Abstract
In developing brains, neural progenitors exhibit cell cycle-dependent nuclear movement within the ventricular zone [interkinetic nuclear migration (INM)] and actively proliferate to produce daughter progenitors and/or neurons, whereas newly generated neurons exit from the cell cycle and begin pial surface-directed migration and maturation. Dysregulation of the balance between the proliferation and the cell cycle exit in neural progenitors is one of the major causes of microcephaly (small brain). Recent studies indicate that cell cycle machinery influences not only the proliferation but also INM in neural progenitors. Furthermore, several cell cycle-related proteins, including p27kip1, p57kip2, Cdk5, and Rb, regulate the migration of neurons in the postmitotic state, suggesting that the growth arrest confers dual functions on cell cycle regulators. Consistently, several types of microcephaly occur in conjunction with neuronal migration disorders, such as periventricular heterotopia and lissencephaly. However, cell cycle re-entry by disturbance of growth arrest in mature neurons is thought to trigger neuronal cell death in Alzheimer's disease. In this review, we introduce the cell cycle protein-mediated regulation of two types of nuclear movement, INM and neuronal migration, during cerebral cortical development, and discuss the roles of growth arrest in cortical development and neurological disorders.
Introduction
The balance between the proliferation and differentiation of progenitors determines the size of many organs, including the brain. The timing of the cell cycle exit of neural progenitors is important for the brain morphology and functions, as the defects result in several neurological disorders, including microcephaly (small brain) (Mochida & Walsh 2004; Bond & Woods 2006; Lizarraga et al. 2010; Miyata et al. 2010; Gruber et al. 2011). Furthermore, recent studies indicate that the regulation of cell cycle and growth arrest may play some roles in subsequent differentiation and maturation steps of postmitotic neurons. Neural progenitors exhibit a cell cycle-dependent nuclear movement within the ventricular zone, named interkinetic nuclear migration (INM), which influences cell fate determination as well as neurogenesis, at least in zebrafish retina (Kosodo 2012). In addition, several cell cycle-related proteins have additional functions in the postmitotic neurons of the developing cerebral cortex (Frank & Tsai 2009). For example, the function of p27kip1, a regulator for cell cycle exit, switches after growth arrest to regulate the migration and morphology of postmitotic neurons through actin cytoskeletal organization (Kawauchi et al. 2006). In mature neurons, the disturbance of growth arrest, which induces cell cycle re-entry, eventually leads to cell death (Herrup & Yang 2007). Thus, growth arrest confers dual functions on cell cycle-related proteins, and disrupting growth arrest may be associated with neurodegenerative diseases. In this review article, we introduce the mechanisms for neurogenesis and neuronal maturation, particularly focusing on INM and neuronal migration, respectively, and discuss the possible roles of growth arrest in brain development and several neurological disorders, such as developmental and neurodegenerative diseases.
Neural progenitor cells in mammalian cerebral cortex
Neural progenitor cells, opposed to their offspring, postmitotic neurons, exhibit cell cycle progression and cell division during brain development. Before the onset of neurogenesis, neural progenitor cells expand their numbers by symmetric, proliferative division, that is, one progenitor cell produces two progenitor cells (also called ‘self-renewal division’). After neurogenesis begins, the division mode switches to asymmetric division, that is, one progenitor cell produces one progenitor and one neuron or other type of progenitor (Gotz & Huttner 2005; Fietz & Huttner 2011). Currently, at least three types of neural progenitor cells have been identified in the developing mammalian cerebral cortex (Fig. 1A): apical progenitor, basal progenitor, and outer subventricular zone (OSVZ) progenitor (Fietz & Huttner 2011; Lui et al. 2011). An apical progenitor [also known as a neuroepithelial cell or radial glial cell (Gotz & Huttner 2005)] is an epithelial cell possessing two long processes along its apico-basal polarity and undergoes both symmetric, proliferative division and asymmetric, neurogenic division at the most apical end (ventricular side) of the ventricular zone (VZ) (Fig. 1A, green). A basal progenitor [also known as an intermediate progenitor (Noctor et al. 2004) or nonsurface dividing cell (Miyata et al. 2004)] lacks obvious processes and undergoes mostly symmetric, neurogenic division at the basal end of the VZ and subventricular zone (SVZ) (Fig. 1A, orange). An OSVZ progenitor [also known as an outer radial glial (oRG) cell (Hansen et al. 2010)] undergoes asymmetric, neurogenic division at the OSVZ, the inner region of brain parenchyma that is partitioned from the SVZ in primate cortex (Smart et al. 2002) (Fig. 1A, magenta). Notably, time-lapse lineage analyses have showed that apical progenitors can produce all three types of progenitor cells, but other types of progenitors do not produce the apical progenitors (Miyata et al. 2001, 2004; Noctor et al. 2001, 2004; Haubensak et al. 2004; Shitamukai et al. 2011; Wang et al. 2011) (Fig. 1A). Thus, apical progenitors can be considered as the stem of all neural progenitor subtypes.
Figure 1.
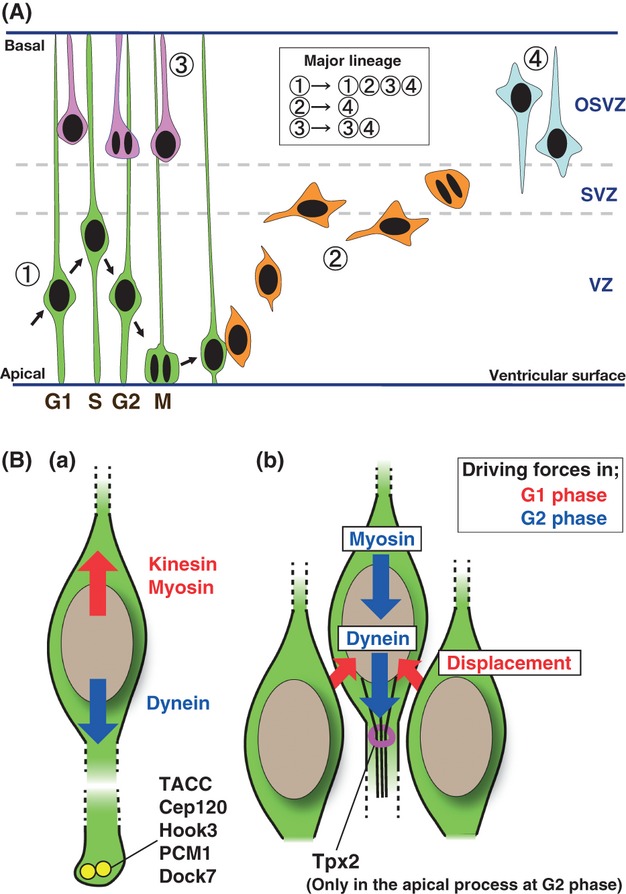
Major lineage of neural progenitors in mammalian cerebral cortex and interkinetic nuclear migration (INM) of apical progenitor. (A) Three kinds of neural progenitors identified in developing mammalian cortex (1–3) and postmitotic neuron (4) are illustrated. 1: Apical progenitor (green). 2: Basal progenitor (orange). 3: OSVZ progenitor (magenta). 4: Postmitotic neuron (light blue). Reported representative lineages from each progenitor (Fietz & Huttner 2011; Lui et al. 2011) are indicated in the square box. Cell cycle phases (G1, S, G2 and M) and the nuclear movement in each phase (arrow) of apical progenitor are described. VZ, ventricular zone; SVZ, subventricular zone; and OSVZ, outer subventricular zone. See text for details. (B) Schematics of mode of nuclear movements and proposed driving forces for INM. Arrows show directions of nuclear movements in each cell cycle phase (red: G1 phase, blue: G2 phase). Proposed driving forces for each direction of nuclear movement are indicated as (a) two opposing driving forces, (b) uni-directed driving force and displacement effect for the other direction from surrounded nuclei. See text for detail. The centrosome (yellow) may play an important role in INM because the functions of many centrosomal proteins are involved in INM. Tpx2 protein (magenta) is required for INM (basal-to-apical movement) and only observed in the apical process at G2 phase during interphase, suggesting that Tpx2 links cell cycle machinery with INM. See text for detail.
Interkinetic nuclear migration
What is INM?
During the progression of cell cycle phases, the nucleus of an apical progenitor conducts a unique mode of movement, named as ‘INM’ or ‘elevator movement’ (Fig. 1). INM is initially proposed by Sauer in 1935 in the embryonic vertebrate neural tube (Sauer 1935). Sauer postulated that the translocation of nuclear position occurs in accordance with the cell cycle progression; the cell division (M phase) of the neural progenitor cells takes place at the apical (ventricular) surface, followed by the nuclear movement from apical to basal during G1 phase. S phase occurs at the most basal end of the VZ and then the nucleus comes back to the apical position in G2 phase for the next cell division. A couple of decades after the first report, an experimental proof of the concept was demonstrated by labeling S-phase nuclei with 3H-thymidine, resulting in the appearance of radioactive label–incorporated chromatids in M-phase cells at the apical surface (Sauer & Walker 1959; Sidman et al. 1959; Fujita 1962). Recent advances in both light microscopy and tissue culturing methods (Miyata et al. 2001; Noctor et al. 2001) allow direct time-lapse imaging of INM.
INM has been identified not only in the embryonic neural tube of vertebrates, but also in other pseudostratified epithelial systems including invertebrates. For instance, retina in the developmental stage is a good model to analyze INM because of its relatively simple structure and accessibility for various experimental approaches, especially live-imaging to track nuclear migrations (Baye & Link 2007; Agathocleous & Harris 2009). Although much knowledge about INM has been derived from studies in the central nervous system (ectodermal origin), it has been demonstrated that INM also occurs in endoderm-originated digestive organs such as epithelia emanating from the liver bud (Bort et al. 2006) or intestin (Grosse et al. 2011) during development. Considering the evolutional aspect, it is important to compare vertebrate and invertebrate systems to clarify the types of molecules originally used for INM. Recent studies demonstrating the existence of INM in the Drosophila wing disc (Meyer et al. 2011) and Nematostella ectoderm (Meyer et al. 2011; Nakanishi et al. 2012) showed that both microtubule and actomyosin motor systems (see below) are required in more phylogenetically primitive organisms, suggesting that it is difficult to presume which motor system was primarily acquired during the nervous system evolution (Kosodo 2012).
