Abstract
Quorum sensing (QS) regulates the onset of bacterial social responses in function to cell density having an important impact in virulence. Autoinducer-2 (AI-2) is a signal that has the peculiarity of mediating both intra- and interspecies bacterial QS. We analyzed the diversity of all components of AI-2 QS across 44 complete genomes of Escherichia coli and Shigella strains. We used phylogenetic tools to study its evolution and determined the phenotypes of single-deletion mutants to predict phenotypes of natural strains. Our analysis revealed many likely adaptive polymorphisms both in gene content and in nucleotide sequence. We show that all natural strains possess the signal emitter (the luxS gene), but many lack a functional signal receptor (complete lsr operon) and the ability to regulate extracellular signal concentrations. This result is in striking contrast with the canonical species-specific QS systems where one often finds orphan receptors, without a cognate synthase, but not orphan emitters. Our analysis indicates that selection actively maintains a balanced polymorphism for the presence/absence of a functional lsr operon suggesting diversifying selection on the regulation of signal accumulation and recognition. These results can be explained either by niche-specific adaptation or by selection for a coercive behavior where signal-blind emitters benefit from forcing other individuals in the population to haste in cooperative behaviors.
Keywords: genome evolution, gene loss, E. coli, balancing selection, social cheater, bacteria signaling
Introduction
There is an increasing awareness of the importance of microbial social interactions (Crespi 2001; West et al. 2006, 2007; Foster et al. 2007). Although unicellular organisms, bacteria can express complex coordinated multicellular behaviors, such as biofilm formation, antibiotic production, and secretion of virulence factors. Some of these behaviors require a large quorum of cooperating bacteria to be effective, that is, high cell density. Quorum sensing (QS) is a key communication system that coordinates cooperative behaviors in bacteria in function of cell density (Crespi 2001; Waters and Bassler 2005; Keller and Surette 2006; West et al. 2006).
QS involves the production, secretion, and recognition of small signal molecules called autoinducers detected by cognate receptors. Most autoinducers are species specific and thus promote intra-specific communication (Waters and Bassler 2005). An important exception is the AI-2 system that uses as a signal a family of small molecules called autoinducer-2 (AI-2). The enzyme that produces AI-2 (LuxS) is present in both Gram-positive and Gram-negative bacteria. Because of the wide taxonomic distribution of LuxS, and the demonstration of the susceptibility of this system to interspecies interference, AI-2 has been proposed to be a signal produced to mediate both intra- and interspecies communication (Surette et al. 1999; Chen et al. 2002; Xavier and Bassler 2005a; Pereira, Thompson, et al. 2012). The substrate for AI-2 synthesis by LuxS is S-ribosylhomocysteine (SRH), which derives from the toxic intermediate S-adenosylhomocysteine (SAH) a product from S-adenosylmethionine (SAM) metabolism, an important and ubiquitous central metabolite of the cell (fig. 1) (Schauder et al. 2001; Winzer et al. 2002, 2003; Xavier and Bassler 2003; De Keersmaecker et al. 2006). For this reason, AI-2 can be considered a recycling product of SAM, and it has been suggested that it might not be a true signaling molecule in all AI-2-producing bacteria (Winzer, Hardie, et al. 2002; Winzer et al. 2002; Vendeville et al. 2005; Hardie and Heurlier 2008).
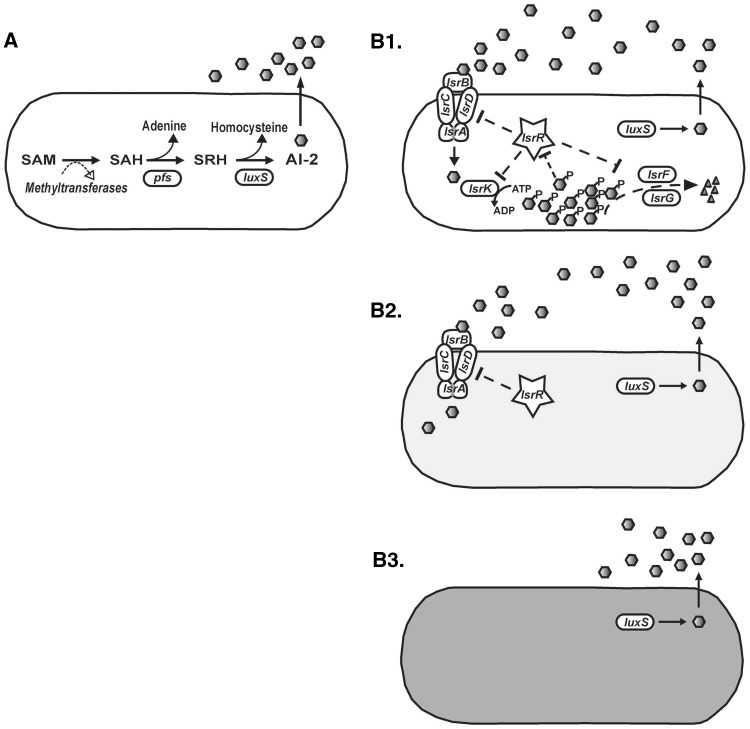
Fig. 1.—
AI-2 biosynthetic pathway and Lsr-mediated transport and processing in Escherichia coli. (A) The precursor of AI-2 biosynthesis is SAM, an essential compound in central metabolism used as a methyl donor for DNA, RNA, and proteins. Following methyl transfer from SAM to its various substrates, the toxic compound SAH is formed. The Pfs enzyme removes adenine from SAH to form SRH. LuxS acts on SRH to produce homocysteine and AI-2 that released into the extracellular environment. (B1) AI-2 is bound by the periplasmic protein LsrB and internalized by the Lsr ATP-binding cassette transporter. Intracellular AI-2 is phosphorylated by LsrK, and the phosphorylated form of the signal (P-AI-2) induces lsr transcription by derepressing the repressor of the lsr operon (LsrR). This results in further assembly of the transporter and rapid AI-2 internalization. LsrF and LsrG proteins are also encoded by the lsr operon and are required for the further processing of intracellular P-AI-2. (B2, B3) Shaded cells represent examples of strains that maintain production of AI-2 although they lack the ability to sequester and process the extracellular AI-2 signal through the Lsr system (B2) or lack the Lsr system completely (B3). Pentagons represent the AI-2 signal.
A major obstacle to understand the role of this molecule as a communication signal has been the lack of information on the molecular mechanisms of AI-2 detection and signal transduction networks in the majority of organisms. Importantly, such mechanisms have now been well characterized in Escherichia coli (reviewed in Pereira, Thompson, et al. 2012). In this bacterium, LuxS produces AI-2 during active growth, which is secreted into the extracellular medium where it accumulates in a cell-density manner until it triggers the activation of the Lsr (for LuxS regulated) system in the receptor cells. The genes of the lsr operon encode an ABC transporter responsible for the internalization of AI-2 into the cells and other enzymes that regulate the expression of the operon and further intracellular metabolic degradation of the AI-2 signal (fig. 1). As a result of the activation of this system, AI-2 levels in the extracellular medium peak in midlate exponential phase and rapidly decline at the transition into stationary phase when the signal is removed from the environment (Wang, Hashimoto, et al. 2005; Wang, Li, et al. 2005; Xavier and Bassler 2005a, 2005b). By mediating the removal of AI-2 from the environment, this process can potentially affect any individual cell in the vicinity with AI-2-dependent gene expression, independently of its species identity (Xavier and Bassler 2005a; Pereira et al. 2008).
