultured alone or 1:1 with syngeneic M regs for 3 days before analysis of proliferation by flow cytometry. Coculture with M regs completely suppressed mitogen-driven T cell proliferation. (b) Suppression of polyclonal C3H and BALB/c T cell proliferation with syngeneic M regs (*P < 0.01; n = 3). (c) B6 M regs suppressed syngeneic T cell proliferation in M reg: T cell ratios of up to 1:5. (d) Relative numbers of T cells surviving 1:1 coculture for three days with B6 M regs. No reduction in the relative number of syngeneic T cells was observed as a consequence of coculture with M regs; however, significantly fewer CD4+ and CD8+ T cells remained when cocultured with allogeneic M regs compared to T cells cultured alone (CD4+ P = 0.04; CD8+ P = 0.02; n = 6). (e) BALB/c T cells were recovered from three-day cocultures with C3H M reg and then restimulated with anti-CD3/28 for 24 hours in order to measure interleukin (IL)-2 secretion (n = 7). (f) BALB/c T cells were either cultured alone or cocultured 1:1 with B6 M regs for 3 days before being reisolated using magnetic beads. Reisolated T cells were restimulated with tumor necrosis factor-α (TNF-α)-matured, irradiated B6 bone marrow-derived dendritic cells (BMDC) in a 3-day MLR at a ratio of 10:1. Subsequently, the frequency of IFN-γ+ CD3+ T cells was assessed by flow cytometry (*P < 0.05; n = 4).
M regs preferentially eliminate allogeneic T cells in direct coculture
Using a flow cytometry-based method, absolute counts of viable CD4+ and CD8+ T cells remaining after three days of direct coculture with M regs at a 1:1 ratio were made (Figure 4d). Coculture of freshly isolated, unstimulated B6 T cells with syngeneic M regs did not affect their survival. In contrast, markedly reduced numbers of CD4+ and CD8+ BALB/c T cells survived coculture with B6 M regs (CD4+ P = 0.04; CD8+ P = 0.02; n = 6). This clear result is interpreted as evidence of an allospecific elimination of T cells by M regs. The fate of T cells eliminated by M regs was investigated by fluorescence microscopy of allogeneic M reg and CFSE-labeled T cells in coculture (Supplementary Figure S5a,b). T cells were seen attached to the surface of M regs and inclusions within M regs contained CFSE; therefore, it appears T cells are removed from direct coculture with M regs by phagocytosis. No change was observed in the cell-surface expression of CD169, CD204, CD206, CD301, Dectin-1, Dectin-2, or MARCO by M regs as a consequence of their interaction with either unstimulated or ConA-stimulated T cells (Supplementary Figure S5c).
To characterize how M regs affect the properties of allogeneic T cells which survive coculture, BALB/c T cells were either cultured alone or reisolated from 3-day direct 1:1 cocultures with C3H M regs before polyclonal activation to quantify their capacity to secrete IL-2. Compared to T cells cultured alone, production of IL-2 by T cells in response to anti-CD3/CD28 stimulation after coculture with M regs was compromised (Figure 4e). The reduced ability to produce IL-2 and nonproliferative condition of T cells cocultured with M regs points to an anergization of allogeneic T cells by M regs.24 To investigate the effect of M reg coculture on the ability of T cells to respond to direct alloantigen stimulation, we assessed the frequency of IFN-γ–producing BALB/c T cells responding to TNF-α–matured, irradiated DCs within populations of T cells which had either been cultured alone or cocultured with B6 M regs for 3 days. A significant decrease in the proportion of IFN-γ+ alloreactive T cells was observed as a consequence of coculture with M regs (Figure 4f).
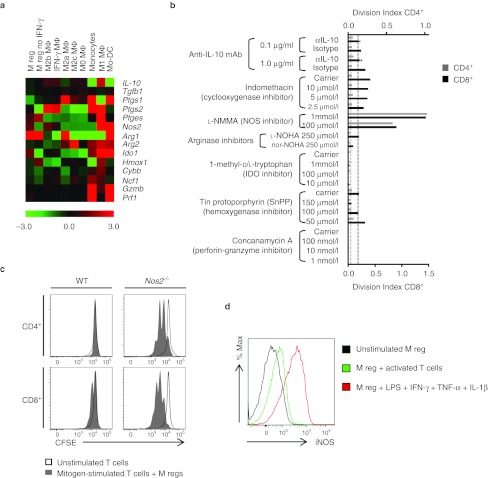
M regs suppress T cell proliferation through the action of iNOS
Diverse mechanisms might account for the T cell-suppressive capacity of M regs in vitro. Examining expression of key mediators of T cell suppression and deletion revealed relative upregulation of Arg1, Nos2, and Ido1 in M regs with respect to their median expression in comparator populations (Figure 5a). Inhibitors of IL-10 (anti-IL-10 mAb), cyclooxygenase (indomethacin), NOS (L-NMMA), arginase (nor-NOHA), indoleamine 2,3-dioxygenase (1-methyl-D/L-tryptophan), hemoxygenase (tin protoporphyrin), and granzyme-mediated killing (concanamycin A) were added to direct 1:1 cocultures of C3H T cells and syngeneic M regs (Figure 5b). Only L-NMMA at a concentration of 1 mmol/l or 100 µmol/l restored mitogen-stimulated T cell proliferation in the presence of M regs, suggesting that the T cell-suppressive effect of M regs is mediated by NOS. This finding was corroborated by demonstrating the inability of iNOS-deficient M regs from Nos2–/– mice to suppress proliferation of wild-type T cells (Figure 5c). To investigate the uniformity and inducibility of iNOS expression in M regs, an intracellular staining method for iNOS was established (Supplementary Figure S6). Using this method, it was shown that direct coculture with activated T cells led to an induction of iNOS in M regs (Figure 5d). Notably, T cell-induced iNOS expression in M regs was at a very much lower level than iNOS expression induced by lipopolysaccharides, IFN-γ, tumor necrosis factor-α, and IL-1β treatment.