Molecular mechanisms of INM
It has been a fascinating trial to uncover the mechanism of INM; how does the direction of nuclear migration correlate to each phase of cell cycle? Using drug treatments to disrupt cellular cytoskeletons, the importance of actin (Messier & Auclair 1974; Murciano et al. 2002) and microtubule (Langman et al. 1966; Karfunkel 1972) organization for INM was determined. Moreover, the molecular machineries controlling several steps of INM were recently revealed by advanced genetic manipulations. For the basal-to-apical nuclear migration, the association of the dynein motor proteins with Lis1 to the microtubule cytoskeleton plays a major role (Gambello et al. 2003; Tsai et al. 2005). Dynactin-1 and NudC, proteins forming a complex with dynein/Lis1, are also required for the basal-to-apical nuclear migration (Del Bene et al. 2008; Cappello et al. 2011). Centrosomes, which localize at the apical surface during interphase (Chenn et al. 1998), act as a microtubule-organizing center. The disruption of centrosomal protein functions, such as TACC, Cep120, Hook3, PCM1, and Dock7, have been found to perturb INM progression (Xie et al. 2007; Ge et al. 2010; Yang et al. 2012) (Fig. 1B). KASH proteins and SUN proteins form a physical link between the nuclear envelope and the dynein complex (Del Bene et al. 2008; Zhang et al. 2009; Yu et al. 2011). In spite of accumulated evidence that the microtubule motor system is important for the basal-to-apical nuclear migration, this is not always the case in INM of all epithelial tissue. It has been reported that in the zebrafish retina (Norden et al. 2009; Leung et al. 2011) and Drosophila wing disc (Meyer et al. 2011), not the dynein/microtubule motor system but the nonmuscle myosin with actin cytoskeleton is the main driver for the basal-to-apical nuclear migration. Interestingly, Rac1, a Rho family small GTPase involved in both microtubule and actin cytoskeletal regulation (Kawauchi 2011), is reported to control the basal-to-apical nuclear migration of neural progenitors (Minobe et al. 2009).
In contrast to what is known basal-to-apical nuclear migration, little information is available for apical-to-basal migration. Recent studies propose significant roles for kinesin, microtubule-associated motor, or actomyosin systems in nuclear apical-to-basal movement (Schenk et al. 2009; Tsai et al. 2010) (Fig. 1Ba). Notably, a critical role for physical displacement as a nonautonomous driving force of INM has been independently demonstrated in two systems (Fig. 1Bb). In developing zebrafish retina, it has been implicated that the trajectories of nuclear movements are largely stochastic, as mathematically postulated to fit nuclear positions (Norden et al. 2009). Subsequently, development of time-lapse quantitative analysis of nuclear movement in retina and hindbrain of zebrafish led to the conclusion that stochastic nuclear movement during phases other than the G2 phases arises passively in response to apical migration in neighboring cells (Leung et al. 2011). In developing mouse cortex, it was demonstrated that apical-to-basal migration is driven by a crowding effect in the epithelial tissue that results from continuous accumulation of nuclei due to the basal-to-apical active nuclear migration. This conclusion is achieved by nonautonomous movement of fluorescent beads from apical to basal, perturbation of basally oriented movement by disruption of basal-to-apical movement of surrounding cells, and simulation analysis (Kosodo et al. 2011). For active basal-to-apical movement, the actomyosin (Norden et al. 2009) or dynein/microtubule (Kosodo et al. 2011) motor system is used (Fig. 1Bb). The uni-directed active movement in INM would help to minimize the imbalance of nuclear density in the apical and basal regions of pseudostratified epithelia so as to preserve the homeostasis of tissue architecture during these developmental stages (Kosodo et al. 2011).
Cell cycle regulations associated with INM
Relationship between cell cycle regulation and INM
As discussed in the previous section, the nuclear movement in INM is tightly coupled to the cell cycle progression. From this standpoint, it arises the following questions: whether cell cycle progression can be a driver of INM or whether the nuclear positions can control the cell cycle progression? Inhibition of INM by the chemical inhibitor-mediated disruption of microtubule or actomyosin has been shown to have essentially no effect on cell cycle progression (Karfunkel 1972; Messier & Auclair 1974; Messier 1978; Gambello et al. 2003). However, treatment with drugs that interfere with several cell cycle steps result in the ectopic accumulation of nuclei in the neuroepithelia of developing mouse and zebrafish (Ueno et al. 2006; Kosodo et al. 2011; Leung et al. 2011). At a molecular resolution, G1 phase arrest, achieved by overexpressing p18Ink4c, an inhibitor protein of cyclin-dependent kinase (CDK) 4 and/or CDK6 (Sherr & Roberts 1999; Thullberg et al. 2000), leads to the accumulation of nuclei at a basal position in the VZ of developing mouse brains (Kosodo et al. 2011). Taken together, these results indicate that cell cycle progression likely regulates the activity of migration machineries.
How then, does the cell cycle progression correlate to the driving force of INM? It has been demonstrated that the function of Tpx2 protein connects cell cycle phases to the organization of the microtubule cytoskeleton required for INM. Tpx2, a microtubule-associated protein, is not observed in G1 phase, but appears during S phase and accumulates during G2 phase and then strongly associates to the mitotic spindle in M phase in HeLa cells (Gruss et al. 2002). In the apical progenitors in mouse brains, Tpx2 localizes on the microtubule in the apical process (but not in the basal process) of G2-phase cells, but not in G1 phase (Kosodo et al. 2011). Microtubule bundles in the apical processes of G2 phase are loosened by knockdown of Tpx2, resulting in a perturbation of basal-to-apical nuclear migration (Kosodo et al. 2011). Another study reported cell cycle control of actomyosin motor systems in zebrafish retina. Visualization of myosin regulatory light chain tagged with fluorescent protein showed its G2 specific recruitment to the basal side of nuclei. This is required for the basal-to-apical nuclear migration, likely by squeezing nuclei toward the apical side of the neuroepithelium (Leung et al. 2011).
Possible involvement of INM in fate determination
As described above, our understanding of INM has greatly expanded, especially with regard to the molecular machineries that generate the forces of nuclear migrations. What remains to be uncovered in the next stage of research is clarification as to whether INM is linked to cell fate determination, particularly in the developing central nervous system (Kosodo 2012). Interestingly, certain correlations between the S-phase positions and the cell fate of neural stem cell exist. In the retina of zebrafish, proliferative cells can be distinguished from neurogenic cells as different populations by the distance of S-phase positions from the apical surface (Baye & Link 2007). One possible scenario to generate this difference in cell fate is the concentration gradient of morphogen or signaling molecule along the axis of apico-basal polarity within the tissue, with nuclei receiving different amounts of neurogenic factor at specific cell cycle phases during INM (Latasa et al. 2009). In support of this hypothesis, Notch signaling–related proteins, whose activity can promote proliferation and cell cycle re-entry of neural stem cells (Pierfelice et al. 2011), show heterogenous apico-basal distributions. INM defects caused by a dynactin mutation result in altered exposure to Notch signals and impair neurogenesis in zebrafish retina (Del Bene et al. 2008).
Provided that S-phase positioning is one of the regulating factors of cell fate in apical progenitors, it is important to consider where nuclei enter into S phase. Using an elegant time-lapse study in the developing zebrafish nervous system, nuclear movement in each stage of cell cycle has been described (Leung et al. 2011); there is a basal drift at the beginning of G1 phase, strong basal-to-apical movement in G2 phase, and complete stochastic movement during S phase. This result essentially matches the nuclear movements observed in the developing mouse cortex (Kosodo et al. 2011).
Given that S-phase nuclei have no underlying directionality, how are the positions of S phase determined? Here, we need to consider the length of G1 phase and the mechanism of apical-to-basal nuclear migration during G1 phase (see previous section). If apical-to-basal nuclear movement is driven by an active motor system, it is likely that the position at the end of G1 phase (just before S-phase entry) from the apical surface toward the basal region changes in proportion to the length of G1 phase. However, if G1 nuclear movement is driven by a passive displacement factor, the position of the S-phase cell is likely to be dependent on both the length of G1 phase and the proportion of G2-phase length to the entire cell cycle. A recent report on accelerating the G1 phase of neural progenitors in the developing mouse brain may answer this question.
Co-over-expression of Cdk4 and cyclinD1 using in utero electroporation in the developing mouse cortex results in a shortened G1 phase, which evokes delayed neurogenesis (Lange et al. 2009). In this study, INM progression with over-expression or down-regulation of Cdk4/cyclinD1, which causes shortening or lengthening of G1 phase, respectively, is examined. Surprisingly, the positions of S-phase entry and exit are essentially the same between untransfected cells and electroporated cells in both shortened and lengthened G1 phase without affecting the number of apical progenitors (Lange et al. 2009). The experimental results show that the length of G1 phase was shortened to 65% by the over-expression of Cdk4 and cyclinD1 (from 9.0 to 5.9 h). As the position of S-phase entry is same in the overexpressed situation, this data do not appear to fit the active migration model unless the velocity of G1-phase nuclei was increased due to a side effect of Cdk4 and cyclinD1 over-expression on the motor system for the apical-to-basal nuclear migration. Next, it was demonstrated that the proportion of G2 phase (including M phase) to the entire cell cycle length increases by 1.36 times (from 14% to 19%) in the Cdk4 and cyclinD1 over-expressed condition. An increased proportion in the G2 phase raises the number of descending nuclei in a unit of time, which results in the higher density of nuclei in the apical region. According to the displacement model (see previous section), increased density of the apical region would raise the pressure to translocate nuclei in G1 phase from apical to basal. This might increase the velocity of apical-to-basal nuclear migration and compensate for a shortened G1-phase length, which would result in no obvious change for the nuclear position of S-phase entry. Perhaps, such a robust mechanism of INM might minimize effects of local disturbances of cell cycle progression on the architecture of the developing brain.
Neuronal migration
Multistep mode of neuronal migration
Newly generated immature neurons begin the pial surface-directed migration from the ventricular (apical) side, which is essential for the formation of architectural and functional cerebral cortex with a six-layered structure (Rakic 2006; Ayala et al. 2007; Kawauchi & Hoshino 2008; Marin et al. 2010; Govek et al. 2011; Kwan et al. 2012). A number of previous studies have indicated that migrating neurons exhibit multistep migration with various morphological changes (Kawauchi & Hoshino 2008) (Fig. 2). Migrating neurons first exhibit multipolar morphologies and subsequently form a leading process and an axon while retracting other neurites (Stensaas 1967; Shoukimas & Hinds 1978; Tamamaki et al. 2001; Tabata & Nakajima 2003; Noctor et al. 2004). The resulting bipolar-shaped neurons, called locomoting neurons, migrate over long distances along radial glial fibers, apical progenitor-derived long processes, with backward elongation of their axons (locomotion mode) (Rakic 1972, 2006; Nadarajah et al. 2001; Hatanaka & Murakami 2002; Noctor et al. 2004). At the final phase of migration, neurons switch from the migration mode into a radial glial fiber-independent terminal translocation mode (Nadarajah et al. 2001; Sekine et al. 2011). During the terminal translocation, dendrite maturation begins. Thus, neuronal migration is required for not only finding the final position but also neuronal maturation (Fig. 2). Defects in neuronal migration cause several neurological disorders, such as periventricular heterotopia and lissencephaly (Gleeson & Walsh 2000; Kawauchi & Hoshino 2008).
