Abstract
Robustness is considered a ubiquitous property of living systems at all levels of organization, and small noncoding RNA (sncRNA) is a genuine model for its study at the molecular level. In this communication, we question whether microRNA precursors (pre-miRNAs) are actually structurally robust, as previously suggested. We found that natural pre-miRNAs are not more robust than expected under an appropriate null model. On the contrary, we found that eukaryotic pre-miRNAs show a significant enrichment in conformational flexibility at the thermal equilibrium of the molecule, that is, in their plasticity. Our results further support the selection for functional diversification and evolvability in sncRNAs.
Keywords: conformational flexibility, evolvability, noncoding RNA, secondary structure, thermodynamics
Introduction
Robustness is the ability of genotypes to display the same phenotype in presence of genetic or environmental perturbations (de Visser et al. 2003; Kitano 2004; Wagner 2005). Robustness is considered a fundamental feature of biological systems at all levels of organization, from single molecules to large networks. Therefore, the study of robustness, the mechanisms by which it evolved, and its implications to adaptation are central topics in nowadays research in evolutionary biology (Draghi et al. 2010; Wagner 2011). In that way, the relationship between the sequence and folding of small noncoding RNAs (sncRNAs) appears as a genuine and biologically grounded model (Eddy 2001) for tackling the above questions. However, whether sncRNAs show the property of structural robustness or not has turned out to be highly controversial. Furthermore, it is also not clear to what extent conformational flexibility (Tokuriki and Tawfik 2009) is significant for natural sncRNAs to modulate structural robustness and to manage their interactions with partners.
Seminal studies already addressed the robustness of RNA molecules (Wagner and Stadler 1999; Ancel and Fontana 2000) by using the predicted secondary structure as a model to link phenotype (structure) and genotype (sequence). Recent work focusing on microRNA precursors (pre-miRNAs) has reported that pre-miRNAs show a significant enrichment of mutational robustness (Borenstein and Ruppin 2006; Shu et al. 2007; Szöllősi and Derényi 2009). In particular, Szöllősi and Derényi (2009) revisited the initial work of Borenstein and Ruppin (2006) attempting to refine the null model, showing that pre-miRNAs are still robust to both single-point mutations and variations in temperature (used to simulate environmental perturbations), then suggesting a pattern of congruent evolution between mutational and environmental robustness (sensu plastogenetic congruence; Ancel and Fontana 2000). However, latest work proposed that pre-miRNAs secondary structure evolved under purifying selection and that these RNAs have not been selected (directly or congruently) for robustness but for function (Price et al. 2011). In this direction, Rodrigo and Fares (2012) reported that bacterial sncRNAs are not more robust than expected from an unbiased null model, advocating further exploration in the case of pre-miRNAs.
Results and Discussion
Here, we follow a computational approach to calculate the structural robustness landscape for the pre-miRNAs from four different model organisms: Epstein–Barr virus (EBV), Caenorhabditis elegans (CEL), Homo sapiens (HSA), and Arabidopsis thaliana (ATH) (table 1 and supplementary data set S1, Supplementary Material online). We distinguish between two types of robustness. Mutational robustness (Rm) accounts for structural changes after single-point mutations in the pre-miRNA sequence, whereas environmental robustness (Re) accounts for structural changes after alterations in the energetic parameters implemented in the thermodynamic model for base pair interactions (Layton and Bundschuh 2005). We also account for plasticity (Pt), which quantifies the variability of structures within the thermodynamic ensemble (conformational flexibility), because one sequence can fold into many different structures (Wuchty et al. 1999). The null model sequences used to assess the statistical significance of natural pre-miRNAs share the minimal free energy (MFE) structure of these molecules (structural analogs) and were obtained by subjecting inverse folded sequences to a random neutral evolution allowing to change single and paired nucleotides (fig. 1 and supplementary fig. S1, Supplementary Material online). Mann–Whitney U tests were carried out to compare the set of natural pre-miRNAs against the whole set of artificial elements, whereas z tests were applied for each pre-miRNA against its particular structural analogs. To perform the computation over RNA secondary structures, we used the ViennaRNA package (Hofacker et al. 1994).