A recent study showed that the ability to bind and internalize AI-2 signal via Lsr is not ubiquitous among E. coli strains. Two E. coli strains were shown to lack many genes in the operon, and phenotypic assays confirmed lack of function (Pereira et al. 2009). The finding of this unexpected polymorphism leads us to investigate the genetic diversity of the AI-2 system among E. coli natural populations. Escherichia coli is an important component of the mammalian gut microbiome, especially during lactation, and is extremely diverse. It comprises both commensal and pathogenic variants, with different tropisms, and even some environmentally adapted strains (Kaper et al. 2004; Tenaillon et al. 2010; Luo et al. 2011). The study of genetic variation in this species can thus provide important information on the role of the interspecies signal, AI-2, in an organism that coexists and interacts with many different species in its natural habitat. In E. coli, AI-2 QS regulates many social traits such as virulence (Zhu et al. 2007), biofilm formation (González-Barrios et al. 2006; Herzberg et al. 2006; Reisner et al. 2006; Lee et al. 2011), and chemotaxis and cell motility (Bansal et al. 2008; Hegde et al. 2011). If the fine tuning of AI-2 concentration via the LuxS production and Lsr system for AI-2 internalization is necessary to regulate the behavior of E. coli and of other species in the mammalian gut, the invasion of individuals that are impaired in signal production or internalization could affect the microbiota species composition and diversity. Such alterations of gut homeostasis can facilitate infections (Garrett et al. 2010; Clemente et al. 2012).
In this study, we analyze the genetic diversity of AI-2 production, detection, internalization, and processing at the gene content and nucleotide levels using all complete sequenced genomes of E. coli and Shigella natural strains. We use this information to determine whether selective processes are implicated in the evolution of this system. Many studies have addressed the biochemical mechanisms or the experimental evolution of QS. Oddly, there have been very few studies on the natural genome diversity of QS. Analyses of natural polymorphisms provide an important tool to understand the selective pressures acting on the evolution of social behaviors in microorganisms. The information provided by comparative genomics of natural organisms, which focus on polymorphisms that have passed the filter of natural selection through millions of generations in their natural habitats, are ideal to study the evolutionary relevance of genes and pathways. Here, we took advantage of the large number of genomes available from natural E. coli and Shigella strains to study from a genome-wide perspective the evolution of polymorphism of the different components of the AI-2 system. Our analysis reveals that the AI-2 system follows a unique pattern of genetic diversification that differs significantly from those of species-specific QS systems.
Materials and Methods
Genome Data
We retrieved all complete genomes of E. coli and Shigella spp. present in the Kegg database (http://www.kegg.jp/kegg/, last accessed December 31, 2012) or in Genbank (http://www.ncbi.nlm.nih.gov/genome/, last accessed December 31, 2012). Shigella spp. genomes were included in the analysis because it is well accepted that these organisms belong to the E. coli species (Ochman et al. 1983; Pupo et al. 2000; Escobar-Páramo et al. 2003; Touchon et al. 2009). We excluded three laboratory strains for the following reasons: 1) E. coli str. K-12 substr. W3110 is a very recent laboratory variant of MG1655 (included in the study); 2) E. coli str. K-12 substr. DH10B; and 3) E. coli “BL21-Gold (DE3) pLysS AG” are genetically modified laboratory strains with closely related nonmodified strains in our data set (E. coli K-12 BW2952 [MC4100] and E. coli B REL606, respectively). We added E. fergusonii as it is an well-established outgroup of E. coli and Shigella strains (Lawrence et al. 1991). In total, we included 45 strains (supplementary table S1, Supplementary Material online), five of which are laboratory strains that were included in the phylogenetic reconstruction with the purpose of putting the natural strains and the natural diversification in the context of the better-studied laboratory strains. However, only natural strains were used in the population genetic analyses. All the genomes were downloaded on September 15, 2011. Gene content of the lsr operon was manually checked in each strain to confirm the presence of functional genes, identify misannotations, and characterize pseudogenes (identification of truncation, stop codon, and inserted sequences).
Phylogenetic Analysis and Character Evolution
The list of orthologs between two genomes was identified using reciprocal best hits with more than 80% similarity in protein sequence and less than 20% difference in length, as in Rocha, Touchon, et al. (2006). The core genome of the clade was built using the intersection of lists of orthologs from the pairwise analyses. Each gene family in the core genome was aligned in protein sequence using MUSCLE (Edgar 2004) and then back-translated to DNA. The multiple alignments were concatenated, and phylogenetic reconstruction was performed on this alignment. A maximum likelihood distance matrix was built by Tree-puzzle 5.2 (Schmidt et al. 2002) under the Hasegawa–Kishino–Yano + Γ model. The tree was inferred from the distance matrix using BIONJ (Gascuel 1997). Support of the topology was estimated by bootstrapping on the core genome concatenated alignment (100×).
To analyze character evolution, we coded each strain in terms of presence/absence of complete operon, pathovar, and the ability to replicate within macrophages. The latter two variables were coded using information from the literature. We traced the history of character change through the phylogeny with the program Mesquite version 2.75, build 566 (Maddison and Maddison 2011). Ancestral state reconstruction was made under maximum likelihood using an asymmetric Markov k-state two-parameter model, with rates estimated from the data. Correlations were carried out with Pagel’s 1994 test of independence between two binary characters (Pagel 1994) This test estimates the log-likelihood difference between a model where the rates of change in the two characters are independent and a model where the rates of change are correlated. The significance value of the log likelihood difference after 1,000 simulations is presented for each correlation.
Character Simulations
We simulated character evolution along the phylogenetic tree to test whether the polymorphism observed for the presence/absence of a complete lsr operon evolves under neutrality. This corresponds to the null model. We evolved 1,000 categorical (binary) characters in Mesquite (Maddison and Maddison 2011) using the aforementioned two-parameter Markov-k model with asymmetric rates of forward and backward changes. Forward changes depict the instantaneous probability of gene inactivating mutations leading to incomplete, thus nonfunctional operon (from state 0 to state 1), and backward changes describe the instantaneous rate probability of regaining a functional operon, by back mutation, by gene conversion, or lateral gene transfer (from state 1 to state 0). To initiate the simulation process, one has to set the character frequencies at the root of the tree. We assumed those frequencies to be at equilibrium (rather than the alternative of equal frequencies). With this option, the expected frequencies at the root are assumed to be consistent with the model's rates, which is a more suitable option when the simulating model contains asymmetrical rates (Schluter et al. 1997). In this analysis, we calibrated the branch lengths of the phylogeny into the same units of the model parameters by considering the coalescent expectation that the time to the most recent common ancestor is in the order of 2Ne generations (Hudson 1990). Considering θ = 2Neµ and for E. coli θ = 0.0187 estimated with the core genome (this study) and µ = 8.9 × 10−11 (Wielgoss et al. 2011) can time to the most recent common ancestor to be approximately 2.11 × 108 generations, which corresponds to 0.0124 total branch lengths as estimated directly from the tree.
The simulations were carried out with rate parameters compatible with neutrality and with rate parameters estimated from the actual data. For each model, we estimated the level of polymorphism generated as the relative frequency of state 0 (functional operon), and the two distributions were tested for significant differences with a two-sample Kolmogorov–Smirnov test in R (http://www.R-project.org/, last accessed December 31, 2012).