Figure 2.
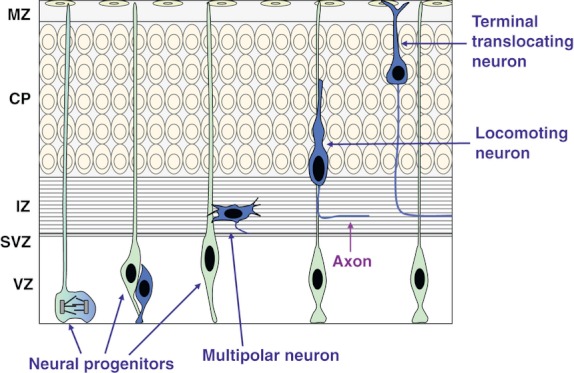
Multistep mode of neuronal migration. Postmitotic excitatory neurons are generated at the ventricular zone (VZ) or subventricular zone (SVZ) (See the enlarged drawing of the VZ and SVZ in Fig. 1) and migrate radially toward the pial surface (Blue cells). Neurons first display multipolar morphology at the lower part of the intermediate zone (IZ) and transform into locomoting neurons. Locomoting neurons possess a leading process and migrate over a long distance along radial glial fibers with elongation of an axon in a reverse direction. The migration mode switches from the locomotion mode into a radial glial fiber-independent terminal translocation mode during the final phase of migration. CP, cortical plate; IZ, intermediate zone; MZ, marginal zone; SVZ, subventricular zone; and VZ, ventricular zone.
c-jun N-terminal kinase pathway and microtubule-associated proteins
The first molecules identified to be involved in the morphological changes of migrating immature neurons were a Rho family small GTPase, Rac1, and its downstream kinase, c-jun N-terminal kinase (JNK) (Kawauchi et al. 2003) (Fig. 3). JNK regulates the transition from multipolar cells into locomoting neurons. JNK phosphorylates several microtubule-associated proteins, such as microtubule-associated protein 1B (MAP1B) and DCX (also known as doublecortin) (Chang et al. 2003; Kawauchi et al. 2003, 2005; Gdalyahu et al. 2004) (Fig. 3). Mutations in DCX gene cause X-linked lissencephaly in males and subcortical band heterotopia (also known as double cortex syndrome) in females (Gleeson et al. 1998; des Portes et al. 1998). Although both MAP1B and DCX promote microtubule stability (Francis et al. 1999; Gleeson et al. 1999; Goold et al. 1999; Horesh et al. 1999; Gordon-Weeks & Fischer 2000; Kawauchi et al. 2005; Trivedi et al. 2005), JNK-mediated phosphorylation diminishes their microtubule-binding affinities, resulting in decreased the microtubule stability (that is, increases the microtubule dynamics) (Chang et al. 2003; Kawauchi et al. 2003, 2005; Gdalyahu et al. 2004). Consistent with the fact that microtubule stability is kept at low levels at the tips of neurites (Shea 1999), phosphorylated MAP1B is strongly observed at the tips of axons (Goold et al. 1999; Gordon-Weeks & Fischer 2000). It has been reported that suppression of JNK or MAP1B disturbs neurite elongation (Takei et al. 2000; Kawauchi et al. 2003; Oliva et al. 2006; Eto et al. 2010). In vivo suppression of JNK disturbs the leading process morphology of migrating neurons and the pial surface-directed neuronal migration (Kawauchi et al. 2003).
Figure 3.
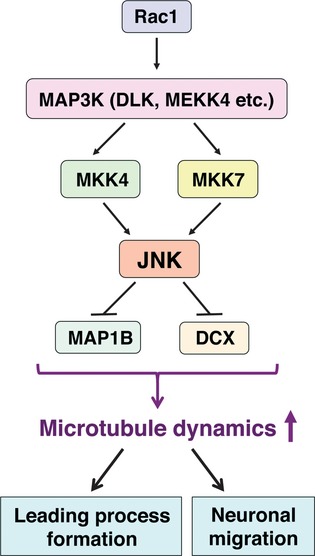
c-jun N-terminal kinase (JNK) pathway in postmitotic migrating neurons. JNK is required for the formation of a leading process (a surface-directed thick neurite of a locomoting neuron, see Fig. 2) and neuronal migration through the regulation of microtubule dynamics. MAP1B and DCX stabilize microtubules, but the phosphorylation by JNK enhances their dissociation from microtubules, resulting in an increase in microtubule dynamics.
As JNK belongs to a MAP kinase family, its activity is controlled by MAPKKs and MAPKKKs (Huang et al. 2004) (Fig. 3). Gene disruption of MKK4 or MKK7, MAPKKs for JNK, delays neuronal migration and disturbs axon formation (Wang et al. 2007; Yamasaki et al. 2011). Although the phosphorylation of MAP1B, but not DCX, is decreased in MKK4-deficient mice, the phosphorylation of both is suppressed in the MKK7 knockout mice. In addition, inhibition of DLK/MUK, a MAPKKK for JNK, results in similar phenotypes (Hirai et al. 2006). Interestingly, gene targeting for MEKK4, another MAPKKK for JNK, shows severe migration defects, resembling periventricular heterotopia (Sarkisian et al. 2006). Filamin A, a causative gene product of periventricular heterotopia (Fox et al. 1998), has also been reported to mediate the JNK signaling pathway in non-neuronal cells (Nomachi et al. 2008; Nakagawa et al. 2010) as well as the morphological changes and migration of cortical neurons (Nagano et al. 2004). Thus, the JNK-mediated pathway has important roles in neuronal migration and axon formation, and its defects may be associated with several cortical malformations.
Cdk5 and cell adhesion
DCX is also phosphorylated by cyclin-dependent kinase 5 (Cdk5) and MAP/microtubule affinity–regulating kinase 2 (MARK2, also known as Par-1) (Schaar et al. 2004; Tanaka et al. 2004) (Fig. 4A). Cdk5 is an unconventional CDK because its activity is mainly observed in postmitotic neurons (Tsai et al. 1993). Cdk5 is activated by p35, p39, and cyclin I, but not cyclin D, E, and A (Lee et al. 1996; Hisanaga & Saito 2003; Brinkkoetter et al. 2009; Su & Tsai 2011). In vivo suppression of Cdk5 activity by gene targeting, in vivo RNA interference and dominant negative experiments, has been shown to lead to severe neuronal migration defects (Ohshima et al. 1996; Gilmore et al. 1998; Kawauchi et al. 2003, 2006) (Fig. 4B). Similar to JNK, Cdk5 is required for the formation of leading process of migrating immature neurons (Kawauchi et al. 2006). However, Cdk5 also regulates multipolar cell morphologies, compared to the lesser effect of JNK on this aspect (Hirai et al. 2006; Kawauchi et al. 2006). A recent study showed that Cdk5 activity is required for the locomotion mode of neuronal migration (Nishimura et al. 2010), indicating that Cdk5 is a central regulator for multistep migration of immature neurons (Fig. 4B).
Figure 4.
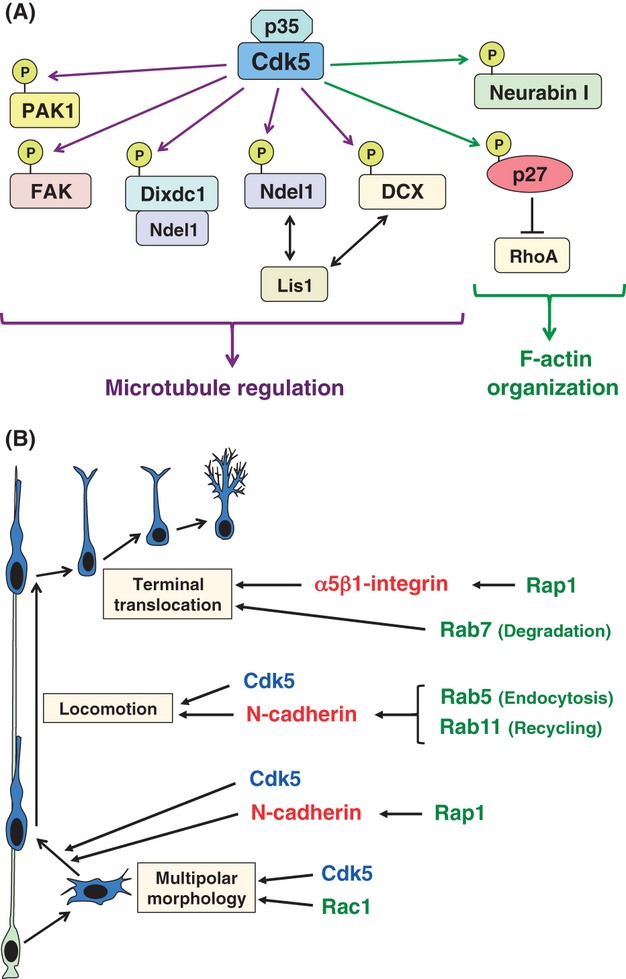
Roles of Cdk5 and cell adhesion molecules in multistep mode of neuronal migration. (A) Cdk5 phosphorylates many substrate molecules, including microtubule- and actin cytoskeleton-regulatory proteins (purple and green arrows, respectively). (B) Cdk5 is required for multiple steps of neuronal migration. Cdk5 (blue) regulates multipolar morphology of migrating neurons in a p27kip1-dependent manner, but its function in the transition into locomoting neurons is independent of p27kip1 as suppression of p27kip1 does not affect this step. Several small GTPases (green) also play important roles in the multistep mode of neuronal migration. Their functions are partly mediated by the regulation of cell adhesion molecules, N-cadherin and α5β1-integrin (red).