Table 1.
Summary of Structural Robustness Landscape (See Values in supplementary data set S1, Supplementary Material online)
| Organism | No. Pre-miRNAs Analyzed | % High Rm | % Low Rm | % High Re | % Low Re | % High Pt | % Low Pt |
|---|---|---|---|---|---|---|---|
| EBV | 25 | 4 | 4 | 0 | 8 | 12 | 0 |
| CEL | 100 | 0 | 10 | 0 | 11 | 17 | 0 |
| HSA | 450 | 2 | 12.9 | 1.3 | 17.6 | 24.4 | 0.7 |
| ATH | 110 | 0 | 16.4 | 0.9 | 16.4 | 50.9 | 0 |
Note.—High or low refers to statistical significance assessed with one-tailed z test (P < 0.05), which was applied for each pre-miRNA against its structural analogs. We took from the online database miRBase (Kozomara and Griffiths-Jones 2011) the sequences of all pre-miRNAs analyzed in this work.
Fig. 1.—
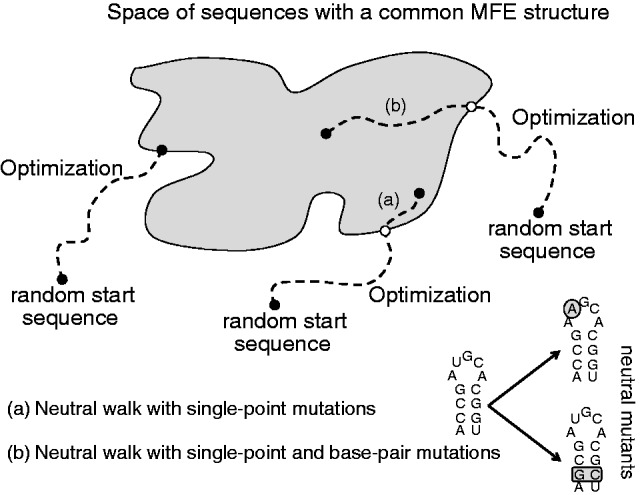
Scheme to illustrate the construction of the null model. From random start sequences, the RNAinverse program (from the ViennaRNA package) can generate sequences with a desired MFE structure by stochastic minimization. However, these sequences present lower than average neutrality. This way, a random walk on the neutral network associated to the MFE structure can be implemented as sort of sequence drift to avoid the optimization bias. This walk can rely, at each step, on just single-point mutations or on both single-point and base pair mutations. The former could deepen not much on the neutral network and then still produce sequences with lower than average neutrality.
We calculated Pt, Rm, and Re for the set of natural pre-miRNAs and the artificial ones. Table 1 presents, for each organism, the percentage of molecules that are significantly more and less robust/plastic (structural robustness landscape) than expected under the null model. The distributions for each variable are shown in figs. 2 and 3. Contrary to previous reports (Borenstein and Ruppin 2006; Shu et al. 2007; Szöllősi and Derényi 2009), we observed that pre-miRNAs are not, on average, significantly more robust to mutations than expected under the null model (P > 0.05 in all cases; fig. 2A), being the fraction of significantly robust molecules lower than 5% in all cases. We neither observed significant enrichment on environmental robustness (P > 0.05; fig. 2B), and the fraction of robust molecules is as low as in the previous case. Therefore, we concluded that natural pre-miRNAs are not more robust than random structural analogs. However, we observed a non-negligible percentage of fragile molecules (with robustness values lower than expected from their structural analogs), being also the pre-miRNAs of HSA and ATH significantly fragile to environmental changes (P < 0.005 in both cases). This discrepancy with previous analyses (see also supplementary fig. S2, Supplementary Material online) is due to the appropriate derivation of the null model, because the structural robustness landscape can indeed vary with this (Szöllősi and Derényi 2009; Rodrigo and Fares 2012). Following our metrics, we can recover similar values of enrichment of robustness as previously reported when using other null models, indicating that the null model of structural analogs results in the Achilles’ heel for determining the structural robustness of sncRNAs. In addition, the pre-miRNAs that exhibit higher/lower levels of Rm also have higher/lower levels of Re (P < 0.005 in all cases except for EBV; supplementary fig. S3, Supplementary Material online). This correlation is still significant when taking into account the phylogenetic relatedness existing among the four organisms (supplementary fig. S3, Supplementary Material online), and it may support an eventual pattern of congruent evolution (Ancel and Fontana 2000; de Visser et al. 2003; Shu et al. 2007; Szöllősi and Derényi 2009; Rodrigo and Fares 2012).