Figure 5.
Suppression of T cell proliferation by M regs is mediated by inducible nitric oxide synthase. (a) Heatmap depicting relative expression of genes (median-centered log2 signal intensities, n = 3) with known T cell-inhibitory functions in macrophages. (b) Screening of inhibitors of known T cell-suppressive mediators to identify the mechanism by which M regs prevent T cell proliferation in vitro. Only L-NMMA, an inhibitor of nitric oxide synthase, was found to inhibit M reg-mediated suppression of T cell division. (c) Using M regs generated from Nos2-deficient mice, inducible nitric oxide synthase (iNOS) was confirmed as the key mediator of T cell suppression by M regs. (d) Constitutive expression of iNOS expression in M regs was low, but was upregulated by coculture with ConA- and T-Stim-activated T cells and to a greater extent by treatment with a cocktail of lipopolysaccharides (LPS), interferon-γ (IFN-γ), interleukin (IL)-1β and TNF-α.
Donor-strain M regs prolong heterotopic heart graft survival
The capacity of M regs to prevent allograft rejection was evaluated in a heterotopic heart transplant model using unconditioned, fully allogeneic, nonimmunosuppressed recipients. A single intravenous administration of 5 × 106 donor-strain M regs given 8 days before transplantation significantly prolonged allograft survival in the stringent C3H-to-BALB/c strain combination (Figure 6a: 32.9 ± 4.5 versus 8.7 ± 0.2 days; P < 0.001). This graft-protective effect was specific to donor cells as recipient M regs did not prolong graft survival compared to untreated controls (Figure 6a: 9.6 ± 0.4 days; ns) and third party-derived M regs only very marginally prolonged graft survival (11.0 ± 0.6 days; P = 0.004). Being capable of indirect presentation of allograft antigens, it was hypothesized that M regs from F1 hybrids of transplant donor and recipient strains would have a greater effect than donor-derived M regs; however, donor M regs were equally as effective in prolonging allograft survival as C3CF1 M regs (Figure 6a: C3CF1 32.8 ± 4.5 days, P = 0.91). Donor strain-derived M regs were similarly effective whether applied 8 or 35 days before transplantation (Figure 6b: 29.3 ± 4.1 days, P = 0.46). Using the alternative strain combination of B6-to-BALB/c, it was shown that prolongation of cardiac allograft survival by donor M regs (Figure 6c: 39.4 ± 15.8 versus 9.7 ± 0.4 days; P < 0.001) and M regs of F1 origin (Figure 6c: CB6F1 31.3 ± 6.4 days, P = 0.68) is a general phenomenon in mice.
Figure 6.
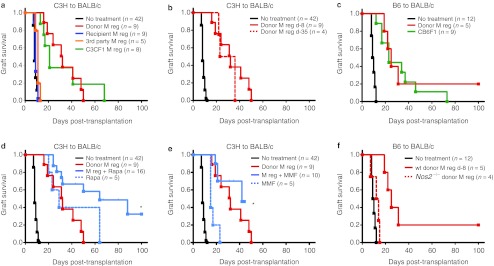
M regs prolong cardiac graft survival in nonimmunosuppressed, fully allogeneic recipients. In all experiments, recipients were treated with 5 × 106 M regs by slow intravenous injection. The curves represent graft survival censored for deaths occurring with a beating heart transplant. For comparison, the relevant control group and donor M reg treatment groups (black and red curves, respectively) are replicated in each panel. (a) Donor-derived M regs (but not recipient or 3rd party M regs) given 8 days before transplantation significantly prolonged allograft survival in the stringent C3H-to-BALB/c strain combination. C3CF1 M regs prolonged C3H allograft survival to a similar extent as donor-derived M regs. (b) Donor-strain M regs were equally protective whether administered on day 8 before transplantation or on day 35 pretransplant. (c) M regs of B6 and CB6F1 origin prolonged graft survival in the B6-to-BALB/c strain combination. (d) A 10-day postoperative course of 1 mg/kg/day rapamycin prolonged allograft survival. A combinatorial beneficial effect of preoperative M reg administration and short-course rapamycin treatment was observed. *Allograft survival was censored for death with a functioning graft. (e) A 10-day postoperative course of 80 mg/kg/day mycophenolate mofetil (MMF) prolonged allograft survival. No addition beneficial effect of preoperative M reg administration and short-course MMF treatment was observed when compared to treatment with M regs alone. *Allograft survival was censored for death with a functioning graft. (f) M regs from Nos2–/– B6-background mice did not prolong B6-to-BALB/c allograft survival, proving that iNOS mediates the graft-protective effect of M regs.
Administration of donor-strain M regs 8 days before transplantation in conjunction with a 10-day postoperative course of 1 mg/kg/day rapamycin treatment significantly prolonged allograft survival with respect to recipients treated with M regs alone (Figure 6d: 66.3 ± 9.0 days, P = 0.005). Graft survival to 100 days was seen in a proportion of recipients cotreated with M regs and rapamycin, whereas no grafts survived to 100 days in recipients treated with either M regs or rapamycin alone. M reg treatment in conjunction with a 10-day postoperative course of 80 mg/kg/day mycophenolate mofetil (MMF) was not superior to M reg treatment alone (Figure 6e: 36.4 ± 3.9 days, P = 0.33). Taken together, these results show that M regs confer a striking alloantigen-specific prolongation of allograft survival, which is enhanced by rapamycin cotreatment.
Only a marginal prolongation of allograft survival was observed using M regs generated from Nos2-deficient B6 mice in BALB/c recipients showing that iNOS is an essential mediator of the in vivo action of M regs (Figure 6f: 12.0 ± 1.8 days, P = 0.049). Very importantly, this experiment proves that the graft-protective effect of M regs cannot be simply attributed to exposure of the recipient to donor alloantigen, but must be mediated by living, metabolically competent cells.