Genetic Diversity and Levels of Selection
Standard analyses of genetic diversity and neutrality tests (Tajima 1989; McDonald and Kreitman 1991; Stoletzki and Eyre-Walker 2011) were carried out for each gene with DnaSP 5.10 (Librado and Rozas 2009) and Mega4 (Tamura et al. 2007). McDonald–Kreitman test detects selection on protein coding sequences by comparing divergence and polymorphism data on synonymous and nonsynonymous sites under the assumption that synonymous substitutions are neutral; the test is robust to complex demography (Nielsen 2005) such as those likely to occur in bacteria. Tajima’s D compares two measures of population genetic diversity that can be used to infer events of selection; but unlike the previous test, Tajima’s D is highly sensitive to demographic effects (Simonsen et al. 1995). The number of synonymous (nonsynonymous) substitutions per synonymous (nonsynonymous) sites was estimated in MEGA4 (Tamura et al. 2007) using the modified Nei–Gojobori method that assumes a transition/transversion rate bias and uses a Jukes–Cantor correction to account for multiple substitutions at the same site. Standard errors were estimated after 1,000 bootstrap replicates. Patristic distances among strains were estimated in Mesquite (Maddison and Maddison 2011) by calculating the path-length distance from one strain to another along branches of core genome tree of figure 2.
Fig. 2.—
The phylogenetic analysis of Escherichia coli and Shigella strains inferred with the core genome (1,524 genes, 1.6 Mb). The tree was inferred by neighbor-joining based on the maximum likelihood distance matrix (see Materials and Methods). Escherichia fergusonii was used as the outgroup. Arrows indicate presence/absence of luxS and genes in the lsr operon. Dashed lines represent pseudogenes. On the right, pathovar and phylogroups are indicated. Pathovars are enteroaggregative E. coli (EAEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteropathogenic E coli (EPEC), adherent invasive E. coli (AIEC), extraintestinal pathogenic E. coli (EXPEC), uropathogenic E coli (UPEC), and avian pathogenic E coli (APEC).
We analyzed codon-specific selection in the two genes with significant McDonald–Kreitman tests. We estimated the rates of nonsynonymous and synonymous changes at each site using likelihood-based approaches as implemented in HYPHY package and made available through the Datamonkey web service (Kosakovsky Pond and Frost 2005). We made separate analyses with the fixed effects likelihood methods (FEL and iFEL) and with the random effect likelihood method (REL) (Pond and Frost 2005). iFEL is the “population-level” version of FEL and applies when one is interested in selective pressures that are restricted to interior branches of the tree (Kosakovsky Pond et al. 2006). REL tends to be more powerful than FEL but has a higher rate of false positives (Pond and Frost 2005). For this reason, we run all methods and compared the results. A Bayes factor of 20 or more in favor of dN > dS is usually considered as providing strong support for adaptive selection at the site (Pond and Frost 2005). For all these analyses, we estimated for each data set the best-fitting nucleotide model and the gene phylogeny using the available options on Datamonkey.
We characterized codon usage of all genes using several statistics and averaging results across genomes: the effective number of codons (Wright 1990), the Codon Bias Index (Morton 1993), and the Codon Adaptation Index (CAI; Sharp and Li 1987). We used as a reference set the codon usage of a set of highly expressed genes in E. coli (Sharp and Li 1986). Expected value of CAI (eCAI) provides a direct threshold value for discerning whether the differences in CAI are statistically significant or whether they are merely artifacts that arise from internal biases in the G+C composition and/or amino acid composition of the query sequences (Puigbò et al. 2008). E-CAI is calculated from a set of query sequences by generating random sequences with G+C and amino acid content similar to those of the input. ENC and CBI were estimated with DNAsp, and eCAI was estimated the E-CAI server available at http://genomes.urv.es/CAIcal/E-CAI (last accessed December 31, 2012).
Phenotypic Characterization of Null Mutants
Wild-type E. coli K-12 strain MG1655 (Blattner et al. 1997) was used as the parental strain (supplementary table S2, Supplementary Material online). To construct chromosomal single-gene deletions, kanamycin resistance cassettes from the Keio collection of single-deletion mutants (obtained from National BioResource Project [Japan] [Baba et al. 2006]) were introduced in the desired gene by phage P1 transduction from the Keio collection to MG1655, as described elsewhere (Silhavy et al. 1984). All deletions were confirmed by polymerase chain reactions using Taq DNA polymerase (New England Biolabs).
The phenotypes of all single-deletion mutants were determined in terms of AI-2 internalization by quantifying the time course of extracellular AI-2 concentration in cell-free supernatants. Bacteria were grown at 37°C with aeration in Luria–Bertani (LB) broth. To measure AI-2 concentrations, we followed the protocol described in Taga and Xavier (2011). Briefly, we diluted overnight cultures (1:100) into fresh LB medium and periodically collected aliquots of which we measured both optical density (OD600) and AI-2 concentration using the in vitro assay based on the CLPY-FRET method (Taga and Xavier 2011).
Results
Diversity of the Gene Repertoire of AI-2 QS
The phylogenetic relationship among E. coli and Shigella strains was inferred with the core genome of all strains including the outgroup (fig. 2). Every node in this phylogeny has 99–100% bootstrap support with the exception of two small internal nodes, one within the phylogroup B1 with 92% and another within B2 with 56%. This phylogeny shows the same phylogroups as previous studies (Touchon et al. 2009). The core genome of the clade (E. coli + E. fergusonii) is only of 1,524 genes corroborating previous conclusions that no single strain can be regarded as highly representative of the species and justifying the need for including gene repertoire variation in population genetic studies of E. coli.
Figure 2 shows a remarkable pattern of presence/absence polymorphism of the AI-2 regulated lsr genes among all sampled 44 strains of E. coli and Shigella. Many strains lack a complete lsr operon, but all natural strains encode the AI-2 synthase (luxS) (these results do not change with the addition of 13 new complete genomes that became available during manuscript revision). The frequency of the complete operon is 38%. This intermediate frequency is very rare in E. coli, where most gene families are at frequencies higher than 90% or lower than 20% (Touchon et al. 2009). The close similarity in topology between the phylogenetic tree built with the sequences of the genes in the lsr operon and the species tree reconstructed with the core genome (supplementary fig. S1, Supplementary Material online) indicates that although recombination and lateral transfer cannot be ruled out, the prevalent phylogenetic signal in this locus is one of the species. Hence, the simplest explanation for the observed polymorphic pattern is that the operon is ancestral to all E. coli, and we can therefore use the presence/absence of genes to infer ancestral states. A maximum-likelihood reconstruction of ancestral states for the presence/absence of a completely functional lsr operon suggests that the operon was present in the last common ancestor of all E. coli (including Shigella) (proportional likelihood = 0.60, this is the scaled likelihood, so that the likelihood of both states add to 1). This is further supported by the taxonomic distribution of the lsr operon, which is found in many Gammaproteobacteria, including most close relatives of E. coli such as E. fergusonii and the new E. coli clades that are outside the "typical" E. coli strains (Luo et al. 2011; Oh et al. 2012). This operon is also found in the close genus of Salmonella, Yersinia, Enterobacter, and Klebsiella. Hence, lsr genes lacking in extant E. coli genomes are most likely the result of gene losses.