Fig. 2.—
Distributions of mutational and environmental robustness for pre-miRNAs of four different organisms. Statistical significance assessed by Mann–Whitney U tests: (A) P = 0.666 (EBV), P = 0.46 (CEL), P = 0.01* (HSA), and P = 0.05* (ATH); (B) P = 0.50 (EBV), P = 0.26 (CEL), P < 0.0001* (HSA), and P = 0.0031* (ATH). Solid lines represent the null models. *Median of natural pre-miRNAs < null model median, so it indicates marginal statistical significance for fragility and not for robustness.
Fig. 3.—
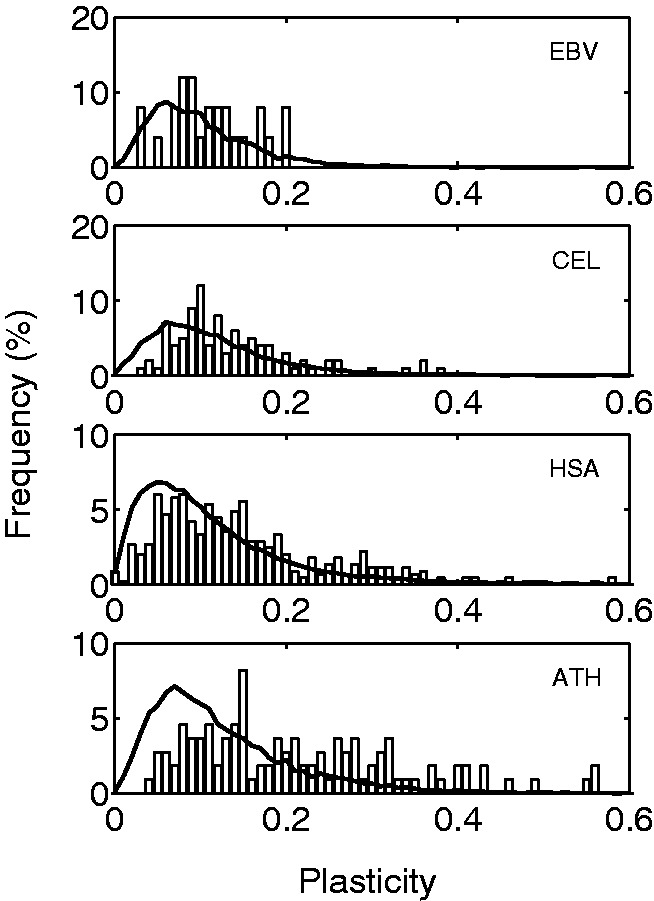
Distributions of plasticity for pre-miRNAs of four different organisms. Statistical significance assessed by Mann–Whitney U tests: P = 0.10 (EBV), P < 0.0001 (CEL), P < 0.0001 (HSA), and P < 0.0001 (ATH). Solid lines represent the null models.