Cdk5 phosphorylates many substrate molecules, including p27kip1 (Kawauchi et al. 2006), Dixdc1 (Singh et al. 2010), Ndel1 (also known as Nudel) (Niethammer et al. 2000), focal adhesion kinase (FAK) (Xie et al. 2003), p21-activated kinase 1 (PAK1) (Rashid et al. 2001), neurabin I (Causeret et al. 2007), as well as DCX (Tanaka et al. 2004) (Fig. 4A). Ndel1 binds to Lis1, a causative gene product for lissencephaly (Reiner et al. 1993), and Ndel1 and Lis1 cooperatively control cytoplasmic dynein functions (Niethammer et al. 2000; Sasaki et al. 2000; Yamada et al. 2008). The Ndel1 phosphorylated by Cdk5 interacts with 14-3-3ε, which regulates the localization of Ndel1 and Lis1 (Toyo-oka et al. 2003). FAK is phosphorylated on Ser732 by Cdk5, and this phosphorylation is required for perinuclear microtubule organization (Xie et al. 2003). However, Cdk5 phosphorylates a neuron-specific F-actin-binding protein, neurabin I (Causeret et al. 2007). Furthermore, Cdk5-mediated phosphorylation of p27kip1 promotes actin reorganization, as described below. In vivo suppression of these Cdk5 substrates, p27kip1, Ndel1, FAK, and Neurabin I, disturbs neuronal migration mainly due to cytoskeletal defects.
In addition to cytoskeletal proteins, Cdk5 is known to regulate cell adhesion. Cell adhesion can be classified into cell-to-cell adhesion and cell-to-extracellular matrix (ECM) adhesion (Kawauchi 2012). Recent studies indicate that N-cadherin-mediated cell-to-cell adhesion plays essential roles in the multipolar and locomotion modes of neuronal migration (Kawauchi et al. 2010; Shikanai et al. 2011), whereas α5β1-integrin, a cell-to-ECM adhesion molecule that binds to fibronectin (Kawauchi 2012), is required for the terminal translocation (Sekine et al. 2012) (Fig. 4B). Rab family small GTPases, Rab5 and Rab11, regulate the intracellular trafficking of N-cadherin, which is required for the locomotion mode of neuronal migration (Kawauchi et al. 2010; Kawauchi 2011). A ras family small GTPase, Rap1, promotes the activities of N-cadherin and integrin at the early and final phases of neuronal migration, respectively (Franco et al. 2011; Jossin & Cooper 2011; Sekine et al. 2012) (Fig. 4B). Interestingly, Cdk5 can control both N-cadherin and integrin in a small GTPase-independent manner in vitro (Kwon et al. 2000; Huang et al. 2009), although it is still unclear whether Cdk5-mediated regulation of cell adhesion is involved in neuronal migration in vivo.
Linking mechanisms of cell cycle exit and neuronal migration
Cdk5 and p27kip1 in cell cycle exit, neuronal differentiation and migration
The cell cycle exit, neuronal differentiation, and migration occur concurrently, along with suppression in the activities of cyclin–CDKs. However, as described above, Cdk5 is strongly activated in postmitotic neurons. Although many studies indicate that Cdk5 is a regulator for cytoskeletal organization and signal transduction, rather than cell cycle, some notable facts remain. One is that Cdk5 directly phosphorylates p27kip1, a CDK inhibitor protein (Kawauchi et al. 2006). In addition, some mature neurons in the cortical plate abnormally re-enter the cell cycle in Cdk5-deficient mice (Cicero & Herrup 2005), similar to what is observed in the brains of p27kip1/p19Ink4d double knockout mice (Zindy et al. 1999), suggesting a functional relationship between Cdk5 and other cell cycle proteins.
It is known that p27kip1 regulates G1 length and cell cycle exit in the ventricular zone of the developing cerebral cortex via suppression of conventional CDK activities (Sherr & Roberts 1999; Mitsuhashi et al. 2001; Tarui et al. 2005). In contrast, Ser10 of p27kip1 is phosphorylated by Cdk5 in postmitotic neurons and this phosphorylation promotes its protein stability through the protection of p27kip1 from proteasome-dependent protein degradation (Ishida et al. 2000; Kotake et al. 2005; Kawauchi et al. 2006), suggesting that Cdk5 is an upstream positive regulator for p27kip1, a CDK inhibitor protein, in G0-arrested neurons, although p27kip1 acts as a negative regulator for conventional CDKs (Fig. 5). Furthermore, the increased protein levels of p27kip1 have essential roles in cortical neuronal migration and the formation of multipolar cell morphologies (Kawauchi et al. 2006). Cdk5-p27kip1 pathway enhances actin reorganization via the suppression of RhoA activity and thereby activation of an actin-binding protein, cofilin (Kawauchi et al. 2006). It has been reported that p27kip1 is also involved in the regulation of microtubule organization (Baldassarre et al. 2005; Godin et al. 2012). Interestingly, a recent study indicates that connexin 43, a component of gap junction involved in both neural progenitor proliferation and neuronal migration (Elias & Kriegstein 2008), acts upstream of p27kip1 to regulate the multipolar morphology of migrating neurons (Liu et al. 2012). Taken together, these findings suggest that p27kip1 acquires additional functions in cytoskeletal regulation and neuronal migration during growth arrest and that this functional switch is mediated at least in part by Cdk5 (Fig. 5).
Figure 5.
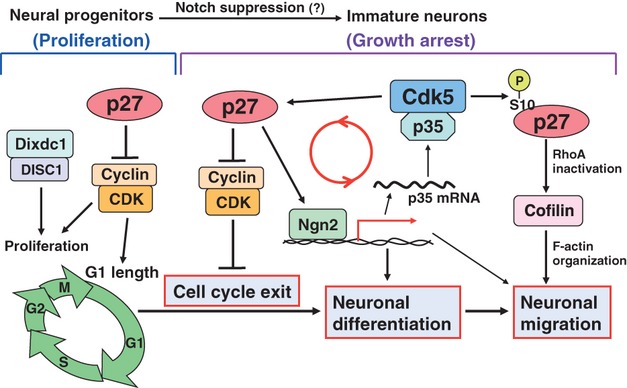
A possible link in mechanisms between cell cycle exit, neuronal differentiation, and neuronal migration. In the developing cerebral cortex, cell cycle exit, neuronal differentiation, and initiation of neuronal migration occur concurrently. A cyclin-dependent kinase (CDK) inhibitor protein, p27kip1, controls the G1 length and cell cycle exit in neural progenitors via the suppression of Cyclin-CDK activities. In addition to these cell cycle regulatory functions, p27kip1 promotes neuronal differentiation via the up-regulation of Ngn2 protein level and neuronal migration through the suppression of RhoA activity and thereby activation of an actin-binding protein, Cofilin. Ngn2 activates the transcription of p35 as well as neuronal differentiation-related genes. In postmitotic neurons, p35 binds to and activates Cdk5, which directly phosphorylates and stabilizes p27kip1 protein and is required for the maintenance of growth arrest. A proposed feedback loop of Cdk5/p35-p27kip1-Ngn2-p35-Cdk5 is shown (red circle).
Cdk5-mediated phosphorylation of Dixdc1 also functions as a molecular switch between neural progenitor proliferation and neuronal migration (Singh et al. 2010). Nonphosphorylated Dixdc1 binds to Disrupted in Schizophrenia-1 (DISC1) and controls neural progenitor proliferation. In contrast, Cdk5 phosphorylates Dixdc1 in postmitotic neurons, resulting in increased interaction between Ndel1 and DISC1 and promotion of neuronal migration.
In addition to the dual functions in neural progenitors and migrating neurons, p27kip1 is involved in neuronal differentiation. A previous report showed that p27kip1 increases the protein levels of Neurogenin 2 (Ngn2), a basic helix-loop-helix-type transcription factor required for neuronal differentiation, and promotes neuronal differentiation (Nguyen et al. 2006). Furthermore, Cdk5 deficiency partially disturbs neuronal differentiation (Cicero & Herrup 2005; Zheng et al. 2010) as well as neuronal migration, and Cdk5-mediated phosphorylation of p27kip1 at Ser10 and Thr187 is involved in the regulation of neuronal differentiation (Zheng et al. 2010). Interestingly, p35, an activator for Cdk5, was identified as a target molecule of Ngn2 (Ge et al. 2006), and it has been reported that Ngn2 is also required for neuronal migration (Hand et al. 2005; Ge et al. 2006; Heng et al. 2008). These findings implicate a positive feedback loop of Cdk5/p35-p27kip1-Ngn2-p35 that has important roles in the growth arrest–associated neuronal differentiation and initiation of migration (Kawauchi & Hoshino 2008) (Fig. 5). The identity of the molecule(s) that turn on the positive feedback loop for the synchronized cellular events of cell cycle exit, neuronal differentiation, and initiation of neuronal migration is still unclear, but there is evidence to indicate that Notch signaling suppresses p27kip1 mRNA and/or protein levels (Sarmento et al. 2005; Vernon et al. 2006; Murata et al. 2009), suggesting that weakened Notch signal may enhance p27kip1 expression and thereby the positive feedback loop.
Other CDK inhibitor proteins and Rb-E2F
Other cell cycle-related proteins have also been reported to have dual functions in proliferating and arrested cells. CDK inhibitor proteins include members of Cip/Kip (p21cip1, p27kip1, and p57kip2) and Ink4 (p16Ink4a, p15Ink4b, p18Ink4c, and p19Ink4d) families (Sherr & Roberts 1999) (Fig. 6A). Although p57kip2 mainly controls the cell cycle exit of early-born neurons (deep layer neurons), p27kip1 preferentially regulates the growth arrest of late-born neurons (upper layer neurons) (Mairet-Coello et al. 2012) (Fig. 6B). In the postmitotic neurons, it has been reported that not only p27kip1 but also p57kip2 is involved in neuronal migration (Itoh et al. 2007). Consistently, both proteins are localized at the leading process and cell soma as well as nucleus in migrating neurons (Kawauchi et al. 2006).
Figure 6.