We observed no significant correlation between the presence/absence of a complete lsr operon and pathogenicity in general (Pagel’s 1994 test, P = 0.449). However, the pathovars that are known to replicate within macrophages—all Shigella and all adherent invasive E. coli (AIEC) strains (Glasser et al. 2001)—lack the full lsr operon (Pagel’s 1994 test, P = 0.048). The majority of EXPEC strains investigated here (seven of eight) also lack the full operon (Pagel’s 1994 test, P = 0.080). EXPEC strains are extraintestinal pathogenic E. coli that are part of a healthy intestinal microbiota but can become virulent in extraintestinal environments (Wold et al. 1992; Nowrouzian et al. 2005; Moreno et al. 2009). These results suggest association between patterns of polymorphism at the lsr operon and relevant phenotypic traits differentiating E. coli strains.
Processes of Pseudogenization
To gain further insight on the process leading to loss of the lsr operon, we studied its patterns of pseudogenization. The maximum-likelihood reconstruction of ancestral states suggests the occurrence of at least eight independent losses of function in the lsr operon during the evolution of the E. coli clade (supplementary fig. S2, Supplementary Material online). Once initiated, the process of pseudogenization is fast and does not follow a fixed gene order. For instance, closely related strains such as the ones in phylogroup B1 can differ substantially in presence/absence of genes within the operon as demonstrated by the two enterohemorragic strains (E. coli O26:H11 11368 and E coli O111:H1 11128, fig. 2). Given the rapidity and complexity of gene degradation upon operon inactivation, it is not always possible to identify the event triggering the pseudogenization process. For instance, a large truncation does not preclude the pre-existence of a frameshift or of a nonsense mutation. Nevertheless, interesting patterns emerge from the comparison of different genomes. Namely, the presence of insertion sequences (ISs, 10 occurrences) and phage-related sequences (two occurrences) in the flanking regions of the lsr operon is only observed in genomes with an inactivated operon that are still in the process of pseudogenization, suggesting that these elements may be involved in rapid changes of gene content (supplementary table S1, Supplementary Material online). Among the 34 observed pseudogenes, 26 (76.5%) correspond to large truncations (>100 bp deletions), 6 (17.6%) to small indels (usually 1 or 2 bp insertions/deletions that cause disruption of the reading frame but also 7-bp deletion), and finally 2 (5.9%) were caused by single point mutations that lead to premature termination codon (supplementary table S1, Supplementary Material online).
We estimated the probability that the polymorphic pattern of presence/absence of the complete operon could have originated by neutral processes alone. To test this hypothesis, we generated 1,000 binary characters under a neutral process on the clade phylogeny and then determined the probability of obtaining the level of polymorphism observed in the data. We considered a model of evolution that assumes asymmetrical transition rates where the forward rate corresponds to the rate of gene inactivation and the backward rate corresponds to the rate of regaining a functional gene that was previously inactivated (see Materials and Methods). Among the many mechanisms that lead to gene inactivation, we considered point mutations, small insertions and deletions, large truncations, and IS transposition. To parameterize our model, we used published data. Single point mutations represented approximately 62.7% of the total mutational events causing gene inactivation in a recent large-scale experimental evolution experiment (Tenaillon et al. 2012). The mutation rate of E. coli was recently estimated to be 8.9 × 10−11 per nucleotide per generation (Wielgoss et al. 2011). Together, these values lead to a rate of gene inactivation (forward rate) of approximately 1.17 × 10−6 mutational events per generation for the lsr operon, considering that the total coding sequence is 8,271 bp.
The backward rate represents the rate of back mutation that overcomes single point mutations, the rate of insertion/deletion that reconstitutes a previous indel, the rate of excision of an IS, and the rate of gene conversion and lateral gene flow. The rate of back mutation will be negligible in relation to the rate of gene inactivation due to stop codons. The rate of excision of transposons is likely to be at least one order of magnitude smaller than the insertion rate (Charlesworth 1985). Lateral gene transfer probably occurs at a highly variable rate and is potentially the most important process that can restore a functional operon. In the absence of estimations of these rates in natural setups, we tested multiple backward rates ranging four orders of magnitude (table 1). With all branches calibrated in units of generations (see Materials and Methods), we tested whether a neutral model could explain the data by contrasting the polymorphism level generated under neutrality and a similar model with rate parameters estimated from the data (forward rate = 6.89 × 10−9, and backward rate = 4.00 × 10−9). From all parameters tested, model 2x is the only model that generates a distribution of polymorphism with a median and a variance that includes the empirical level of polymorphism (0.38); however, this result implies a point mutation rate that is three orders of magnitude smaller than what was estimated empirically (Wielgoss et al. 2011). This is consistent with the interpretation that there is ongoing selection to maintain a functional operon in some strains of E. coli. Importantly, the level of polymorphism obtained by simulating 1,000 characters under a model with parameters estimated from the data is significantly different from the distribution generated with model 2x (forward = 1.17 × 10−6, backward = 5.80 × 10−7), mainly due to the large variance of the former (Kolmogorov–Smirnov text, D = 0.2352, P value < 0.001, table 1).
Table 1.
Distributions of Presence/Absence Polymorphism Generated under Diverse Evolutionary Models of Character Evolution
| Forward Rate | Backward Rate | Ratio | Median | Variance | Kolmogorov–Smirnov Test | |
|---|---|---|---|---|---|---|
| Empirical data | 6.89 × 10−9 | 4.00 × 10−9 | 1.72 | 0.356 | 0.0223 | — |
| Model 0.01x | 1.17 × 10−6 | 1.17 × 10−4 | 0.01 | 1.000 | 0.0002 | D = 1, P < 0.001 |
| Model 0.1x | 1.17 × 10−6 | 1.17 × 10−5 | 0.1 | 0.911 | 0.0019 | D = 0.994, P < 0.001 |
| Model 1x | 1.17 × 10−6 | 1.17 × 10−6 | 1 | 0.511 | 0.0057 | D = 0.5245, P < 0.001 |
| Model 2x | 1.17 × 10−6 | 5.80 × 10−7 | 2 | 0.333 | 0.0054 | D = 0.2352, P < 0.001 |
| Model 5x | 1.17 × 10−6 | 2.34 × 10−7 | 5 | 0.178 | 0.0033 | D = 0.6617, P < 0.001 |
| Model 10x | 1.17 × 10−6 | 1.17 × 10−7 | 10 | 0.089 | 0.0021 | D = 0.8498, P < 0.001 |
| Model 100x | 1.17 × 10−6 | 1.17 × 10−8 | 100 | 0.000 | 0.0003 | D = 0.987, P < 0.001 |
Note.—Forward and backward rates are model parameters (see text), and the generated distributions are characterized by their median and variance of the relative frequency of strains with complete operon. These distributions were tested against the distribution generated with empirical parameters with a two-sample Kolmogorov–Smirnov test. The only model that generates a distribution of polymorphism that includes the empirical value (0.38) is in bold.