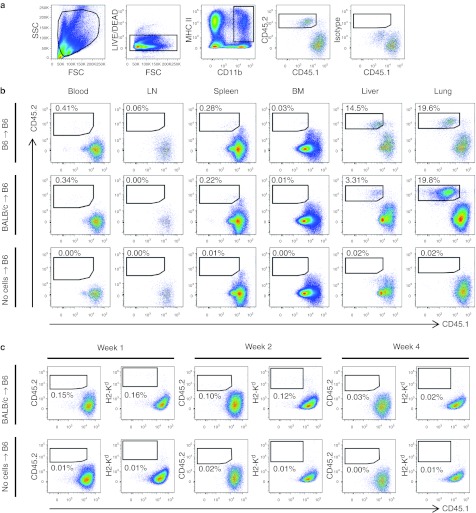
Both syngeneic and allogeneic M regs are short-lived in recipient animals
The anatomical distribution of M regs after injection into CD45.1+ H2-Kb B6 recipients was assessed using M regs from congeneic CD45.2+ H2-Kb B6 mice or allogeneic CD45.2+ H2-Kd BALB/c M regs (Figure 7a). One day after administration, M regs were readily detected in the blood, spleen, liver, and lung of both congeneic and allogeneic recipients, but were not reliably detected in lymph nodes or bone marrow (Figure 7b). At weeks 1 and 2, relatively fewer CD45.2+ H2-Kd M regs were detected in allogeneic recipients compared to day 1 and these cells were most evident in the lung (Figure 7c). However, a similar decrease in the proportion of M regs detected in all tissues of congeneic recipients was also observed (data not shown). By the fourth week, no M regs could be reliably detected. Although it cannot be strictly excluded that the apparent disappearance of M regs was due to a failure of detection (either because they changed phenotype or redistributed too widely) it seems most likely that M regs are short-lived after transfer, surviving only days to weeks. Trans-presentation of H2-Kd by CD45.1+ H2-Kb-expressing recipient antigen-presenting cells was not observed (Figure 7c).
Figure 7.
Anatomical distribution of M regs after administration to syngeneic or allogeneic recipients. 5 × 106 CD45.2+ M regs were administered to recipients by iv injection and their presence in various tissues was detected by flow cytometry at 24 hours, 1, 2, and 4 weeks postinfusion. Data are representative of three independent experiments. (a) Gating strategy to identify live CD45.2+ M regs within the pool of CD11b+ MHC II+ cells. The position of the CD45.2 gate was set according to the fluorescence-minus-one (FMO) isotype control in each case. (b) Tissue distribution of CD45.2+ B6 M regs and CD45.2+ BALB/c M regs in CD45.1+ B6 recipients at 24 hours postinfusion. Both syngeneic and allogeneic M regs were found in the blood, spleen, liver, and lung. The “no cell” control group received an injection of 1 ml DPBS-containing 62 U/ml heparin. Data are representative of three independent experiments. (c) BALB/c M regs were readily detectable for up to 2 weeks in B6 recipients, albeit at a much lower frequency than at 24 hours postinfusion. Results representative of three independent experiments identifying M regs present in lung are shown.
Does a naturally occurring counterpart of the M reg exist?
In developing M regs as a cell-based adjunct immunosuppressive treatment, the paramount considerations are whether administration of M regs is safe and has a consistent, clinically relevant effect. From this strictly technological perspective, it is unimportant whether or not mouse M regs correspond to a physiological macrophage population. Nevertheless, macrophages exhibiting an M reg-like phenotype could be identified in the spleen of B6 mice. These CD11b+ CD11c+ F4/80+ MHC class II+ CD169+ Dectin-1+ cells represent 12.1 ± 3.7% of MHC class II+ CD11b+ splenocytes, so are a relatively minor macrophage subpopulation (Figure 8). Although it cannot be claimed that these M reg-like splenic macrophages are naturally occurring counterparts of in vitro-derived M reg, their presence does suggest that in vitro-derived M regs are not of a wholly artificial phenotype.
Figure 8.
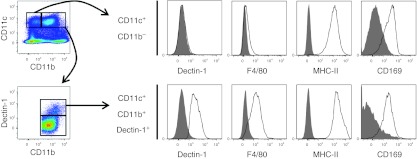
CD11b+ CD11c+ Dectin-1+ F4/80+ MHC II CD169+ macrophages can be found in the spleen of B6 mice. Data are representative of n = 6 animals.
Discussion
This work seeks to establish two principal conclusions, namely, that IFN-γ stimulation in the absence of other proinflammatory signals drives mouse macrophages to a suppressor phenotype and that treatment of transplant recipients with M regs extends allograft survival. The primary importance of these conclusions lies in the utility of the mouse M reg as an experimental and preclinical model for studying the biology and potential clinical benefits of human M reg therapy.10 Our group has recently reported the first application of human M regs as an adjunct immunosuppressive therapy in two living-donor renal transplant recipients.9 This concept is now being carried forward within The ONE Study, a multinational clinical trial of cell therapy in renal transplantation supported by the European Union 7th Framework Programme (www.onestudy.org). Consequently, the detailed phenotypic and functional characterization of the mouse M reg provided here is of both scientific and immediate translational relevance.
Acting within grafts, as well as distant sites, macrophages profoundly influence the development of various transplant pathologies. Macrophages are involved both in acute and chronic allograft injuries, acting not only as simple effectors of innate responses, but also amplifying the adaptive responses.25,26 On the other hand, not all macrophage-mediated processes are detrimental to allografts. In particular, re-establishment of tissue homeostasis by anti-inflammatory macrophages appears to be a major determinant of long-term transplant outcomes.27,28,29 Three general strategies for therapeutically manipulating the behavior of macrophages can be envisaged: depletion or prevention of monocyte migration into inflamed sites; in situ induction of macrophages with an anti-inflammatory or tissue-repair phenotype; and the use of ex vivo-generated regulatory macrophages as a cell-based medicinal product.30 Although using cell preparations as therapeutic agents is a novel and technologically challenging approach, directly administering M regs to patients circumvents many of the difficulties of pharmacologically targeting macrophages with monoclonal antibodies, recombinant proteins or conventional drugs.