Moreover, we found that the pre-miRNAs of CEL, HSA, and ATH are on average significantly more plastic than expected from the null model (P < 0.0001; fig. 3) and that the fraction of significantly plastic and fragile molecules increases in the same way, reflecting a negative association between plasticity and robustness. An analysis of covariance indicates that this association depends on the organism (P < 0.0001; supplementary fig. S4, Supplementary Material online), and the overall trend is still significant after considering the underlying phylogenetic relationship between species (supplementary fig. S4, Supplementary Material online). Nevertheless, we did not observe a significant enrichment of plasticity in the case of EBV (P > 0.05; fig. 3). The fraction of significantly plastic molecules increases from 12% for EBV, to 17% for CEL, to 24.4% for HSA, which might indicate a trend with organism complexity (measured as the total number of genes). This fraction is even higher, 50.9%, for ATH. Because the GC content among organisms is variable (P < 0.0001; supplementary fig. S5, Supplementary Material online), the fact that ATH has the lowest one might entail minor thermal stability or, in other words, major levels of Pt (Fang et al. 2001), but we found no correlation between the GC content and Pt. We also observed that the average lengths of the pre-miRNAs of EBV, CEL, and HSA are 83, 87, and 84 nucleotides, respectively, whereas the pre-miRNAs of ATH are much longer (173 nucleotides on average), and this difference in length may capture, at least in part, the elevated levels of Pt found for ATH (supplementary fig. S6, Supplementary Material online). In addition, we can compare robustness among organisms to show that, although the pre-miRNAs of ATH are not overall highly robust to mutations with respect to their structural analogs, they appear to be more robust than the pre-miRNAs of HSA (P < 0.0001; fig. 1A). The difference in length can explain, as for bacterial sncRNAs, the higher levels of Rm in that case (Rodrigo and Fares 2012).
Early work (Borenstein and Ruppin 2006) challenged the current population genetics theory (de Visser et al. 2003; Orr 2005) by pointing to directional selection for mutational robustness in populations of small effective size as the mechanism for the evolution of robustness. However, directional selection for robustness, in theory, requires high mutation rates, as it occurs with viruses (Sanjuán et al. 2007) but not with higher eukaryotes. We now report the enrichment of plasticity in populations of eukaryotic pre-miRNAs (table 1) and that robustness (neither mutational nor environmental) did not evolve in these populations, which agrees with the theoretical prediction (de Visser et al. 2003). Consistently, one could suggest that plasticity, which is a trait that could promote evolvability in sncRNAs (Ancel and Fontana 2000) and which is also extensible for proteins (Tokuriki and Tawfik 2009), evolves to counteract the low genetic variability in complex organisms (Lynch and Conery 2003). This could also entail that eukaryotic pre-miRNAs have the potential for producing diverse mature miRNA sequences (Starega-Roslan et al. 2011) after a flexible interaction (Tokuriki and Tawfik 2009) with Dicer proteins.
In this work, we have relied 1) on the use of the secondary structure as a fitness-related magnitude, which certainly is an oversimplification to the problem and 2) on the ability of the ViennaRNA package (Hofacker et al. 1994) to produce reliable structures, which may be a limitation. Future work could aim at determining the robustness to changes, instead of in the pre-miRNA structure, in the maturation rate in the cytoplasm by accounting for the interaction between the pre-miRNA and Dicer proteins (Lee et al. 2002), and even use a more accurate model, although at a high computational cost, with the three-dimensional structure of RNA molecules (Parisien et al. 2009). A biologically more relevant fitness function that incorporates both the binding rate to the target transcript and its degradation rate (the final biological function of the mature miRNA molecule) might be considered as well, because some pre-miRNAs with altered structures could still be processed and active for targeting their transcripts with high affinity. To compare natural RNAs against structural analogs, we could also incorporate into the null model the nucleotide composition of the natural pool (Clote et al. 2005). Even though randomly generated sequences of pre-miRNAs do not account for the evolution in short sequence distance (Nozawa et al. 2010). Price et al. (2011) have already dealt with this situation and have shown, on average, a small, yet marginally significant, decrease in mutational robustness and a likewise small increase in plasticity for Drosophila pre-miRNAs over millions of years of evolution. According to our own results, we would expect an overall increase of Pt from ancestors, which may result in side effects on Rm and Re.