Phenotypic Characterization of Gene Losses
We made all single-gene deletions of the lsr operon as well as for two genes in the biosynthetic pathway of AI-2. We then determined the phenotypic effects for all deletions in terms of AI-2 accumulation and internalization in E. coli K-12 MG1655. AI-2 internalization profiles clearly show that every gene knockout leads to a measurable phenotypic effect, albeit some cause stronger phenotypic effects than others (fig. 3). In particular, mutants in the genes from the ABC transporter (lsrB, lsrA, lsrC, and lsrD) are less efficient in removing AI-2 from the extracellular medium, whereas the kinase mutant (lsrK) does not internalize the signal. The lsrG mutant, which is less efficient in degrading the inducer of the system (AI-2-P), and the repressor mutant (lsrR) are the only mutants that ensue a premature AI-2 internalization. These results are consistent with the previous studies on the characterization of the lsr operon and its regulation (Ren et al. 2004; Xavier and Bassler 2005a, 2005b; Li et al. 2007; Pereira et al. 2009), but here we show the phenotype of extracellular AI-2 accumulation for all the lsr single mutants. Our results show that extracellular AI-2 concentration is affected by every single-gene deletion. This can impact on how the different cells in the population sense the signal and therefore will impact on the selective pressure acting on each gene deletion.
Fig. 3.—
Extracellular AI-2 profiles of single-mutant knockouts cultured in LB at 37°C on the left, and respective absolute growth curves, on the right. On the top are results for single-gene knockout mutants of the lsr operon and below are results from knockout mutants of the genes involved in the AI-2 production pathway (pfs and luxS). WT, wild type.
Extrapolating the phenotypic effects of single knockout mutants obtained by genetic manipulation to the genotypes observed in the natural isolates suggests that gene deletions in natural strains decrease or abolish AI-2 internalization. The former corresponds to the two enterotoxigenic E. coli strains (E. coli O139:H28 E24377A and E. coli UMNK88). These strains have an impaired Lsr transporter, but due to the presence of functional kinase and repressor, it is expected that these strains are still capable of internalizing the signal, albeit at a lower rate via a less efficient system (Pereira et al. 2012). On the other hand, all other natural strains without a complete lsr operon lack a functional lsrK, which is sufficient to prevent any decrease of the extracellular AI-2 concentration. LsrK is responsible for producing the inducer of the system (AI-2-P) and in its absence, even if the receptor gene (lsrB) is present, the protein is not expressed, hence lsrK mutants are impaired not only in AI-2 internalization but also in AI-2 sensing. The loss of lsrR leads to a constitutive expression of the lsr operon and thus to a very low accumulation of AI-2 in the extracellular medium because all AI-2 that is produced is also internalized. Interestingly, the lsrR gene is only absent in the genomes that also lack a functional ABC transporter (LsrACDB) and the corresponding signal kinase (LsrK). Therefore, also in these genomes, we would predict an overall phenotypic effect that is similar to a complete absence of the lsr operon and to no internalization of AI-2 by those cells. Hence, we do not expect a more efficient removal of the AI-2 signal from the extracellular medium for any of the natural isolates analyzed.
Interestingly, in monocultures, and under the experimental conditions tested, the luxS and all lsr mutants show small, if any, differences in growth rates (fig. 3). This suggests a weak metabolic fitness cost for the QS and contrasts with a deletion in the pfs gene, the enzyme upstream of luxS responsible for the degradation of SAH (fig. 1) that carries major fitness costs to the cell (fig. 3). This is important given that both pfs and luxS mutants have the same key effect of not producing AI-2 (flat curves of fig. 3C).
Genetic Diversity and Levels of Selection
Pairwise nucleotide diversity (π) varies across the genes in the operon (fig. 4 and table 2). It is particularly high in the lsrA gene encoding the ABC-ATPase enzyme that provides the energy necessary for the internalization of AI-2 into the cell, and in its two flanking genes lsrR and lsrC, but in every case, the levels of diversity are within the range of those observed in the core genome, even when controlling for similar codon usage bias (fig. 4). The level of diversity for nonsynonymous substitutions is much lower than for synonymous in all genes, consistent with some degree of evolutionary constrain (table 2). This is corroborated by the pattern of intermediate levels of codon usage bias recovered from the several measures used (table 2).
Fig. 4.—
Genetic diversity at the lsr operon. Black squares indicate total nucleotide diversity (mean and standard deviation) for each gene in the operon. Triangles are nucleotide diversity at nonsynonymous sites estimated in complete genes for strains with the full lsr operon (black) and for strains with partial lsr operons (white). For comparison, the total nucleotide diversity and nonsynonymous diversity of core genes with similar codon usage was added.
Table 2.
Genetic Diversity and Codon Usage
| Gene | L | S | N | h | πtotal |
πS | πNS | No. of Codons | ENC | CBI | CAI | eCAI | CAI/eCAI | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | ||||||||||||||
| Strains with complete lsr operon | lsrK | 1,593 | 85 | 15 | 10 | 0.014 | 0.0023 | 0.047 | 0.003 | 530 | 51.728 | 0.325 | 0.315 | 0.337 | 0.935 |
| lsrR | 954 | 58 | 15 | 9 | 0.018 | 0.0017 | 0.067 | 0.002 | 317 | 53.143 | 0.375 | 0.302 | 0.325 | 0.929 | |
| lsrA | 1,533 | 116 | 15 | 12 | 0.022 | 0.0026 | 0.059 | 0.009 | 511 | 46.361 | 0.433 | 0.322 | 0.328 | 0.982 | |
| lsrC | 1,029 | 74 | 15 | 11 | 0.019 | 0.0025 | 0.058 | 0.005 | 342 | 48.842 | 0.440 | 0.333 | 0.310 | 1.075 | |
| lsrD | 993 | 35 | 15 | 11 | 0.011 | 0.0013 | 0.035 | 0.002 | 330 | 45.614 | 0.433 | 0.295 | 0.302 | 0.976 | |
| lsrB | 1,023 | 40 | 15 | 12 | 0.011 | 0.0014 | 0.040 | 0.002 | 340 | 49.022 | 0.331 | 0.363 | 0.349 | 1.040 | |
| lsrF | 876 | 47 | 15 | 12 | 0.014 | 0.0017 | 0.047 | 0.004 | 291 | 44.400 | 0.518 | 0.344 | 0.343 | 1.003 | |
| lsrG | 291 | 15 | 15 | 8 | 0.011 | 0.0026 | 0.043 | 0.002 | 96 | 48.112 | 0.504 | 0.489 | 0.398 | 1.228 | |
| All strains | luxS | 516 | 21 | 40 | 20 | 0.010 | 0.0005 | 0.029 | 0.003 | 171 | 45.907 | 0.517 | 0.497 | 0.366 | 1.359 |
Note.—L is sequence length, s is the number of segregating sites, N is sample size, and h is the number of unique haplotypes, and π is nucleotide diversity (mean and standard deviation). Nucleotide diversity is presented for total sequence, synonymous sites (S), and nonsynonymous sites (NS) only. ENC is the effective number of codons, CBI is the Codon Bias Index, CAI is the Codon Adaptation Index, and eCAI is the effective number of codons. Genes in the lsr operon are ordered following position on the chromosome.
Nonsynonymous substitution patterns are not significantly different between complete genes in functional operons and complete genes in partly inactivated operons as would be expected for genes under relaxed purifying selection (fig. 4). This suggests that either the cell is still using these genes or that the process of pseudogenization is too recent to have left a signature in the patterns of substitution. The gene lsrG, which encodes one of the two enzymes responsible for processing of AI-2-P, is an exception as it shows high nucleotide diversity caused by an increased number of nonsynonymous substitutions (fig. 4, white triangles). This gene seems to be always the last to pseudogenize and hence would have more time to accumulate changes.