Administration of a single intravenous dose of donor-strain M regs substantially prolonged allograft survival in fully mismatched, nonimmunosuppressed recipients without lymphodepletive preconditioning or conventional immunosuppressive treatment. This therapeutic effect was alloantigen-specific and depended upon expression of iNOS by M regs, implying that allograft protection is conferred by living cells. Several mutually redundant mechanisms might be invoked to explain the in vivo effect of M regs and, in our view, it appears that more than one mechanism is in operation. It is firmly established that preoperative exposure to donor alloantigen predisposes recipients to allograft acceptance31 and that delivery of donor alloantigen as apoptotic cell debris enhances this effect.32,33 Both CD8α+ DCs34 and F4/80+ PD-L1+ IL-10-producing macrophages35 of the splenic marginal zone appear to be important for the tolerogenic effects of complement-opsonized apoptotic bodies.36 Because M regs in syngeneic recipients disappear with similar kinetics to M regs in MHC-disparate recipients, it appears that M regs have an inherently limited lifespan; therefore, one pathway of M reg action may be systemic provision of apoptotic donor alloantigen-expressing material in the absence of inflammation. Such a mechanism operates in tolerance induction protocols using donor-specific transfusion and αCD154 treatment, in which indirect presentation of donor alloantigen results in a predominantly deletional tolerance.37 And yet, the graft-protective effect of donor alloantigen exposure in the absence of costimulatory blockade38 or lymphodepletive conditioning39 is rarely as profound as that achieved with M reg treatment, even in less stringent transplant models.40
It is difficult to reconcile the requirement for iNOS expression by M regs with the suggestion that M regs act merely as a passive source of alloantigen, so the question arises as to how M regs exert an active immunosuppressive effect in vivo. Conceivably, M regs might act rapidly before dying or, as our tracking experiments do not entirely exclude the possibility, a small number of M regs might persist in recipients and exercise long-lived effects. In B6 mice tolerized to BALB/c cardiac allografts by donor-specific transfusion and αCD154 treatment, Ochando's group recently demonstrated the need for CD11b+ CSF1R+ endogenous macrophages for the establishment of allograft acceptance; further, they showed that IFN-γ receptor expression was indispensible for the tolerogenic activity of these cells.41 Our gene expression profiling experiments revealed the central influence of IFN-γ in M reg development, its major effect being to drive M regs toward a state of incomplete activation, such that they expressed high levels of MHC class II, iNOS and PD-L1, but little CD40 or CD86. Since T cells encountering partially matured antigen-presenting cells may become anergized, it is conceivable that directly alloreactive recipient T cells recognising M regs could undergo abortive activation in vivo.42 That allogeneic T cells cocultured with M regs were rendered nonproliferative and produced less IL-2 provides some support for this suggestion.
If donor M regs play an active immunosuppressive role in allogeneic recipients, is it possible to account for the antigenic specificity of their in vivo effect? The profound influence of M regs on cocultured T cells cannot be explained by a single mechanism: M regs act by suppressing T cell proliferation through iNOS, inhibiting IL-2 production and by directly eliminating alloantigen-reactive T cells. Presently, the relative contribution of each of these mechanisms to the in vivo effects of M regs is not clear. Preferential elimination of allogeneic T cells from coculture points to a truly allospecific suppressive activity of M regs, which may be very important given reports that deletion of alloreactive T cells is essential for long-term transplant tolerance;43 however, evidence for direct M reg-mediated T cell deletion in vivo is not yet available. Although iNOS-mediated suppression of T cell proliferation in vitro was not MHC-restricted, T cell activation and suppression were artificially divorced by polyclonal T cell stimulation in our experimental system.44 By contrast, in our transplantation experiments, specific activation of recipient T cells and their suppression by donor M regs may have been the selfsame event. Induction of T cell anergy by costimulation-deficient M regs might also be expected to be an antigen-specific phenomenon. Therefore, it is plausible that M regs could induce donor-specific unresponsiveness in T cells reacting in the direct pathway, as well as the indirect pathway.
Expression of iNOS is readily induced in mouse macrophages upon activation, especially in response to IFN-γ.45 iNOS-mediated NO production was originally characterized as a protective cytotoxic response of activated macrophages to various pathogens, but it is now clear that iNOS also plays immunoregulatory roles in malignancy and resolution of autoimmune disease.46 iNOS is responsible for the conversion of L-arginine to L-citrulline and NO, so iNOS-expressing macrophages both deprive T cells of arginine and expose them to the direct toxic effects of NO. NO itself serves as an intercellular signalling molecule, but its reaction products (including peroxynitrite formed through the reaction of NO with superoxide) also exert important biological effects.47 Which of these mechanisms of NO action predominates in the M reg-mediated suppression of T cell proliferation is as yet unknown; however, differences in NO downstream signalling pathways might help to explain why M regs are suppressive, but iNOS-expressing M1 macrophages are not. As well as affecting T cells, the same mechanisms of NO action can also alter the behavior of neighbouring macrophages and DCs.48,49 In this context, it is important to recognise that iNOS-dependent suppression of T cells by M regs in vitro does not imply iNOS-dependent allograft protection is primarily a direct effect of M regs on recipient T cells, as M regs expressing iNOS might equally act via recipient antigen-presenting cells. Accordingly, one potentially important fate for M regs may be to migrate into tissues and modulate resident antigen-presenting cell populations, before dying in a suitably self-conditioned local environment.