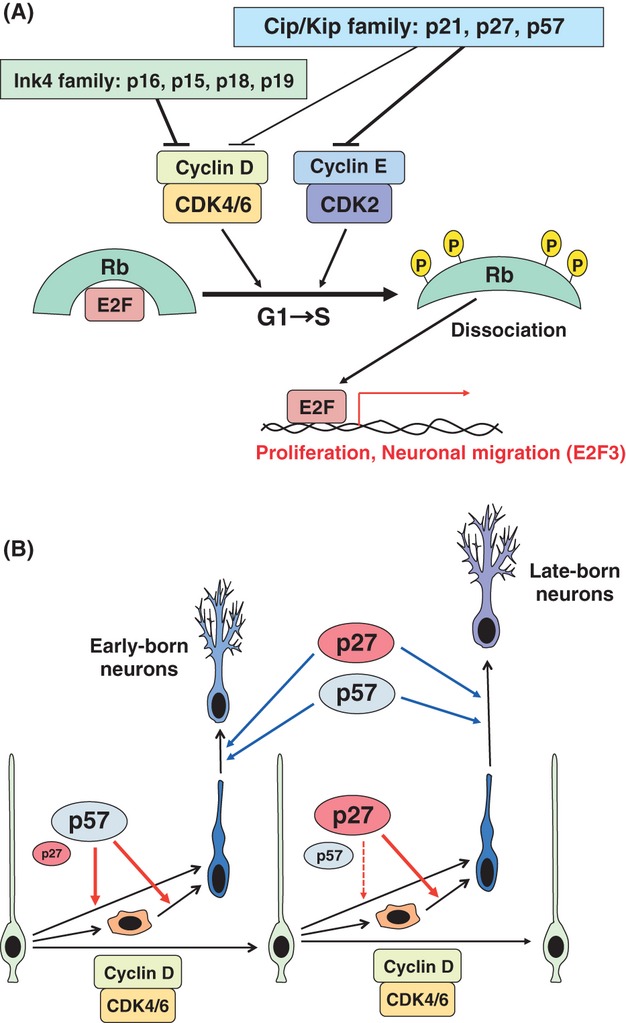
Cyclin-dependent kinase (CDK) inhibitor proteins regulate cell cycle progression, growth arrest, and postmitotic neuronal migration. (A) Molecular mechanisms for G1/S transition. The transition from G1 to S phase is dependent on CyclinD-Cdk4/6 and CyclinE-Cdk2 activities, which phosphorylate Rb protein. The phosphorylated Rb protein dissociates E2F family transcription factors. Both E2F1 and E2F3 promote G1/S transition in neural progenitors, whereas E2F3, but not E2F1, regulates neuronal positioning. The activities of Cyclin-CDK complexes are suppressed by CDK inhibitor proteins, which are composed of a Cip/Kip family (p21cip1, p27kip1, and p57kip2) and Ink4 family (p16Ink4a, p15Ink4b, p18Ink4c, and p19Ink4d). (B) Roles of CDK inhibitor proteins, p27kip1 and p57kip2, in cell cycle exit and subsequent neuronal migration. p57kip2 and p27kip1 preferentially control the cell cycle exit of neural progenitors for early-born (deep layer) and late-born (upper layer) neurons, respectively. p27kip1 mainly functions in basal progenitors (orange cells) rather than apical progenitors (green cells). Both p27kip1 and p57kip2 have been shown to regulate the migration of postmitotic neurons as well as the cell cycle exit.
Furthermore, retinoblastoma (Rb) protein and E2F family transcription factors are reported to regulate both cell cycle in neural progenitors and migration in postmitotic neurons. Rb protein binds to and represses the E2F functions, whereas Cdk-dependent phosphorylation of Rb dissociates E2Fs from the Rb protein, allowing E2Fs to interact with target DNA sequences (Giacinti & Giordano 2006) (Fig. 6A). Knockout of the Rb gene perturbs the neuronal positioning in cerebral cortex, and the phenotypes are rescued by double knockout of Rb and E2F3, but not E2F1 (Ferguson et al. 2005; McClellan et al. 2007). Although the switching mechanism of Rb-E2F functions is unclear, a recent study shows that Cdk5 has the ability to phosphorylate Rb protein (Futatsugi et al. 2012). In addition to the regulators for G1/S transition, Aurora A and anaphase-promoting complex/cyclosome (APC/C), both of which mainly function at M phase, are reported to regulate neuronal migration and axon/dendrite formation (Konishi et al. 2004; Kim et al. 2009; Mori et al. 2009; Takitoh et al. 2012). Therefore, growth arrest signals may provide additional functions beyond cell cycle regulation for some cell cycle-related proteins.
Growth arrest and developmental neurological disorders
Disruption of the balance between progenitor self-renewal and cell cycle exit (neuronal differentiation) leads to several neurological disorders. For example, abnormally enhanced cell cycle exit of neural progenitors leads to premature differentiation and thereby exhaustion of neural progenitors, resulting in microcephaly (small brain) (Mochida & Walsh 2004; Bond & Woods 2006; Lizarraga et al. 2010; Miyata et al. 2010; Buchman et al. 2011; Gruber et al. 2011). Interestingly, microcephaly is sometimes accompanied by neuronal migration disorders. Mutation in ArfGEF2 causes microcephaly and periventricular heterotopia (Sheen et al. 2004). ArfGEF2 encodes Big2/ArfGEF2 protein, which regulates membrane trafficking from Golgi apparatus via the activation of Arf family small GTPases. Furthermore, it is reported that Big2 is also localized at recycling endosomes (Shin et al. 2004). Consistent with this, endocytosis and recycling of a cell-cell adhesion molecule, N-cadherin, are known to play essential roles in the locomotion mode of neuronal migration (Kawauchi et al. 2010; Shikanai et al. 2011). Interestingly, N-cadherin is also required for the maintenance of neuroepithelial (ventricular zone) structures (Kadowaki et al. 2007), whose disruption is observed in the brains with periventricular heterotopia (Ferland et al. 2009). Therefore, the regulation of membrane trafficking may be another mechanism that links neural progenitor proliferation and neuronal migration.
Human mutations in the Nde1 gene result in microcephaly with lissencephaly (referred to as ‘microlissencephaly’) (Feng & Walsh 2004; Alkuraya et al. 2011). Furthermore, knockdown of abnormal spindle microcephaly (ASPM), a causative gene for autosomal recessive primary microcephaly (MCPH, for microcephaly primary hereditary), disturbs neuronal migration as well as neural progenitor proliferation in mice (Fish et al. 2006; Buchman et al. 2011). In addition to human neurological disorder–related genes, many molecules, including Lis1, dynein, SUN proteins, and Rac1, are required for both INM and neuronal migration (Hirotsune et al. 1998; Gambello et al. 2003; Kawauchi et al. 2003; Tsai et al. 2005, 2007; Yoshizawa et al. 2005; Minobe et al. 2009; Zhang et al. 2009; Kawauchi 2011; Yu et al. 2011). Because most of these proteins function in both neural progenitors and postmitotic neurons, neural progenitor proliferation and neuronal migration share several common intracellular pathways in centrosome and/or microtubule regulation. Considering that Cdk5 acts upstream of Lis1, dynein, and Rac1 (Niethammer et al. 2000; Xin et al. 2004; Govek et al. 2011) and that p27kip1 is involved in the regulation of microtubules as well as actin cytoskeleton (Baldassarre et al. 2005; Kawauchi et al. 2006; Godin et al. 2012), the growth arrest-mediated Cdk5 activation by the up-regulation of p35 protein may alter the function of several cell cycle-related proteins, which exert different cellular events in part using common machineries.
Growth arrest in postmitotic mature cells
In adulthood, many cells, including mature neurons, maintain a quiescent state throughout life. It has been reported that cyclin E binds to and suppresses the activity of Cdk5, resulting in the enhancement of synapse formation (Odajima et al. 2011). This suggests that some cell cycle-related proteins also function in mature neurons. Thus, alternative functions for cell cycle-related proteins are important for growth-arrested cells. However, several studies have indicated that cell cycle re-entry by perturbing growth arrest is a trigger for cell death.
Mammalian auditory epithelium, composed of hair cells and supporting cells, has limited capability for regeneration, which remains an obstacle for the development of therapeutics for sensorineural hearing loss (Roberson & Rubel 1994; Forge et al. 1998; White et al. 2006). In contrast, in the avian auditory epithelium, the loss of hair cells leads to re-entry of supporting cells into the cell cycle, giving rise to both hair cells and supporting cells (Corwin & Cotanche 1988; Ryals & Rubel 1988). For the purpose of promoting regeneration of the cochlea in mammals, knockdown of p27kip1 in the postmitotic supporting cells of mouse auditory epithelia was performed (Ono et al. 2009). That study reported the successful re-activation of the proliferative capacities of the auditory supporting cells, but induction of the apoptotic pathway occurred several days later (Fig. 7A).
Figure 7.
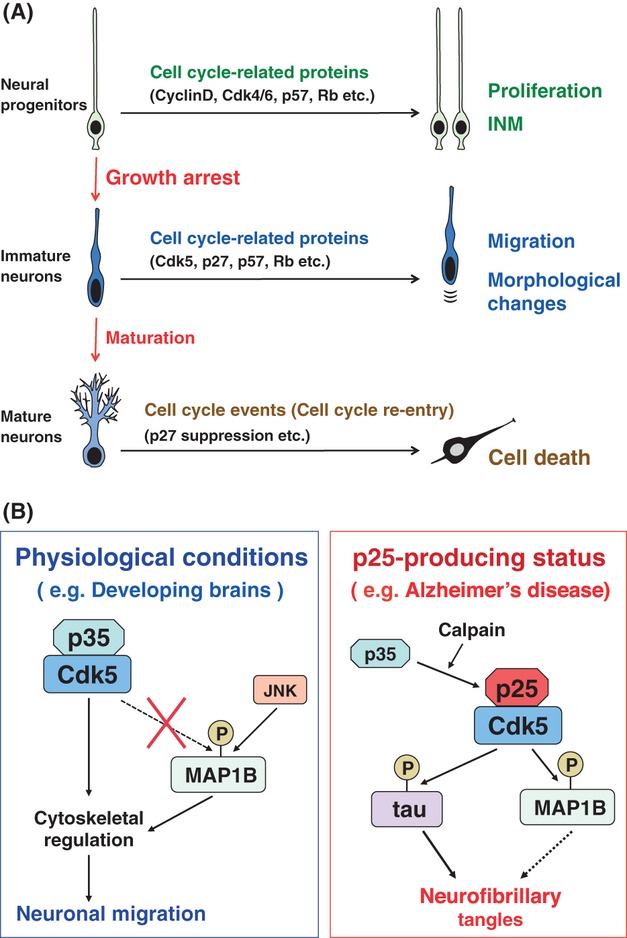
Alternative functions of cell cycle-related proteins in the construction and maintenance of brains throughout life. (A) Cell cycle-related proteins function in not only the proliferation of neural progenitors but also various aspects of brain construction and its maintenance throughout life. Cell cycle machinery controls interkinetic nuclear migration (INM) in neural progenitors, and after growth arrest, several cell cycle-related proteins change their functions to control the migration and morphology of postmitotic neurons. However, cell cycle re-entry by disturbance of growth arrest is thought to trigger cell death. (B) Cdk5 functions in brain development and neurodegenerative diseases. Cdk5, binding to its activator, p35, phosphorylates many substrate molecules and controls the multistep mode of neuronal migration in developing brains (see Fig. 4). In contrast, p35 is cleaved into the more stable p25 in pathogenic conditions, including Alzheimer's disease. Cdk5/p25, but not Cdk5/p35, strongly phosphorylates tau and MAP1B, which may be associated with the formation of neurofibrillary tangles in neurodegenerative diseased brains.