To test for signatures of selection on shaping levels and patterns of diversity, we performed McDonald–Kreitman and Tajima’s D tests (table 3). Tajima’s D statistic was not significantly different from zero in any of the genes analyzed. Tajima’s D is highly sensitive to population demography (Simonsen et al. 1995), and it tends to assume negative values in expanding populations. Because it was proposed that E. coli population enjoyed a large recent population expansion (Wirth et al. 2006), expected value of D is negative (and not zero) based on demographic effects. We then decided to use McDonald–Kreitman tests to analyze the patterns of selection in this work because this test is much more robust to demographic effects (Nielsen 2005). Both LuxS (AI-2 synthase) and LsrA have a pattern of diversity that rejects the null model of neutrality (McDonald–Kreitman test, table 3) but for very different reasons. The gene luxS shows levels of nonsynonymous polymorphism close to the average gene but much smaller nonsynonymous divergence (after normalization by synonymous substitutions, fig. 5). This is suggestive of strong purifying selection on nonsynonymous polymorphisms. On the other hand, nonsynonymous polymorphisms are much more abundant in lsrA than in all the others genes after normalization by synonymous rates (PS and DS). The pattern for lsrA suggests the maintenance of high nonsynonymous polymorphism segregating in the population.
Table 3.
Neutrality Tests
| Tajima’s D | McDonald and Kreitman test |
DOS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DN | DS | PN | PS | P | α | NI | ||||
| Strains with complete lsr operon | lsrK | −0.7462 | 9 | 80 | 14 | 71 | 0.265 | −0.753 | 1.753 | −0.064 |
| lsrR | −0.2277 | 3 | 23 | 6 | 53 | 1.000 | 0.132 | 0.868 | 0.014 | |
| lsrA | −0.5846 | 11 | 84 | 34 | 87 | 0.004** | −1.984 | 2.984 | −0.165 | |
| lsrC | −0.6240 | 4 | 36 | 15 | 60 | 0.198 | −1.250 | 2.250 | −0.100 | |
| lsrD | 0.0367 | 5 | 41 | 4 | 31 | 1.000 | −0.058 | 1.058 | −0.006 | |
| lsrB | −0.4743 | 6 | 28 | 9 | 32 | 0.775 | −0.313 | 1.313 | −0.043 | |
| lsrF | −0.7434 | 1 | 8 | 12 | 36 | 0.668 | −1.667 | 2.667 | −0.139 | |
| lsrG | −1.3815 | 0 | 2 | 2 | 14 | 1.000 | — | — | −0.125 | |
| All strains | luxS | −0.1734 | 0 | 14 | 7 | 15 | 0.029* | — | — | −0.318 |
Note.—DS and DN are fixed synonymous and nonsynonymous substitutions, Ps and PN are segregating synonymous and nonsynonymous substitutions. P value (two tailed) is the significant level of the Fisher exact test that DN/DS = PN/PS. α = 1 − NI, and NI is the neutrality index, α is the proportion of evolutionary change that is due to positive selection, these values cannot be computed when DN is zero. DOS is direction of selection. Divergence was estimated between E. coli strains against the closest outgroup (E. fergusonii).
Fig. 5.—
Nonsynonymous to synonymous rates across genes in the lsr operon and the luxS. Intraspecific comparisons correspond to nucleotide diversity for synonymous and nonsynonymous sites (black squares), whereas comparisons with the outgroup (Escherichia fergusonii) correspond to synonymous and nonsynonymous nucleotide divergence (triangles). Black triangles are average nucleotide differences between species, whereas white triangles correspond to net divergence, which subtracts to the previous statistic the average nucleotide differences for polymorphic sites. Error bars are standard errors estimated after 1,000 bootstrap replicates.
The patterns of high dN/dS between closely related strains for lsrA might arise simply from the presence of newly created slightly deleterious mutations that have not yet had time to be purged (Rocha et al. 2006). However, the high effective population size of E. coli should lead to a rapid elimination of these mutations. Indeed, we found that dN/dS falls rapidly to low values in the core genome (supplementary fig. S3, Supplementary Material online). When we compare the average trend of dN/dS over patristic distances with that of lsrA, we find much higher values for the latter, suggesting that slow purifying selection of slightly deleterious changes is not driving the differences between the two sets. This trend could result from a lower number of deleterious nonsynonymous changes in this gene relative to the core. However, when we compared E. coli with E. fergusonii, there was no difference in dN/dS between lsrA and the core genome. The observation of an excess of dN/dS in lsrA relative to core genome within the species and identical dN/dS in the two sets when comparisons are made between species fits very well the hypothesis of diversifying selection for lsrA.
Figure 6 shows for lsrA and for luxS a sliding window analysis of the ratio of nonsynonymous to synonymous substitutions segregating within E. coli and between E. coli and E. fergusonii. The asterisks indicate the approximate gene location of codons where a significant signal of selection was detected with the codon-based approach. Three significant codons distributed throughout the gene (positions 76, 181, and 482) were detected for lsrA, whereas only one codon (position 145) was identified for luxS.
Fig. 6.—
Sliding window analysis of the ratio of nonsynonymous (N) to synonymous (S) substitutions segregating within Escherichia coli (P) and between E. coli and E. fergusonii (D) across lsrA and luxS genes. Window size is 30 bp with 15-bp steps. Sample size is 15 for lsrA and 40 for luxS corresponding to strains with complete operons and all natural strains, respectively. Asterisks indicate approximate position of codons identified as targets of selection (codons 76, 181, and 482 in LsrA, and codon 145 in LuxS).
Discussion
We have found that E. coli exhibits a gene repertoire polymorphism in the lsr operon. We have experimentally shown that such polymorphism leads to cells lacking the ability to bind, internalize, and/or process the QS signal. However, all strains maintain a functional LuxS, the synthase of the QS signal, even though the fitness cost of this deletion in monocultures is as low as that of many lsr genes. Overall, the evolution of the genes essential for regulating AI-2 concentration was shown here to be complex and non-neutral. Fifty-eight percent of E. coli strains cannot regulate AI-2 extracellular concentrations, 23 of the 40 strains analyzed lack a functional LsrK, these strains produce AI-2 but do not have the ability to sense or remove AI-2 from itself or others. We did not find any natural strain that lacks the LsrR repressor and still have a potentially functional operon. This suggests counter selection of strains that could be more efficient at removing AI-2. Hence, the overall phenotypic effect of the observed operon pseudogenization is always toward the decrease or total abolishment of AI-2 internalization and removal from the environment. Importantly, the comparative genomic analyses indicate that this functional polymorphism is maintained by natural selection.
We find both signatures of selection to lose the operon and selection to maintain it, creating a balanced polymorphism at the level of gene content. This leads to a frequency of the lsr operon intermediate between that of persistent and of volatile genes (van Passel et al. 2008; Kuo and Ochman 2009; Touchon et al. 2009) The selective pressure to lose the operon is supported by the inference of at least eight independent events of operon inactivation and the observation that pseudogenization, when it occurs, is too fast to be a neutral process. This fast gene extinction dynamics was already observed in the genomes of Salmonella enterica vs. Gallinarum (Kuo and Ochman 2010) and occurs through the same general mechanisms described for bacterial pseudogene formation (e.g., large truncations, small frameshift indels, and stop codons) (Lerat and Ochman 2005; Ochman and Davalos 2006). Although our data differ from Kuo and Ochman (2010) in that all genes are inferred to be ancestral and pseudogenization is shared with many strains, some of which very distantly related. In that study, the authors analyze 147 pseudogenes of which only five were shared with the closest related strain and only three are inferred to be ancestral. Our data fit better the models of balancing selection than a random model of gene loss, even though we cannot exclude the possibility that pseudogenization of one gene accelerates the loss of the other genes in the operon. Selection to maintain the ability to respond to extracellular AI-2 is suggested by balancing selection patterns in lsrA gene, as well as the polymorphism observed in all other lsr genes of complete operons that are typical of functional genes. In addition, we inferred through simulation that the majority of the E. coli strains should have already lost the operon unless there is ongoing selective pressure to maintain it.