A number of classically proinflammatory factors can induce suppressive activities in macrophages, including repetitive TLR stimulation and PGE2 treatment.30 This may reflect the physiological behavior of macrophages in strongly inflammatory environments, which act to limit the extent of inflammation by providing a crucial level of negative feedback. IFN-γ appears to be another example of a proinflammatory cytokine with paradoxical effects in macrophages: On one hand, IFN-γ can enhance the microbicidal activity and antigen-presenting capacity of macrophages; on the other, our data shows that IFN-γ is essential for the development of suppressive M regs. It is interesting to note that IFN-γ conditioning promotes the expansion of regulatory T cells by inducing the conversion of non-T reg precursors into T regs, promoting activation-induced cell death in non-T regs and suppressing the development of Th2 and Th17 cells.50
In synopsis, this study identifies a mouse counterpart of the human M reg, which defies ready classification as an M1-polarized or M2-polarized macrophage. Administration of mouse M regs to fully allogeneic transplant recipients has a therapeutically relevant graft-protective effect, which was shown to depend on iNOS activity, proving that the beneficial action of M regs cannot be solely attributed to recipient exposure to alloantigen. We conclude that the reproducible therapeutic benefit of M reg therapy in mice is extremely encouraging and augers well for the human M reg treatment group of The ONE Study.
Materials and Methods
Mice. Animal experiments were performed with the approval of the Regierung der Oberpfalz (54-2532.1-23/10) in accordance with the German Animal Welfare Act. C57BL/6 (H-2b), C3H (H-2k), BALB/c (H-2d), and CB6F1 mice were purchased from Charles River (Sulzfeld, Germany). B6.129P2-Nos2tm/Lau/J (iNOS-knockout mice) and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1 congeneic mice) were purchased from The Jackson Laboratory (Bar Harbor, ME). C3CF1 mice were bred in-house. Mice in all experiments were male and 8–12-weeks old.
Generation of mouse M reg and other macrophage populations. M reg were generated from mononuclear cells obtained from blood, spleen, and bone marrow in an process analogous to that used to produce human M reg.51 Cells were cultured for 7 days in 100 mm untreated petri dishes with 5 ml of RPMI 1640 containing 100 U/ml penicillin-streptomycin, 25 mmol/l HEPES, 2 mmol/l glutamine, and 2 mmol/l GlutaMAX (Invitrogen, Karlsruhe, Germany), and supplemented with 10% FCS (Biochrom, Berlin, Germany), 10% human AB serum (Lonza, Cologne, Germany) and 5 ng/ml M-CSF (Sigma, Munich, Germany). Every other day, medium was exchanged without disrupting the adherent cell layer. On day 6, cultures were stimulated with 25 ng/ml IFN-γ (Chemicon, Billerica, MA). On day 7, cells were washed in DPBS and harvested by pipetting and gentle scraping. For comparative analyses, all macrophage types (including M regs) were generated from CD11b+ magnetic bead-sorted monocytes from bone marrow, which were plated in 35-mm petri dishes at 2 × 105 cells/ml/cm2 in RPMI 1640-based medium and stimulated as indicated in Figure 2a. For culture of M2b MΦ, petri dishes were coated overnight with 1 mg/ml human immunoglobulin (Sandoglobulin; CSL Behring, Hattersheim am Main, Germany) in Na-Bicarbonate buffer at 4 °C, and washed three times with DPBS before use. For gene expression profiling experiments, the input cells were FACS-sorted CD11b+ Ly6C+ Ly6G− monocytes from bone marrow.
Flow cytometry. Samples were prepared for flow cytometry as described elsewhere.52 Dead cells were excluded using 7-AAD (BD Biosciences, Heidelberg, Germany) or LIVE/DEAD Aqua (Invitrogen). Antibodies are specified in Supplementary Table S1. Commercial reagents were used for fixation and permeabilization of M regs for detection of CD68 and iNOS (BD Biosciences). Flow cytometry was performed on a FACS Canto II (Becton Dickinson, Heidelberg, Germany) and analysed with FlowJo (Tree Star, Ashland, OR).
In vitro suppression assay. 5 × 105 M reg and 5 × 105 CFSE-labeled T cells were plated into 24-well plates in 1 ml complete RPMI 1640 supplemented with 10% FCS, 1 mmol/l sodium pyruvate, 100 µmol/l nonessential aminoacids and 100 µmol/l 2-ME (Invitrogen). T cells used in this assay were isolated from spleen, labeled with 2 µmol/l CFSE (Invitrogen) for 15 minutes and enriched with CD90 MicroBeads (Miltenyi, Bergisch-Gladbach, Germany). Cultures were stimulated with 5 µg/ml concanavalin A (Sigma) and 5% T-STIM (BD). The following inhibitors were prepared as follows: anti-IL-10 or rat IgG2b isotype control in DPBS (BD); indomethacin in ethanol (Sigma); L-NMMA in water (NG-methyl-L-arginine acetate salt, Sigma); NG-Hydroxil-L-arginine acetate (L-NOHA) in water (Sigma); Nω-Hydroxy-nor-L-arginine diacetate (nor-NOHA) in water (Calbiochem, Darmstadt, Germany); 1-methyl-D/L-tryptophan (1-MT) in HCl before pH adjustment (Sigma); tin protoporphyrin (SnPP) in NaOH before pH adjustment (Tocris, Bristol, UK); Concanamycin A in DMSO (Sigma). CD4+ and CD8+ T cell proliferation and absolute cell counts were assessed by flow cytometry using Countbright beads (Invitrogen).
Cytokine secretion assay. 106 CD4+ spleen T cells were plated alone or with 106 C3H M reg in 6-well plates containing 3-ml complete RPMI 1640 supplemented with 10% FCS, sodium pyruvate, nonessential aminoacids and 2-ME (Invitrogen). After 3 days, T cells were harvested, sorted with CD90 MicroBeads, and 0.2 × 106 cells in 200 µl medium were seeded into wells of a 96-well round bottom plate. T cells were then stimulated with plate-bound anti-CD3 and soluble anti-CD28 (BD). After 24 hours, supernatants were collected and IL-2 concentration was determined by ELISA (R&D Systems, Wiesbaden Nordenstadt, Germany).