In conclusion, our study provides a quantitative, new recharacterization of the robustness landscape of pre-miRNAs. We have shown that pre-miRNAs are not as robust as previously stated when properly compared with unbiased structural analogs obtained by combining inverse folding and neutral walk. However, pre-miRNAs are significantly enriched in plasticity, supporting the hypothesis that they have been predominantly selected for functional diversification and evolvability. By virtue of a particular evolutionary history, certain pre-miRNAs will be more plastic than others. These results for pre-miRNAs are in agreement with those reported for bacterial sncRNAs (Rodrigo and Fares 2012), where plasticity appears as a fundamental variable. Accordingly, we suggest that plasticity in pre-miRNAs could be a mechanism to promote phenotypic variability, either to enlarge the functional repertoire of a single molecule (e.g., isomiRs; Neilsen et al. 2012) or to promote evolvability (Ancel and Fontana 2000) in organisms that have small effective population sizes. Our results can strengthen the understanding of the evolution of robustness and plasticity in sncRNAs and warrant further experimental exploration in the field.
Materials and Methods
Thermodynamic Model
For a given pre-miRNA sequence (of length L), there is a thermodynamic ensemble (Ω) that contains the optimal (MFE) and several suboptimal structures, each with a given free energy (Gi). Thus, the probability that the pre-miRNA folds into the structure i is given by  , where Z is the partition function and reads
, where Z is the partition function and reads 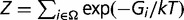 . We took T = 37°C, then kT = 0.616 kcal/mol. For comparing two different sequences, we balanced the two ensembles of structures, instead of just comparing the MFE structures. In addition, to evaluate the difference between two structures, we used the base pair distance (dBP) (Gruber et al. 2008), given by the number of base pairs not shared by them. We also considered the magnitude introduced in that report accounting for the structural variability within
. We took T = 37°C, then kT = 0.616 kcal/mol. For comparing two different sequences, we balanced the two ensembles of structures, instead of just comparing the MFE structures. In addition, to evaluate the difference between two structures, we used the base pair distance (dBP) (Gruber et al. 2008), given by the number of base pairs not shared by them. We also considered the magnitude introduced in that report accounting for the structural variability within  (Si denotes structure i) given by
(Si denotes structure i) given by  (i.e., how heterogeneous is
(i.e., how heterogeneous is  ). To calculate d0, we used ViennaRNA (Hofacker et al. 1994), which implements a dynamic programming algorithm for efficient computation of
). To calculate d0, we used ViennaRNA (Hofacker et al. 1994), which implements a dynamic programming algorithm for efficient computation of  and Z (McCaskill 1990). Using the ViennaRNA function to calculate dBP between two ensembles of structures, we calculated d0, as well as d1 and de (see later).
and Z (McCaskill 1990). Using the ViennaRNA function to calculate dBP between two ensembles of structures, we calculated d0, as well as d1 and de (see later).