The mechanisms of selection maintaining these polymorphisms are an important line of future research. Theoretical models have shown that balancing selection may occur for diverse reasons and could potentially be quite common (Gillespie 2004). We propose two nonmutually exclusive hypotheses for the maintenance of polymorphisms in this system. We observed that all Shigella and AIEC, which are strains known to replicate within macrophages, lack the lsr operon. The loss of the lsr operon in these strains could thus be a consequence of adaptation to a specific pathovar. This fits the hypothesis that cooperative processes regulated by QS are less important in bacteria with low infectious dose and able to replicate in professional phagocytes (Gama et al. 2012). However, this intracellular niche adaptation hypothesis cannot explain all losses observed in our data because many other strains lack lsr.
The other hypothesis relates to the social consequences of mutations in the genes regulating AI-2 QS. We found no measurable growth cost for the loss of AI-2 QS mechanism (fig. 3), but in E. coli, AI-2 regulates costly group behaviors such as virulence and biofilm formation (González-Barrios et al. 2006; Herzberg et al. 2006; Reisner et al. 2006; Zhu et al. 2007; Lee et al. 2011). Hence, although QS mutations have little direct metabolic effects, as growth is not affected in monocultures, they are likely to have ecological benefits by providing the cells the ability to exploit social processes in microbiomes. In most QS systems of Gram-negative bacteria, high cell densities are associated with high concentration of the signal. Elements that have lost the ability of producing the signal but still benefit from the information of producers (emitters) are thus noncooperating and have a fitness advantage (Diggle et al. 2007), in these systems receptors are much more abundant than emitters (Patankar and González 2009). In contrast, in the E. coli AI-2 QS system, we observed gene repertoire polymorphism at the level of the signal receptor (lsr operon) and not at the signal emitter (luxS). All these gene losses reduce or abolish AI-2 reception and internalization but do not reduce AI-2 production. This can be interpreted as coercive behavior, which is a particular type of social cheating if demonstrated that these cells would benefit from forcing the nearby cells into cooperative behaviors, while themselves refraining from cooperating (Diggle et al. 2007; Foster et al. 2007). Consistently, it was shown that E. coli lsr mutants can induce the onset of cooperative behaviors of Vibrio harveyi and V. cholerae even when they are at low quorum (Xavier and Bassler 2005a)
Because AI-2 has been shown to regulate biofilm formation and virulence traits in E. coli, it is expectable that the cells that do not internalize AI-2 but still contribute to their increased concentration in the extracellular medium will promote the remaining AI-2-sensitive cells in the vicinity to hasten the onset of the behavior they regulate with AI-2. It was recently predicted by Van Dyken and Wade (2012) that when social cheaters are maintained in natural populations as an evolutionary stable strategy, then it should also be expected that cheaters would be characterized by large insertion/deletions, frameshift mutations, or premature STOP codons; these are features that characterize the lsr operons of E. coli natural variants that do not regulate extracellular AI-2.
The interpretation that gene repertoire polymorphism in the lsr operon is maintained through a process of social evolution is further strengthened by the observation that luxS, the signal synthase, is present in all strains (fig. 2) even though we detect no fitness effect in the single-gene knockout mutant monocultures (fig. 3). Because of its enzymatic role in recycling products of SAM metabolism, it was suggested that the selective pressure to maintain luxS was primarily to detoxify the cell and recycle the products of SAM metabolism (Schauder et al. 2001; Winzer et al. 2003; Vendeville et al. 2005; De Keersmaecker et al. 2006; Hardie and Heurlier 2008). Importantly, we show that a mutant in Pfs, the enzyme immediately upstream of LuxS in the metabolism of SAM, does show a marked growth defect (fig. 3) probably due to the toxic accumulation of SAH (Schauder et al. 2001; Winzer et al. 2002). This strongly indicates that Pfs, not LuxS, is the major enzyme responsible for preventing the toxic consequences of SAH accumulation. The similarity in the phenotypic effects and their extreme difference in fitness cost highlight the importance of pfs in central metabolism, and it suggests that the selective pressure to maintain a functional luxS in the cell is not metabolic but social. Naturally, this conclusion has to be contextualized in the whole discussion of the AI-2 as a QS signal; a social cost cannot be attributed to any gene that does not present a strong metabolic cost.
We still lack a direct experimental demonstration of such social benefit. This is difficult to show for inter-specific QS because it requires experimentation in complex environments. Nevertheless, it is known that in the vertebrate gut, E. coli experiences a complex multispecies environment where the ability to interact (or interfere) with other cells, of the same or of different species, may influence the evolution of its AI-2 regulation system (McNab et al. 2003). Interestingly, extraintestinal virulence of an E. coli AI-2 QS-negative strain (E. coli B2S) was shown to be boosted in mix strains infections compared with pure culture infections when mixed with an AI-2 QS positive regarded as commensal (E. coli MG1655) (Tourret et al. 2011). Hence, QS polymorphisms might lead to exploitation of commensals by pathogens to increase virulence.
Overall, our findings suggest that complex adaptations of species with polyclonal interactions, such as E. coli, can be due to genes maintained at intermediary frequencies rather than ubiquitous or pathovar-specific genes.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S3 are available at Genome Biology and Evolution online (http://gbe.oxfordjournals.org/).
Acknowledgments
The authors thank João B. Xavier for critically reading the manuscript and providing suggestions. They also acknowledge Paulo B. Correia for value experimental help. They acknowledge National BioResource Project (Japan): E. coli for providing the Keio Collection strains. This work was supported by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. 260421 - ECOADAPT; by an International Early Career Scientist grant from the Howard Hughes Medical Institute (HHMI 55007436); by the Institut Pasteur, the CNRS, and the European Research Council under the ERC grant agreement no. 281605 - EVOMOBILOME to E.P.C.R.; and by FCT award to P.H.B. (SFRH/BPD/26852/2006). I.G. and K.B.X. also acknowledge the salary support of LAO/ITQB & FCT.