RNA isolation, amplification, and labeling for microarrays. RNA was isolated from three independent series of FACS-sorted comparator macrophage types using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). RNA quality was determined using the Agilent 2100 Bioanalyzer platform and only RNAs with calculated RNA Integrity Numbers (RIN) higher than 6 were accepted. Thirty six microarray datasets were obtained from single-color hybridization of murine RNAs to Agilent Whole Mouse Genome Oligo Microarrays (4 × 44). Sample labeling was performed as detailed in the “One-Color Microarray-Based Gene Expression Analysis protocol (version 5.7, part number G4140-90040).” Briefly, 0.5 µg of each total RNA samples was used for the amplification and labeling step using the Agilent Quick Amp Labeling Kit (Agilent Technologies, Bobingen, Germany). Yields of cRNA and the dye-incorporation rate were measured with the ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Hybridization of agilent whole mouse genome oligo microarrays. The hybridization procedure was performed according to the the “One-Color Microarray-Based Gene Expression Analysis protocol (version 5.7, part number G4140-90040) using the Agilent Gene Expression Hybridization Kit (Agilent Technologies). Briefly, 1.65 µg Cy3-labeled fragmented cRNA in hybridization buffer was hybridized overnight (17 hours, 65 °C) to Agilent Whole Mouse Genome Oligo Microarrays 4x44K using Agilent's recommended hybridization chamber and oven. Following hybridization, the microarrays were washed once with the Agilent Gene Expression Wash Buffer 1 for 1 minute at room temperature followed by a second wash with preheated Agilent Gene Expression Wash Buffer 2 (37 °C) for 1 minute. The last washing step was performed with acetonitrile.
Scanning and data analysis. Fluorescence signals of the hybridized Agilent Microarrays were detected using Agilent's Microarray Scanner System (Agilent Technologies). The Agilent Feature Extraction Software 10.5.1.1 was used to read out and process the microarray image files. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE32690. Background corrected intensity values were quantile normalized and log2 transformed. Only reporters with at least two valid signal intensity values in at least one cell type were considered. For visualization in heat map format, the log2 intensity values were median-centered for each reporter. One-way ANOVA was conducted using GeneSpring GX (v.11.0.2) (Agilent Technologies). Differentially expressed genes were identified by SAM (estimated FDR = 0) as implemented in the MultiExperiment Viewer of the TM4 microarray software suite (v.4.6.1).
Heterotopic heart transplantation. Abdominal heterotopic heart transplants were performed as previously described.53 Graft rejection was defined as cessation of palpable cardiac contractions with verification by direct inspection of the allograft via laparotomy. Recipient mice either received no additional treatment, or received 5 × 106 donor, recipient, or third-party M regs on day 8 before transplantation unless otherwise stated. M regs were resuspended in 1 ml DPBS-containing 62 U heparin and then administered by slow injection into the tail vein. Some experimental groups received 1 mg/kg/day i.p. rapamycin (Rapamune; Pfizer, Berlin, Germany) diluted in saline or 80mg/kg/day i.p. mycophenolate mofetil (Cellcept; Roche, Grenzach, Germany) diluted in 5% glucose from the day of transplantation until day 10.
Tracking of M regs in vivo. 5 × 106 B6 M reg, BALB/c M reg or a “no-cell control” were injected in the tail vein of B6-CD45.1 congeneic mice. Blood, LN, spleen, BM, liver, and lung were harvested on day 1, week 1, 2, and 4 after cell injection. Single cell suspensions of blood, LN, spleen, and BM were made by mechanical disruption and erythrocyte lysis with ACK buffer. Liver and lung were homogenized using gentleMACS (Miltenyi) according to recommended protocols before enrichment of CD11b+ cells by AutoMACS Pro. Cells were subsequently stained for markers discriminating donor M regs from recipient cells and analysed by flow cytometry.
Statistics. Statistical analysis was performed with GraphPad and SigmaPlot software. Values given in histograms represent mean ± SEM unless otherwise stated. As appropriate, paired one-tailed or two-tailed t-tests, or Mann–Whitney U-tests, were used for all tests of significance. The LogRank test was applied to compare cardiac graft survival between groups. Statistical treatment of microarray data is described above.
SUPPLEMENTARY MATERIAL Figure S1. Mouse M regs do not correspond to previously described macrophage populations. Figure S2. Interexperiment correlation analysis of macrophage samples belonging to three independent experimental series. Figure S3. Hierarchically clustered heatmaps (Pearson uncentered, average linked) depicting reporters returned by SAM as significantly upregulated (a) and downregulated (b) in M regs compared to M regs without IFN-γ treatment (M reg no IFN-γ). Figure S4. M regs suppress allogeneic T cell proliferation. Figure S5. M regs interact with activated T cells in coculture. Figure S6. Establishment of intracellular FACS-staining protocols for mouse iNOS. Table S1. Antibodies used for flow cytometry.
Acknowledgments
The work presented in this report was funded by the Deutsche Forschungsgemeinschaft (award GE-1188/1-1) and the European Union 7th Framework Programme under the auspices of The ONE Study consortium (award 260687). PR received an intramural ReFoRM A award from UKR to conduct this work. EKG was the recipient of an unrestricted basic research grant from Pfizer to study rapamycin. The authors are grateful to Hr. Prof. J.S. Schröder for his expert assistance with the EM studies. The authors wish to thank Anke Hofmann, Judith Bausenwein, Anna Höhn and Joachim Herman for their invaluable technical support. The gene expression data presented in this article have been submitted to the National Center for Biotechnology Information/Gene Expression Omnibus: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE32690
Supplementary Material
Mouse M regs do not correspond to previously described macrophage populations.
Interexperiment correlation analysis of macrophage samples belonging to three independent experimental series.
Hierarchically clustered heatmaps (Pearson uncentered, average linked) depicting reporters returned by SAM as significantly upregulated (a) and downregulated (b) in M regs compared to M regs without IFN-γ treatment (M reg no IFN-γ).
M regs suppress allogeneic T cell proliferation.