Re-activation of cell cycle machinery in mature neurons is also associated with cell death. In the brains of Alzheimer's disease mouse models, re-expression of cell cycle proteins, such as cyclin A and PCNA, and DNA replication are observed before neuronal cell death (Yang et al. 2001, 2003; Varvel et al. 2008). These ‘cell cycle events’ themselves do not seem to directly induce neuronal cell death, but are thought to be important priming phenomena for neurodegenerative diseases (Yang & Herrup 2007). Furthermore, it has been reported that the abnormal activation of Cdk5 is involved in neurodegeneration. Inhibition of Cdk5 induces cell cycle events, suggesting that Cdk5 suppresses the cell cycle in mature neurons (Cicero & Herrup 2005; Zhang et al. 2008). The activator for Cdk5 is changed from p35 into a more stable isoform, p25, through a calpain-mediated cleavage in brains with neurodegenerative diseases (Patrick et al. 1999; Kusakawa et al. 2000; Lee et al. 2000). It is known that Cdk5/p35 and Cdk5/p25 exhibit different substrate specificities. Unlike Cdk5/p35, Cdk5/p25 strongly phosphorylates tau and MAP1B (Patrick et al. 1999; Kawauchi et al. 2005), and their hyperphosphorylation is observed in Alzheimer's diseased brains (Hasegawa et al. 1990; Ulloa et al. 1994; Cruz et al. 2003; Hisanaga & Saito 2003; Tsai et al. 2004; Su & Tsai 2011). Cdk5/p25 interacts with and inhibits the activity of histone deacetylase 1 (HDAC1), and suppression of HDAC1 induces double-stranded DNA breaks and cell cycle activity in neurons (Kim et al. 2008). These results indicate that the re-activation of cell cycle machinery, including DNA replication, in mature postmitotic cells induces cell death and further suggest that the growth arrest of mature neurons plays essential roles in neuronal survival and normal brain functions.
Conclusion remarks
The tight regulation of cell cycle proteins is essential for the proliferation and cell cycle exit of neural progenitors during brain development. Recent studies also indicate that cell cycle-related proteins contribute to much broader events beyond the cell cycle regulation in the developing and adult brains (Fig. 7A). In neural progenitors, the cell cycle machinery is closely associated with and actively controls INM at least in part through Tpx2-mediated organization of microtubules. Even after growth arrest, cell cycle-related proteins, such as p27kip1 and Rb, exhibit alternative functions that affect the migration and changes in morphology of postmitotic neurons. Interestingly, although these alternative functions are essential for brain development, disruption of growth arrest in mature neurons or other postmitotic cells is closely associated with cell death, suggesting that re-activation of cell cycle progression itself may be harmful to postmitotic neurons. As a large proportion of cells in adulthood are in a postmitotic state, it is possible that growth arrest contributes to the maintenance of cellular homeostasis in the whole body.
Acknowledgments
The authors thank Dr Ruth T. Yu for critical reading of the manuscript. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, and Science and Technology, Japan and by grants from the JST PRESTO program.
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Alkuraya FS, Cai X, Emery C, Mochida GH, Al-Dosari MS, Felie JM, Hill RS, Barry BJ, Partlow JN, Gascon GG, Kentab A, Jan M, Shaheen R, Feng Y, Walsh CA. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am. J. Hum. Genet. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7:51–63. doi: 10.1016/j.ccr.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Woods CG. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Bort R, Signore M, Tremblay K, Martinez Barbera JP, Zaret KS. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 2006;290:44–56. doi: 10.1016/j.ydbio.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Brinkkoetter PT, Olivier P, Wu JS, Henderson S, Krofft RD, Pippin JW, Hockenbery D, Roberts JM, Shankland SJ. Cyclin I activates Cdk5 and regulates expression of Bcl-2 and Bcl-XL in postmitotic mouse cells. J. Clin. Invest. 2009;119:3089–3101. doi: 10.1172/JCI37978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman JJ, Durak O, Tsai LH. ASPM regulates Wnt signaling pathway activity in the developing brain. Genes Dev. 2011;25:1909–1914. doi: 10.1101/gad.16830211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Monzo P, Vallee RB. NudC is required for interkinetic nuclear migration and neuronal migration during neocortical development. Dev. Biol. 2011;357:326–335. doi: 10.1016/j.ydbio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causeret F, Jacobs T, Terao M, Heath O, Hoshino M, Nikolic M. Neurabin-I is phosphorylated by Cdk5: implications for neuronal morphogenesis and cortical migration. Mol. Biol. Cell. 2007;18:4327–4342. doi: 10.1091/mbc.E07-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Jones Y, Ellisman MH, Goldstein LS, Karin M. JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell. 2003;4:521–533. doi: 10.1016/s1534-5807(03)00094-7. [DOI] [PubMed] [Google Scholar]
- Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol. Cell. Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- Cicero S, Herrup K. Cyclin-dependent kinase 5 is essential for neuronal cell cycle arrest and differentiation. J. Neurosci. 2005;25:9658–9668. doi: 10.1523/JNEUROSCI.1773-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias LA, Kriegstein AR. Gap junctions: multifaceted regulators of embryonic cortical development. Trends Neurosci. 2008;31:243–250. doi: 10.1016/j.tins.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K, Kawauchi T, Osawa M, Tabata H, Nakajima K. Role of dual leucine zipper-bearing kinase (DLK/MUK/ZPK) in axonal growth. Neurosci. Res. 2010;66:37–45. doi: 10.1016/j.neures.2009.09.1708. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Ferguson KL, McClellan KA, Vanderluit JL, McIntosh WC, Schuurmans C, Polleux F, Slack RS. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 2005;24:4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Batiz LF, Neal J, et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum. Mol. Genet. 2009;18:497–516. doi: 10.1093/hmg/ddn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Huttner WB. Cortical progenitor expansion, self-renewal and neurogenesis-a polarized perspective. Curr. Opin. Neurobiol. 2011;21:23–35. doi: 10.1016/j.conb.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Paabo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl Acad. Sci. USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forge A, Li L, Nevill G. Hair cell recovery in the vestibular sensory epithelia of mature guinea pigs. J. Comp. Neurol. 1998;397:69–88. [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- Francis F, Koulakoff A, Boucher D, Chafey P, Schaar B, Vinet MC, Friocourt G, McDonnell N, Reiner O, Kahn A, McConnell SK, Berwald-Netter Y, Denoulet P, Chelly J. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Muller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Tsai LH. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron. 2009;62:312–326. doi: 10.1016/j.neuron.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. Kinetics of cellular proliferation. Exp. Cell Res. 1962;28:52–60. doi: 10.1016/0014-4827(62)90311-7. [DOI] [PubMed] [Google Scholar]
- Futatsugi A, Utreras E, Rudrabhatla P, Jaffe H, Pant HC, Kulkarni AB. Cyclin-dependent kinase 5 regulates E2F transcription factor through phosphorylation of Rb protein in neurons. Cell Cycle. 2012;11:1603–1610. doi: 10.4161/cc.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J. Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdalyahu A, Ghosh I, Levy T, Sapir T, Sapoznik S, Fishler Y, Azoulai D, Reiner O. DCX, a new mediator of the JNK pathway. EMBO J. 2004;23:823–832. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl Acad. Sci. USA. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Gilmore EC, Ohshima T, Goffinet AM, Kulkarni AB, Herrup K. Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J. Neurosci. 1998;18:6370–6377. doi: 10.1523/JNEUROSCI.18-16-06370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, Walsh CA. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Walsh CA. Neuronal migration disorders: from genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- Godin JD, Thomas N, Laguesse S, et al. p27(Kip1) is a microtubule-associated protein that promotes microtubule polymerization during neuron migration. Dev. Cell. 2012;23:729–744. doi: 10.1016/j.devcel.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Goold RG, Owen R, Gordon-Weeks PR. Glycogen synthase kinase 3beta phosphorylation of microtubule-associated protein 1B regulates the stability of microtubules in growth cones. J. Cell Sci. 1999;112:3373–3384. doi: 10.1242/jcs.112.19.3373. [DOI] [PubMed] [Google Scholar]
- Gordon-Weeks PR, Fischer I. MAP1B expression and microtubule stability in growing and regenerating axons. Microsc. Res. Tech. 2000;48:63–74. doi: 10.1002/(SICI)1097-0029(20000115)48:2<63::AID-JEMT2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Govek EE, Hatten ME, Van Aelst L. The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol. 2011;71:528–553. doi: 10.1002/dneu.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse AS, Pressprich MF, Curley LB, Hamilton KL, Margolis B, Hildebrand JD, Gumucio DL. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development. 2011;138:4423–4432. doi: 10.1242/dev.065789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber R, Zhou Z, Sukchev M, Joerss T, Frappart PO, Wang ZQ. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1-Cdc25 pathway. Nat. Cell Biol. 2011;13:1325–1334. doi: 10.1038/ncb2342. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Wittmann M, Yokoyama H, Pepperkok R, Kufer T, Sillje H, Karsenti E, Mattaj IW, Vernos I. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 2002;4:871–879. doi: 10.1038/ncb870. [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, Schuurmans C, Guillemot F, Polleux F. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Ihara Y. Immunochemical evidence that fragments of phosphorylated MAP5 (MAP1B) are bound to neurofibrillary tangles in Alzheimer's disease. Neuron. 1990;4:909–918. doi: 10.1016/0896-6273(90)90144-5. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Murakami F. In vitro analysis of the origin, migratory behavior, and maturation of cortical pyramidal cells. J. Comp. Neurol. 2002;454:1–14. doi: 10.1002/cne.10421. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, Guillemot F. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hirai SI, Feng Cui D, Miyata T, Ogawa M, Kiyonari H, Suda Y, Aizawa S, Banba Y, Ohno S. The c-Jun N-terminal kinase activator dual leucine zipper kinase regulates axon growth and neuronal migration in the developing cerebral cortex. J. Neurosci. 2006;26:11992–12002. doi: 10.1523/JNEUROSCI.2272-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Hisanaga S, Saito T. The regulation of cyclin-dependent kinase 5 activity through the metabolism of p35 or p39 Cdk5 activator. Neurosignals. 2003;12:221–229. doi: 10.1159/000074624. [DOI] [PubMed] [Google Scholar]