Defining Plasticity and Robustness
We here define plasticity (Pt) as conformational flexibility, given by the probabilistically averaged distance between all possible structures in which an RNA molecule can fold. Higher plasticity also indicates higher temperature sensitivity. This way, Pt quantifies the inherent ability to fluctuate at the equilibrium between several phenotypes (in this work, structural conformations), which can turn out into functional promiscuity (Tokuriki and Tawfik 2009). For defining mathematically Pt, we used d0 (Rodrigo and Fares 2012), because higher values of d0 correspond to systems in which there are many different, possible states in  (folds), whereas lower values indicate that
(folds), whereas lower values indicate that  is predominantly governed by the MFE structure. More plastic is a sequence, more structural fluctuations present at the equilibrium, then we defined plasticity as
is predominantly governed by the MFE structure. More plastic is a sequence, more structural fluctuations present at the equilibrium, then we defined plasticity as  . On the other hand, mutational robustness (Rm) accounts for the ability of maintaining the ensemble of structures (not only the MFE structure) after mutations in the sequence. Using an analogous formulation as before, the average distance between structural ensembles after one single-point mutation (d1) reads
. On the other hand, mutational robustness (Rm) accounts for the ability of maintaining the ensemble of structures (not only the MFE structure) after mutations in the sequence. Using an analogous formulation as before, the average distance between structural ensembles after one single-point mutation (d1) reads  (where
(where  1 is the ensemble of mutants and Πj is calculated using the partition function of
1 is the ensemble of mutants and Πj is calculated using the partition function of  1, denoted by Z1). d0 is subtracted to eliminate the intrinsic variability of the ensemble. Thus,
1, denoted by Z1). d0 is subtracted to eliminate the intrinsic variability of the ensemble. Thus,  is the average structural distance after one single-point mutation computed by stochastic sampling (L calculations of d1), and then we defined mathematically mutational robustness as
is the average structural distance after one single-point mutation computed by stochastic sampling (L calculations of d1), and then we defined mathematically mutational robustness as 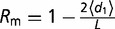 . In addition, environmental robustness (Re) quantifies the ability of maintaining the ensemble of structures, as Rm, but after perturbations that model changes in the environment where the cell that expresses the pre-miRNAs lives. These changes could be physical, chemical, or thermal. We calculated the distance between ensembles after one environmental perturbation (de) simulating a random Gaussian variation (up to 20%) over the value of all the energetic parameters that define the model for base pair interactions (i.e., base pairing and stacking). Hence, being
. In addition, environmental robustness (Re) quantifies the ability of maintaining the ensemble of structures, as Rm, but after perturbations that model changes in the environment where the cell that expresses the pre-miRNAs lives. These changes could be physical, chemical, or thermal. We calculated the distance between ensembles after one environmental perturbation (de) simulating a random Gaussian variation (up to 20%) over the value of all the energetic parameters that define the model for base pair interactions (i.e., base pairing and stacking). Hence, being 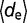 the average structural distance after an environmental perturbation computed by stochastic sampling (1,000 calculations of de), we defined environmental robustness as
the average structural distance after an environmental perturbation computed by stochastic sampling (1,000 calculations of de), we defined environmental robustness as  . All three d0,
. All three d0,  and
and  were rescaled by L/2 to have dimensionless variables and then define Pt, Rm, and Re, respectively.
were rescaled by L/2 to have dimensionless variables and then define Pt, Rm, and Re, respectively.
Generating the Null Model
Structural robustness and plasticity were tested for significance by comparing them with a distribution of these values generated from a large set of artificially constructed sequences. The natural and artificial sequences shared the property of yielding the same MFE structure, although their thermodynamic ensembles were different. This allows comparing robustness and plasticity between sequences that are supposed to be equally fit. For each pre-miRNA, we generated 100 random sequences with the same phenotype (i.e., MFE structure) as a null model. For that, we first solved the corresponding inverse folding problems using different initial sequences with the ViennaRNA package (default energetic parameters, dangles = 2, MFE objective; Hofacker et al. 1994). However, Szöllősi and Derényi (2009) identified lower than average neutrality in sequences obtained by minimization, then proposing a random neutral walk in structure as sort of sequence drift to obtain a null model with more relevant values of neutrality (fig. 1). Subsequently, to minimize the bias introduced by the optimization method, we performed a neutral evolution, introducing L mutations that did not change the MFE structure. If the nucleotide was not paired in the MFE structure, a neutral single-point mutation was applied. On the contrary, if it was paired, a neutral base pair mutation (changing the selected nucleotide and its pair) was applied. This allowed enlarging considerably the sequence space and avoiding efficiently the bias produced by inverse folding methods. A walk with only single-point mutations could deepen not much on the neutral network and then still produce sequences with lower than average neutrality (Rodrigo and Fares 2012). The difference in statistical significance of robustness when using a null model obtained with a neutral walk with base pair mutations or not is shown in supplementary figure S2, Supplementary Material online.