M regs interact with activated T cells in coculture.
Establishment of intracellular FACS-staining protocols for mouse iNOS.
Antibodies used for flow cytometry.
REFERENCES
- Halloran PF. Immunosuppressive drugs for kidney transplantation. N Engl J Med. 2004;351:2715–2729. doi: 10.1056/NEJMra033540. [DOI] [PubMed] [Google Scholar]
- Meier-Kriesche HU, Schold JD, Srinivas TR., and, Kaplan B. Lack of improvement in renal allograft survival despite a marked decrease in acute rejection rates over the most recent era. Am J Transplant. 2004;4:378–383. doi: 10.1111/j.1600-6143.2004.00332.x. [DOI] [PubMed] [Google Scholar]
- Orlando G, Hematti P, Stratta RJ, Burke GW 3rd, Di Cocco P, Cocco PD.et al. (2010Clinical operational tolerance after renal transplantation: current status and future challenges Ann Surg 252915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P.et al. (2010Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans J Clin Invest 1201848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, Immune Tolerance Network ST507 Study Group et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120:1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie M. Immunology. Regulatory T cells get their chance to shine. Science. 2011;332:1020–1021. doi: 10.1126/science.332.6033.1020. [DOI] [PubMed] [Google Scholar]
- Bluestone JA. Mechanisms of tolerance. Immunol Rev. 2011;241:5–19. doi: 10.1111/j.1600-065X.2011.01019.x. [DOI] [PubMed] [Google Scholar]
- Edinger M., and, Hoffmann P. Regulatory T cells in stem cell transplantation: strategies and first clinical experiences. Curr Opin Immunol. 2011;23:679–684. doi: 10.1016/j.coi.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Hutchinson JA, Riquelme P, Sawitzki B, Tomiuk S, Miqueu P, Zuhayra M.et al. (2011Cutting Edge: Immunological consequences and trafficking of human regulatory macrophages administered to renal transplant recipients J Immunol 1872072–2078. [DOI] [PubMed] [Google Scholar]
- Hutchinson JA, Riquelme P., and, Geissler EK. Human regulatory macrophages as a cell-based medicinal product. Curr Opin Organ Transplant. 2012;17:48–54. doi: 10.1097/MOT.0b013e32834ee64a. [DOI] [PubMed] [Google Scholar]
- Hutchinson JA, Riquelme P, Geissler EK., and, Fändrich F. Human regulatory macrophages. Methods Mol Biol. 2011;677:181–192. doi: 10.1007/978-1-60761-869-0_13. [DOI] [PubMed] [Google Scholar]
- Gordon S., and, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Biswas SK., and, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Helming L., and, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- Mosser DM., and, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M., and, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A., and, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- Reis e Sousa Dendritic cells in a mature age. Nat Rev Immunol. 2006;6:476–483. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD., and, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- Lees JR, Azimzadeh AM., and, Bromberg JS. Myeloid derived suppressor cells in transplantation. Curr Opin Immunol. 2011;23:692–697. doi: 10.1016/j.coi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Liu K, Helft J, Bogunovic M, Greter M, Hashimoto D.et al. (2009The origin and development of nonlymphoid tissue CD103+ DCs J Exp Med 2063115–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M.et al. (2010Dissecting interferon-induced transcriptional programs in human peripheral blood cells PLoS ONE 5e9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme P, Gövert F, Geissler EK, Fändrich F., and, Hutchinson JA. Human transplant acceptance-inducing cells suppress mitogen-stimulated T cell proliferation. Transpl Immunol. 2009;21:162–165. doi: 10.1016/j.trim.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Chappert P., and, Schwartz RH. Induction of T cell anergy: integration of environmental cues and infectious tolerance. Curr Opin Immunol. 2010;22:552–559. doi: 10.1016/j.coi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ., and, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyburn KR, Jose MD, Wu H, Atkins RC., and, Chadban SJ. The role of macrophages in allograft rejection. Transplantation. 2005;80:1641–1647. doi: 10.1097/01.tp.0000173903.26886.20. [DOI] [PubMed] [Google Scholar]
- Jang HS, Kim J, Park YK., and, Park KM. Infiltrated macrophages contribute to recovery after ischemic injury but not to ischemic preconditioning in kidneys. Transplantation. 2008;85:447–455. doi: 10.1097/TP.0b013e318160f0d1. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Abu-Dahab R, Menger MD, Schäfer U, Vollmar B, Wada H.et al. (2005Depletion of alveolar macrophages by clodronate-liposomes aggravates ischemia-reperfusion injury of the lung J Heart Lung Transplant 2438–45. [DOI] [PubMed] [Google Scholar]
- Devey L, Ferenbach D, Mohr E, Sangster K, Bellamy CO, Hughes J.et al. (2009Tissue-resident macrophages protect the liver from ischemia reperfusion injury via a heme oxygenase-1-dependent mechanism Mol Ther 1765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broichhausen C, Riquelme P, Geissler EK., and, Hutchinson JA. Regulatory macrophages as therapeutic targets and therapeutic agents in solid organ transplantation. Curr Opin Organ Transplant. 2012;17:332–342. doi: 10.1097/MOT.0b013e328355a979. [DOI] [PubMed] [Google Scholar]
- Bushell A, Karim M, Kingsley CI., and, Wood KJ. Pretransplant blood transfusion without additional immunotherapy generates CD25+CD4+ regulatory T cells: a potential explanation for the blood-transfusion effect. Transplantation. 2003;76:449–455. doi: 10.1097/01.TP.0000083043.84630.99. [DOI] [PubMed] [Google Scholar]