- Horesh D, Sapir T, Francis F, Wolf SG, Caspi M, Elbaum M, Chelly J, Reiner O. Doublecortin, a stabilizer of microtubules. Hum. Mol. Genet. 1999;8:1599–1610. doi: 10.1093/hmg/8.9.1599. [DOI] [PubMed] [Google Scholar]
- Huang C, Jacobson K, Schaller MD. MAP kinases and cell migration. J. Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- Huang C, Rajfur Z, Yousefi N, Chen Z, Jacobson K, Ginsberg MH. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat. Cell Biol. 2009;11:624–630. doi: 10.1038/ncb1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N, Kitagawa M, Hatakeyama S, Nakayama K. Phosphorylation at serine 10, a major phosphorylation site of p27(Kip1), increases its protein stability. J. Biol. Chem. 2000;275:25146–25154. doi: 10.1074/jbc.M001144200. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Masuyama N, Nakayama K, Nakayama KI, Gotoh Y. The cyclin-dependent kinase inhibitors p57 and p27 regulate neuronal migration in the developing mouse neocortex. J. Biol. Chem. 2007;282:390–396. doi: 10.1074/jbc.M609944200. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Cooper JA. Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 2011;14:697–703. doi: 10.1038/nn.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M. N-cadherin mediates cortical organization in the mouse brain. Dev. Biol. 2007;304:22–33. doi: 10.1016/j.ydbio.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Karfunkel P. The activity of microtubules and microfilaments in neurulation in the chick. J. Exp. Zool. 1972;181:289–301. doi: 10.1002/jez.1401810302. [DOI] [PubMed] [Google Scholar]
- Kawauchi T. Regulation of cell adhesion and migration in cortical neurons: not only Rho but also Rab family small GTPases. Small GTPases. 2011;2:36–40. doi: 10.4161/sgtp.2.1.15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T. Cell adhesion and its endocytic regulation in cell migration during neural development and cancer metastasis. Int. J. Mol. Sci. 2012;13:4564–4590. doi: 10.3390/ijms13044564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat. Cell Biol. 2006;8:17–26. doi: 10.1038/ncb1338. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nishimura YV, Nabeshima Y, Hoshino M. MAP1B phosphorylation is differentially regulated by Cdk5/p35, Cdk5/p25, and JNK. Biochem. Biophys. Res. Commun. 2005;331:50–55. doi: 10.1016/j.bbrc.2005.03.132. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev. Neurosci. 2008;30:36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Sekine K, Shikanai M, Chihama K, Tomita K, Kubo K, Nakajima K, Nabeshima Y, Hoshino M. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron. 2010;67:588–602. doi: 10.1016/j.neuron.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Frank CL, Dobbin MM, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell. Mol. Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosodo Y, Suetsugu T, Suda M, Mimori-Kiyosue Y, Toida K, Baba SA, Kimura A, Matsuzaki F. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakayama K, Ishida N, Nakayama KI. Role of serine 10 phosphorylation in p27 stabilization revealed by analysis of p27 knock-in mice harboring a serine 10 mutation. J. Biol. Chem. 2005;280:1095–1102. doi: 10.1074/jbc.M406117200. [DOI] [PubMed] [Google Scholar]
- Kusakawa G, Saito T, Onuki R, Ishiguro K, Kishimoto T, Hisanaga S. Calpain-dependent proteolytic cleavage of the p35 cyclin-dependent kinase 5 activator to p25. J. Biol. Chem. 2000;275:17166–17172. doi: 10.1074/jbc.M907757199. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Gupta A, Zhou Y, Nikolic M, Tsai LH. Regulation of N-cadherin-mediated adhesion by the p35-Cdk5 kinase. Curr. Biol. 2000;10:363–372. doi: 10.1016/s0960-9822(00)00411-5. [DOI] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Langman J, Guerrant RL, Freeman BG. Behavior of neuro-epithelial cells during closure of the neural tube. J. Comp. Neurol. 1966;127:399–411. doi: 10.1002/cne.901270308. [DOI] [PubMed] [Google Scholar]
- Latasa MJ, Cisneros E, Frade JM. Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis. Int. J. Dev. Biol. 2009;53:895–908. doi: 10.1387/ijdb.082721ml. [DOI] [PubMed] [Google Scholar]
- Lee MH, Nikolic M, Baptista CA, Lai E, Tsai LH, Massague J. The brain-specific activator p35 allows Cdk5 to escape inhibition by p27Kip1 in neurons. Proc. Natl Acad. Sci. USA. 1996;93:3259–3263. doi: 10.1073/pnas.93.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kwon YT, Li M, Peng J, Friedlander RM, Tsai LH. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature. 2000;405:360–364. doi: 10.1038/35012636. [DOI] [PubMed] [Google Scholar]
- Leung L, Klopper AV, Grill SW, Harris WA, Norden C. Apical migration of nuclei during G2 is a prerequisite for all nuclear motion in zebrafish neuroepithelia. Development. 2011;138:5003–5013. doi: 10.1242/dev.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun L, Torii M, Rakic P. Connexin 43 controls the multipolar phase of neuronal migration to the cerebral cortex. Proc. Natl Acad. Sci. USA. 2012;109:8280–8285. doi: 10.1073/pnas.1205880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Tury A, Van Buskirk E, Robinson K, Genestine M, DiCicco-Bloom E. p57(KIP2) regulates radial glia and intermediate precursor cell cycle dynamics and lower layer neurogenesis in developing cerebral cortex. Development. 2012;139:475–487. doi: 10.1242/dev.067314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Valiente M, Ge X, Tsai LH. Guiding neuronal cell migrations. Cold Spring Harb. Perspect. Biol. 2010;2:a001834. doi: 10.1101/cshperspect.a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol. 2007;27:4825–4843. doi: 10.1128/MCB.02100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier PE. Microtubules, interkinetic nuclear migration and neurulation. Experientia. 1978;34:289–296. doi: 10.1007/BF01922992. [DOI] [PubMed] [Google Scholar]
- Messier PE, Auclair C. Effect of cytochalasin B on interkinetic nuclear migration in the chick embryo. Dev. Biol. 1974;36:218–223. doi: 10.1016/0012-1606(74)90206-1. [DOI] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr. Biol. 2011;21:485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Minobe S, Sakakibara A, Ohdachi T, Kanda R, Kimura M, Nakatani S, Tadokoro R, Ochiai W, Nishizawa Y, Mizoguchi A, Kawauchi T, Miyata T. Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neurosci. Res. 2009;63:294–301. doi: 10.1016/j.neures.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi T, Aoki Y, Eksioglu YZ, Takahashi T, Bhide PG, Reeves SA, Caviness VS., Jr Overexpression of p27Kip1 lengthens the G1 phase in a mouse model that targets inducible gene expression to central nervous system progenitor cells. Proc. Natl Acad. Sci. USA. 2001;98:6435–6440. doi: 10.1073/pnas.111051398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi D, Kawaguchi A, Gotoh Y. Mechanisms that regulate the number of neurons during mouse neocortical development. Curr. Opin. Neurobiol. 2010;20:22–28. doi: 10.1016/j.conb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Mochida GH, Walsh CA. Genetic basis of developmental malformations of the cerebral cortex. Arch. Neurol. 2004;61:637–640. doi: 10.1001/archneur.61.5.637. [DOI] [PubMed] [Google Scholar]
- Mori D, Yamada M, Mimori-Kiyosue Y, Shirai Y, Suzuki A, Ohno S, Saya H, Wynshaw-Boris A, Hirotsune S. An essential role of the aPKC-Aurora A-NDEL1 pathway in neurite elongation by modulation of microtubule dynamics. Nat. Cell Biol. 2009;11:1057–1068. doi: 10.1038/ncb1919. [DOI] [PubMed] [Google Scholar]
- Murata J, Ohtsuka T, Tokunaga A, Nishiike S, Inohara H, Okano H, Kageyama R. Notch-Hes1 pathway contributes to the cochlear prosensory formation potentially through the transcriptional down-regulation of p27Kip1. J. Neurosci. Res. 2009;87:3521–3534. doi: 10.1002/jnr.22169. [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol. Cell. Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Nagano T, Morikubo S, Sato M. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J. Neurosci. 2004;24:9648–9657. doi: 10.1523/JNEUROSCI.2363-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K, Sugahara M, Yamasaki T, Kajiho H, Takahashi S, Hirayama J, Minami Y, Ohta Y, Watanabe T, Hata Y, Katada T, Nishina H. Filamin associates with stress signalling kinases MKK7 and MKK4 and regulates JNK activation. Biochem. J. 2010;427:237–245. doi: 10.1042/BJ20091011. [DOI] [PubMed] [Google Scholar]
- Nakanishi N, Renfer E, Technau U, Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development. 2012;139:347–357. doi: 10.1242/dev.071902. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20:1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M, Smith DS, Ayala R, Peng J, Ko J, Lee MS, Morabito M, Tsai LH. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- Nishimura YV, Sekine K, Chihama K, Nakajima K, Hoshino M, Nabeshima Y, Kawauchi T. Dissecting the factors involved in the locomotion mode of neuronal migration in the developing cerebral cortex. J. Biol. Chem. 2010;285:5878–5887. doi: 10.1074/jbc.M109.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nomachi A, Nishita M, Inaba D, Enomoto M, Hamasaki M, Minami Y. Receptor tyrosine kinase Ror2 mediates Wnt5a-induced polarized cell migration by activating c-Jun N-terminal kinase via actin-binding protein filamin A. J. Biol. Chem. 2008;283:27973–27981. doi: 10.1074/jbc.M802325200. [DOI] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odajima J, Wills ZP, Ndassa YM, et al. Cyclin E constrains Cdk5 activity to regulate synaptic plasticity and memory formation. Dev. Cell. 2011;21:655–668. doi: 10.1016/j.devcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna Pant HC, Brady RO, Martin LJ, Kulkarni AB. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl Acad. Sci. USA. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Jr, Atkins CM, Copenagle L, Banker GA. Activated c-Jun N-terminal kinase is required for axon formation. J. Neurosci. 2006;26:9462–9470. doi: 10.1523/JNEUROSCI.2625-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Nakagawa T, Kojima K, Matsumoto M, Kawauchi T, Hoshino M, Ito J. Silencing p27 reverses post-mitotic state of supporting cells in neonatal mouse cochleae. Mol. Cell. Neurosci. 2009;42:391–398. doi: 10.1016/j.mcn.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Patrick GN, Zukerberg L, de la Nikolic M, Monte S, Dikkes P, Tsai LH. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, Catala M, Kahn A, Beldjord C, Chelly J. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- Rakic P. A century of progress in corticoneurogenesis: from silver impregnation to genetic engineering. Cereb. Cortex. 2006;16(Suppl 1):i3–i17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- Rashid T, Banerjee M, Nikolic M. Phosphorylation of Pak1 by the p35/Cdk5 kinase affects neuronal morphology. J. Biol. Chem. 2001;276:49043–49052. doi: 10.1074/jbc.M105599200. [DOI] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Roberson DW, Rubel EW. Cell division in the gerbil cochlea after acoustic trauma. Am. J. Otol. 1994;15:28–34. [PubMed] [Google Scholar]
- Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, Torii M, Flavell RA, Rakic P. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, Miele L, Cardoso AA, Classon M, Carlesso N. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J. Exp. Med. 2005;202:157–168. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Sauer FC. Mitosis in the neural tube. J. Comp. Neurol. 1935;62:377–405. [Google Scholar]
- Sauer ME, Walker BE. Radioautographic study of interkinetic nuclear migration in the neural tube. Proc. Soc. Exp. Biol. Med. 1959;101:557–560. doi: 10.3181/00379727-101-25014. [DOI] [PubMed] [Google Scholar]
- Schaar BT, Kinoshita K, McConnell SK. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 2004;41:203–213. doi: 10.1016/s0896-6273(03)00843-2. [DOI] [PubMed] [Google Scholar]
- Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc. Natl Acad. Sci. USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Honda T, Kawauchi T, Kubo K, Nakajima K. The outermost region of the developing cortical plate is crucial for both the switch of the radial migration mode and the Dab1-dependent “inside-out” lamination in the neocortex. J. Neurosci. 2011;31:9426–9439. doi: 10.1523/JNEUROSCI.0650-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Kawauchi T, Kubo K, Honda T, Herz J, Hattori M, Kinashi T, Nakajima K. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin alpha5beta1. Neuron. 2012;76:353–369. doi: 10.1016/j.neuron.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea TB. Selective stabilization of microtubules within the proximal region of developing axonal neurites. Brain Res. Bull. 1999;48:255–261. doi: 10.1016/s0361-9230(98)00019-7. [DOI] [PubMed] [Google Scholar]
- Sheen VL, Ganesh VS, Topcu M, Sebire G, Bodell A, Hill RS, Grant PE, Shugart YY, Imitola J, Khoury SJ, Guerrini R, Walsh CA. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat. Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shikanai M, Nakajima K, Kawauchi T. N-cadherin regulates radial glial fiber-dependent migration of cortical locomoting neurons. Commun. Integr. Biol. 2011;4:326–330. doi: 10.4161/cib.4.3.14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HW, Morinaga N, Noda M, Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol. Biol. Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukimas GM, Hinds JW. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J. Comp. Neurol. 1978;179:795–830. doi: 10.1002/cne.901790407. [DOI] [PubMed] [Google Scholar]
- Sidman RL, Miale IL, Feder N. Cell proliferation and migration in the primitive ependymal zone: an autoradiographic study of histogenesis in the nervous system. Exp. Neurol. 1959;1:322–333. doi: 10.1016/0014-4886(59)90024-x. [DOI] [PubMed] [Google Scholar]
- Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, Tsai LH. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stensaas LJ. The development of hippocampal and dorsolateral pallial regions of the cerebral hemisphere in fetal rabbits. II. Twenty millimeter stage, neuroblast morphology. J. Comp. Neurol. 1967;129:71–84. doi: 10.1002/cne.901300204. [DOI] [PubMed] [Google Scholar]
- Su SC, Tsai LH. Cyclin-dependent kinases in brain development and disease. Annu. Rev. Cell Dev. Biol. 2011;27:465–491. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Multipolar migration: the third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 2003;23:9996–10001. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J. Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takitoh T, Kumamoto K, Wang CC, Sato M, Toba S, Wynshaw-Boris A, Hirotsune S. Activation of Aurora-A is essential for neuronal migration via modulation of microtubule organization. J. Neurosci. 2012;32:11050–11066. doi: 10.1523/JNEUROSCI.5664-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci. Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Tseng HC, Kulkarni AB, Tsai LH, Gleeson JG. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/s0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- Tarui T, Takahashi T, Nowakowski RS, Hayes NL, Bhide PG, Caviness VS. Overexpression of p27 Kip 1, probability of cell cycle exit, and laminar destination of neocortical neurons. Cereb. Cortex. 2005;15:1343–1355. doi: 10.1093/cercor/bhi017. [DOI] [PubMed] [Google Scholar]
- Thullberg M, Bartkova J, Khan S, Hansen K, Ronnstrand L, Lukas J, Strauss M, Bartek J. Distinct versus redundant properties among members of the INK4 family of cyclin-dependent kinase inhibitors. FEBS Lett. 2000;470:161–166. doi: 10.1016/s0014-5793(00)01307-7. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai LH, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14–3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- Trivedi N, Marsh P, Goold RG, Wood-Kaczmar A, Gordon-Weeks PR. Glycogen synthase kinase-3beta phosphorylation of MAP1B at Ser1260 and Thr1265 is spatially restricted to growing axons. J. Cell Sci. 2005;118:993–1005. doi: 10.1242/jcs.01697. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Lee MS, Cruz J. Cdk5, a therapeutic target for Alzheimer's disease? Biochim. Biophys. Acta. 2004;1697:137–142. doi: 10.1016/j.bbapap.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Tsai LH, Takahashi T, Caviness VS, Jr, Harlow E. Activity and expression pattern of cyclin-dependent kinase 5 in the embryonic mouse nervous system. Development. 1993;119:1029–1040. doi: 10.1242/dev.119.4.1029. [DOI] [PubMed] [Google Scholar]
- Ueno M, Katayama K, Yamauchi H, Nakayama H, Doi K. Cell cycle progression is required for nuclear migration of neural progenitor cells. Brain Res. 2006;1088:57–67. doi: 10.1016/j.brainres.2006.03.042. [DOI] [PubMed] [Google Scholar]
- Ulloa L, Montejo de Garcini E, Gomez-Ramos P, Moran MA, Avila J. Microtubule-associated protein MAP1B showing a fetal phosphorylation pattern is present in sites of neurofibrillary degeneration in brains of Alzheimer's disease patients. Brain Res. Mol. Brain Res. 1994;26:113–122. doi: 10.1016/0169-328x(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Varvel NH, Bhaskar K, Patil AR, Pimplikar SW, Herrup K, Lamb BT. Abeta oligomers induce neuronal cell cycle events in Alzheimer's disease. J. Neurosci. 2008;28:10786–10793. doi: 10.1523/JNEUROSCI.2441-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, Movassagh M, Horan I, Wise H, Ohnuma S, Philpott A. Notch targets the Cdk inhibitor Xic1 to regulate differentiation but not the cell cycle in neurons. EMBO Rep. 2006;7:643–648. doi: 10.1038/sj.embor.7400691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nadarajah B, Robinson AC, McColl BW, Jin JW, Dajas-Bailador F, Boot-Handford RP, Tournier C. Targeted deletion of the mitogen-activated protein kinase kinase 4 gene in the nervous system causes severe brain developmental defects and premature death. Mol. Cell. Biol. 2007;27:7935–7946. doi: 10.1128/MCB.00226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat. Neurosci. 2011;14:555–561. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441:984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Xin X, Ferraro F, Back N, Eipper BA, Mains RE. Cdk5 and Trio modulate endocrine cell exocytosis. J. Cell Sci. 2004;117:4739–4748. doi: 10.1242/jcs.01333. [DOI] [PubMed] [Google Scholar]
- Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, Setou M, Wynshaw-Boris A, Torisawa T, Toyoshima YY, Hirotsune S. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Kawasaki H, Arakawa S, Shimizu K, Shimizu S, Reiner O, Okano H, Nishina S, Azuma N, Penninger JM, Katada T, Nishina H. Stress-activated protein kinase MKK7 regulates axon elongation in the developing cerebral cortex. J. Neurosci. 2011;31:16872–16883. doi: 10.1523/JNEUROSCI.1111-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer's disease. J. Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Herrup K. Cell division in the CNS: protective response or lethal event in post-mitotic neurons? Biochim. Biophys. Acta. 2007;1772:457–466. doi: 10.1016/j.bbadis.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Mufson EJ, Herrup K. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J. Neurosci. 2003;23:2557–2563. doi: 10.1523/JNEUROSCI.23-07-02557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, Wang CL, Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat. Neurosci. 2012;15:1201–1210. doi: 10.1038/nn.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Kawauchi T, Sone M, Nishimura YV, Terao M, Chihama K, Nabeshima Y, Hoshino M. Involvement of a Rac activator, P-Rex1, in neurotrophin-derived signaling and neuronal migration. J. Neurosci. 2005;25:4406–4419. doi: 10.1523/JNEUROSCI.4955-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum. Mol. Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cicero SA, Wang L, Romito-Digiacomo RR, Yang Y, Herrup K. Nuclear localization of Cdk5 is a key determinant in the postmitotic state of neurons. Proc. Natl Acad. Sci. USA. 2008;105:8772–8777. doi: 10.1073/pnas.0711355105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YL, Li BS, Rudrabhatla P, Shukla V, Amin ND, Maric D, Kesavapany S, Kanungo J, Pareek TK, Takahashi S, Grant P, Kulkarni AB, Pant HC. Phosphorylation of p27Kip1 at Thr187 by cyclin-dependent kinase 5 modulates neural stem cell differentiation. Mol. Biol. Cell. 2010;21:3601–3614. doi: 10.1091/mbc.E10-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Cunningham JJ, Sherr CJ, Jogal S, Smeyne RJ, Roussel MF. Postnatal neuronal proliferation in mice lacking Ink4d and Kip1 inhibitors of cyclin-dependent kinases. Proc. Natl Acad. Sci. USA. 1999;96:13462–13467. doi: 10.1073/pnas.96.23.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]