Selecting the Pre-miRNA Sequences
We took from the online database miRBase (Kozomara and Griffiths-Jones 2011) the sequences of all pre-miRNAs for EBV, CEL, HSA, and ATH. Among all sequences available, we randomly selected a subset of them to carry out our analyses. For EBV, we took 25 pre-miRNAs (100%), for CEL 100 (47%), for HSA 450 (32%), and for ATH 110 (47%).
Supplementary Material
Supplementary figures S1–S6 and data set S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank R.B.R. Azevedo for useful comments. This work was supported by an EMBO long-term fellowship co-funded by Marie Curie actions (ALTF-1177-2011) to G.R. and by grant BFU2012-30805 from the Spanish Secretaría de Estado de Investigación, Desarrollo e Innovación, to S.F.E.
Literature Cited
- Ancel LW, Fontana W. Plasticity, evolvability, and modularity in RNA. J Exp Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Borenstein E, Ruppin E. Direct evolution of genetic robustness in microRNA. Proc Natl Acad Sci U S A. 2006;103:6593–6598. doi: 10.1073/pnas.0510600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clote P, Ferré F, Kranakis E, Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JAGM, et al. Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–355. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Fang XW, et al. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc Natl Acad Sci U S A. 2001;98:4355–4360. doi: 10.1073/pnas.071050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Bernhart SH, Hofacker IL, Washietl S. Strategies for measuring evolutionary conservation of RNA secondary structures. BMC Bioinformatics. 2008;9:122. doi: 10.1186/1471-2105-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, et al. Fast folding and comparison of RNA secondary structures. Monatsch Chem. 1994;125:167–188. [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton DM, Bundschuh R. A statistical analysis of RNA folding algorithms through thermodynamic parameter perturbation. Nucleic Acids Res. 2005;33:519–524. doi: 10.1093/nar/gkh983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- Neilsen CT, Goodall GJ, Bracken CP. IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Miura S, Nei M. Origins and evolution of microRNA genes in Drosophila species. Genome Biol Evol. 2010;2:180–189. doi: 10.1093/gbe/evq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Parisien M, Almeida Cruz J, Westhof E, Major F. New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA. 2009;15:1875–1885. doi: 10.1261/rna.1700409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N, Cartwright RA, Sabath N, Graur D, Azevedo RBR. Neutral evolution of robustness in Drosophila microRNA precursors. Mol Biol Evol. 2011;28:2115–2123. doi: 10.1093/molbev/msr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo G, Fares MA. Describing the structural robustness landscape of bacterial small RNAs. BMC Evol Biol. 2012;12:52. doi: 10.1186/1471-2148-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Cuevas JM, Furió V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Bo X, Ni M, Zheng Z, Wang S. In silico genetic robustness analysis of microRNA secondary structures: potential evidence of congruent evolution in microRNA. BMC Evol Biol. 2007;7:223. doi: 10.1186/1471-2148-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega-Roslan J, et al. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szöllősi GJ, Derényi I. Congruent evolution of genetic and environmental robustness in micro-RNA. Mol Biol Evol. 2009;26:867–874. doi: 10.1093/molbev/msp008. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- Wagner A. Robustness and evolvability in living systems. Princeton (NJ): Princeton University Press; 2005. [Google Scholar]
- Wagner A. The role of robustness in phenotypic adaptation and innovation. Proc Biol Sci. 2011;279:1249–1258. doi: 10.1098/rspb.2011.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Stadler PF. Viral RNA and evolved mutational robustness. J Exp Zool. 1999;285:119–127. [PubMed] [Google Scholar]
- Wuchty S, Fontana W, Hofacker IL, Schuster P. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers. 1999;49:145–165. doi: 10.1002/(SICI)1097-0282(199902)49:2<145::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