- Morelli AE., and, Larregina AT. Apoptotic cell-based therapies against transplant rejection: role of recipient's dendritic cells. Apoptosis. 2010;15:1083–1097. doi: 10.1007/s10495-010-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Turley S, Mellman I., and, Inaba K. The induction of tolerance by dendritic cells that have captured apoptotic cells. J Exp Med. 2000;191:411–416. doi: 10.1084/jem.191.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu CH, Miyake Y, Kaise H, Kitamura H, Ohara O., and, Tanaka M. Novel subset of CD8{alpha}+ dendritic cells localized in the marginal zone is responsible for tolerance to cell-associated antigens. J Immunol. 2009;182:4127–4136. doi: 10.4049/jimmunol.0803364. [DOI] [PubMed] [Google Scholar]
- Getts DR, Turley DM, Smith CE, Harp CT, McCarthy D, Feeney EM.et al. (2011Tolerance induced by apoptotic antigen-coupled leukocytes is induced by PD-L1+ and IL-10-producing splenic macrophages and maintained by T regulatory cells J Immunol 1872405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli AE, Larregina AT, Shufesky WJ, Zahorchak AF, Logar AJ, Papworth GD.et al. (2003Internalization of circulating apoptotic cells by splenic marginal zone dendritic cells: dependence on complement receptors and effect on cytokine production Blood 101611–620. [DOI] [PubMed] [Google Scholar]
- Quezada SA, Fuller B, Jarvinen LZ, Gonzalez M, Blazar BR, Rudensky AY.et al. (2003Mechanisms of donor-specific transfusion tolerance: preemptive induction of clonal T-cell exhaustion via indirect presentation Blood 1021920–1926. [DOI] [PubMed] [Google Scholar]
- Hancock WW, Sayegh MH, Zheng XG, Peach R, Linsley PS., and, Turka LA. Costimulatory function and expression of CD40 ligand, CD80, and CD86 in vascularized murine cardiac allograft rejection. Proc Natl Acad Sci USA. 1996;93:13967–13972. doi: 10.1073/pnas.93.24.13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson TC, Madsen JC, Larsen CP, Morris PJ., and, Wood KJ. Induction of transplantation tolerance in adults using donor antigen and anti-CD4 monoclonal antibody. Transplantation. 1992;54:475–483. doi: 10.1097/00007890-199209000-00018. [DOI] [PubMed] [Google Scholar]
- Niimi M, Roelen DL, Witzke O, van Rood JJ, Claas FH., and, Wood KJ. The importance of H2 haplotype sharing in the induction of specific unresponsiveness by pretransplant blood transfusions. Transplantation. 2000;69:411–417. doi: 10.1097/00007890-200002150-00018. [DOI] [PubMed] [Google Scholar]
- Garcia MR, Ledgerwood L, Yang Y, Xu J, Lal G, Burrell B.et al. (2010Monocytic suppressive cells mediate cardiovascular transplantation tolerance in mice J Clin Invest 1202486–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JK. Costimulatory blockade with monoclonal antibodies to induce alloanergy in donor lymphocytes. Int J Hematol. 2011;93:594–601. doi: 10.1007/s12185-011-0819-6. [DOI] [PubMed] [Google Scholar]
- Andreola G, Chittenden M, Shaffer J, Cosimi AB, Kawai T, Cotter P.et al. (2011Mechanisms of donor-specific tolerance in recipients of haploidentical combined bone marrow/kidney transplantation Am J Transplant 111236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton AM., and, Shevach EM. Suppressor effector function of CD4+CD25+ immunoregulatory T cells is antigen nonspecific. J Immunol. 2000;164:183–190. doi: 10.4049/jimmunol.164.1.183. [DOI] [PubMed] [Google Scholar]
- Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A., and, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- Bogdan C. Regulation of lymphocytes by nitric oxide. Methods Mol Biol. 2011;677:375–393. doi: 10.1007/978-1-60761-869-0_24. [DOI] [PubMed] [Google Scholar]
- Pacher P, Beckman JS., and, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Bonham CA, Chambers FG, Watkins SC, Hoffman RA, Simmons RL.et al. (1996Induction of nitric oxide synthase in mouse dendritic cells by IFN-gamma, endotoxin, and interaction with allogeneic T cells: nitric oxide production is associated with dendritic cell apoptosis J Immunol 1573577–3586. [PubMed] [Google Scholar]
- Ren G, Su J, Zhao X, Zhang L, Zhang J, Roberts AI.et al. (2008Apoptotic cells induce immunosuppression through dendritic cells: critical roles of IFN-gamma and nitric oxide J Immunol 1813277–3284. [DOI] [PubMed] [Google Scholar]
- Feng G, Gao W, Strom TB, Oukka M, Francis RS, Wood KJ.et al. (2008Exogenous IFN-gamma ex vivo shapes the alloreactive T-cell repertoire by inhibition of Th17 responses and generation of functional Foxp3+ regulatory T cells Eur J Immunol 382512–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JA, Brem-Exner BG, Riquelme P, Roelen D, Schulze M, Ivens K.et al. (2008A cell-based approach to the minimization of immunosuppression in renal transplantation Transpl Int 21742–754. [DOI] [PubMed] [Google Scholar]
- Hutchinson JA, Riquelme P, Brem-Exner BG, Schulze M, Matthäi M, Renders L.et al. (2008Transplant acceptance-inducing cells as an immune-conditioning therapy in renal transplantation Transpl Int 21728–741. [DOI] [PubMed] [Google Scholar]
- Corry RJ, Winn HJ., and, Russell PS. Primarily vascularized allografts of hearts in mice. The role of H-2D, H-2K, and non-H-2 antigens in rejection. Transplantation. 1973;16:343–350. doi: 10.1097/00007890-197310000-00010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse M regs do not correspond to previously described macrophage populations.
Interexperiment correlation analysis of macrophage samples belonging to three independent experimental series.
Hierarchically clustered heatmaps (Pearson uncentered, average linked) depicting reporters returned by SAM as significantly upregulated (a) and downregulated (b) in M regs compared to M regs without IFN-γ treatment (M reg no IFN-γ).
M regs suppress allogeneic T cell proliferation.
M regs interact with activated T cells in coculture.
Establishment of intracellular FACS-staining protocols for mouse iNOS.
Antibodies used for flow cytometry.