Literature Cited
- Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78:811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The population genetics of transposable elements. In: Ohta T, Aoki K, editors. Population genetics and molecular evolution. Berlin (Germany): Springer-Verlag; 1985. pp. 213–232. [Google Scholar]
- Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi BJ. The evolution of social behavior in microorganisms. Trends Ecol Evol. 2001;16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker SCJ, Sonck K, Vanderleyden J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006;14:114–119. doi: 10.1016/j.tim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Gardner A, West SA, Griffin AS. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Philos Trans R Soc Lond B Biol Sci. 2007;362:1241–1249. doi: 10.1098/rstb.2007.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Páramo PP, Giudicelli CC, Parsot CC, Denamur EE. The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J Mol Evol. 2003;57:140–148. doi: 10.1007/s00239-003-2460-3. [DOI] [PubMed] [Google Scholar]
- Foster KR, Parkinson K, Thompson CRL. What can microbial genetics teach sociobiology? Trends Genet. 2007;23:74–80. doi: 10.1016/j.tig.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama JA, Abby SS, Vieira-Silva S, Dionisio F, Rocha EPC. Immune subversion and quorum-sensing shape the variation in infectious dose among bacterial pathogens. PLoS Pathog. 2012;8:e1002503. doi: 10.1371/journal.ppat.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. Population genetics: a concise guide. 2nd ed. Baltimore (MD): Johns Hopkins University Press; 2004. [Google Scholar]
- Glasser AL, et al. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Barrios AF, et al. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie K, Heurlier K. Establishing bacterial communities by "word of mouth": LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008;6:635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- Hegde M, et al. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol. 2011;193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J, editors. Oxford surveys in evolutionary biology. Vol. 7. Oxford (UK): Oxford University Press; 1990. pp. 1–44. [Google Scholar]
- Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, et al. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comp Biol. 2006;2:e62. doi: 10.1371/journal.pcbi.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Ochman H. The fate of new bacterial genes. FEMS Microbiol Rev. 2009;33:38–43. doi: 10.1111/j.1574-6976.2008.00140.x. [DOI] [PubMed] [Google Scholar]
- Kuo C-H, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6:e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H, Hartl DL. Molecular and evolutionary relationships among enteric bacteria. J Gen Microbiol. 1991;137:1911–1921. doi: 10.1099/00221287-137-8-1911. [DOI] [PubMed] [Google Scholar]
- Lee J-HJ, Kim Y-GY, Cho MHM, Wood TKT, Lee JJ. Transcriptomic analysis for genetic mechanisms of the factors related to biofilm formation in Escherichia coli O157:H7. Curr Microbiol. 2011;62:1321–1330. doi: 10.1007/s00284-010-9862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 2005;33:3125–3132. doi: 10.1093/nar/gki631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J Bacteriol. 2007;189:6011–6020. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Luo C, et al. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A. 2011;108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011 Available from: http://mesquiteproject.org (last accessed January 2, 2013) [Google Scholar]
- McDonald J, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- McNab R, et al. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E, et al. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect. 2009;11:274–280. doi: 10.1016/j.micinf.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Morton BRB. Chloroplast DNA codon use: evidence for selection at the psb A locus based on tRNA availability. J Mol Evol. 1993;37:273–280. doi: 10.1007/BF00175504. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nowrouzian FL, Wold AE, Adlerberth I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis. 2005;191:1078–1083. doi: 10.1086/427996. [DOI] [PubMed] [Google Scholar]
- Ochman H, Davalos L. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- Ochman H, Whittam TS, Caugant DA, Selander RK. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Oh SS, Buddenborg SS, Yoder-Himes DRD, Tiedje JMJ, Konstantinidis KTK. Genomic diversity of Escherichia isolates from diverse habitats. PLoS One. 2012;7:e47005–e47005. doi: 10.1371/journal.pone.0047005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc Lond B Biol Sci. 1994;255:37–45. [Google Scholar]
- Patankar AV, González JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33:739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- Pereira CS, de Regt AK, Brito PH, Miller ST, Xavier KB. Identification of functional LsrB-like autoinducer-2 receptors. J Bacteriol. 2009;191:6975–6987. doi: 10.1128/JB.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, McAuley JR, Taga ME, Xavier KB, Miller ST. Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol Microbiol. 2008;70:1223–1235. doi: 10.1111/j.1365-2958.2008.06477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. Forthcoming 2012 doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- Pereira CS, et al. Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol Microbiol. 2012;84:93–104. doi: 10.1111/j.1365-2958.2012.08010.x. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Puigbò P, Bravo IG, Garcia-Vallvé S. E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI) BMC Bioinformatics. 2008;9:65. doi: 10.1186/1471-2105-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GMG, Lan RR, Reeves PRP. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol. 2006;188:3572–3581. doi: 10.1128/JB.188.10.3572-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Bedzyk L, Ye R, Thomas S, Wood T. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl Environ Microbiol. 2004;70:2038–2043. doi: 10.1128/AEM.70.4.2038-2043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, Touchon M, Feil EJ. Similar compositional biases are caused by very different mutational effects. Genome Res. 2006;16:1537–1547. doi: 10.1101/gr.5525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers A, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, Haeseler von A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Li WH. Codon usage in regulatory genes in Escherichia coli does not reflect selection for "rare" codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. The Codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. New York: Cold Spring Harbor Press; 1984. [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Estimation of the neutrality index. Mol Biol Evol. 2011;28:63–70. doi: 10.1093/molbev/msq249. [DOI] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taga ME, Xavier KB. Methods for analysis of bacterial autoinducer-2 production. Curr Protoc Microbiol. 2011 doi: 10.1002/9780471729259.mc01c01s23. Chapter 1:Unit1C.1. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura KK, Dudley JJ, Nei MM, Kumar SS. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- Touchon M, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourret J, et al. The interaction between a non-pathogenic and a pathogenic strain synergistically enhances extra-intestinal virulence in Escherichia coli. Microbiology. 2011;157:774–785. doi: 10.1099/mic.0.037416-0. [DOI] [PubMed] [Google Scholar]
- Van Dyken JD, Wade MJ. Detecting the molecular signature of social conflict: theory and a test with bacterial quorum sensing genes. Am Nat. 2012;179:436–450. doi: 10.1086/664609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MWJ, Marri PR, Ochman H. The emergence and fate of horizontally acquired genes in Escherichia coli. PLoS Comp Biol. 2008;4:e1000059. doi: 10.1371/journal.pcbi.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making "sense" of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Wang L, Hashimoto Y, Tsao C-Y, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J Bacteriol. 2005;187:2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li J, March JC, Valdes JJ, Bentley WE. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J Bacteriol. 2005;187:8350–8360. doi: 10.1128/JB.187.24.8350-8360.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- West SA, Diggle SP, Buckling A, Gardner A, Griffins AS. The social lives of microbes. Annu Rev Ecol Evol S. 2007;38:53–77. [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Wielgoss S, et al. Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3. Genes Genomes Genet. 2011;1:183–186. doi: 10.1534/g3.111.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr Opin Microbiol. 2002;5:216–222. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- Winzer K, Hardie KR, Williams P. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv Appl Microbiol. 2003;53:291–396. doi: 10.1016/s0065-2164(03)53009-x. [DOI] [PubMed] [Google Scholar]
- Winzer K, et al. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology. 2002;148:909–922. doi: 10.1099/00221287-148-4-909. [DOI] [PubMed] [Google Scholar]
- Wirth T, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold AEA, Caugant DAD, Lidin-Janson GG, de Man PP, Svanborg CC. Resident colonic Escherichia coli strains frequently display uropathogenic characteristics. J Infect Dis. 1992;165:46–52. doi: 10.1093/infdis/165.1.46. [DOI] [PubMed] [Google Scholar]
- Wright F. The "effective number of codons" used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005a;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005b;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CC, et al. The possible influence of LuxS in the in vivo virulence of rabbit enteropathogenic Escherichia coli. Vet Microbiol. 2007;125:313–322. doi: 10.1016/j.vetmic.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.