Abstract
Half of the world’s population is infected with Helicobacter pylori and approximately 20% of infected individuals develop overt clinical disease such as ulcers and stomach cancer. Paradoxically, despite its classification as a class I carcinogen, H. pylori has been shown to be protective against development of asthma, allergy, and esophageal disease. Given these conflicting roles for H. pylori, researchers are attempting to define the environmental, host, and pathogen interactions that ultimately result in severe disease in some individuals. From the bacterial perspective, the toxins, CagA and VacA, have each been shown to be polymorphic and to contribute to disease in an allele-dependent manner. Based on the notable advances that have recently been made in the CagA field, herein we review recent studies that have begun to shed light on the role of CagA polymorphism in H. pylori disease. Moreover, we discuss the potential interaction of CagA and VacA as a mediator of gastric disease.
Keywords: Helicobacter pylori, CagA, VacA
Introduction
In 1982, the Gram-negative, spiral shaped microaerophilic bacterium Helicobacter pylori was first isolated from the stomach of a patient with gastritis.1 Thirty years later, H. pylori has been shown to colonize over half of the world’s population. In approximately 20% of infected individuals, infection with H. pylori may result in clinical disease that manifests as peptic or duodenal ulcers, gastric adenocarcinoma, or mucosa-associated lymphoid tissue (MALT) lymphoma2-5; these last two manifestations have led H. pylori to be the only bacterium classified as a class I carcinogen by the World Health Organization.6 For the remaining 80% of infected individuals, the chronic inflammatory response induced by H. pylori in the gastric mucosa results in no overt clinical symptoms.
H. pylori, is the predominant bacterial species known to colonize the stomach, and the bacterium is typically acquired early in childhood; in the absence of antibiotic treatment, lifetime colonization occurs.7H. pylori association with humans is hypothesized to have originated in Africa preceding the human migration that is believed to have occurred approximately 58,000 years ago.8,9 As humans migrated from Africa to various continents, H. pylori was carried with them and evolved into present day strains; this close association has been used to study human migration patterns.10-14 Due to the large number of colonized individuals, the majority of which do not develop disease, and because H. pylori has evolved with humans for centuries, there is currently a movement to classify H. pylori as normal human flora (discussed below).
H. pylori Classification and Role in Health and Disease
Since its discovery, H. pylori has been shown to be one of the most variable species of bacteria. This heterogeneity has been the basis for a series of classification systems that have been used to discriminate between strains. Initially, H. pylori strains were categorized as type I or type II based on the presence or absence of both the cytotoxin-associated gene-pathogenicity island (cag-PAI) and the vacuolating cytotoxin, VacA, respectively.15-17 Following the identification of allelic variation in cagA, which is carried on the island and encodes for the CagA toxin, strains were next classified as either East Asian or Western based on the particular cagA allele that they carried. This system specifically utilizes variation in the C-terminus of the protein to distinguish strains. East Asian strains are predominantly from Japan, South Korea, and China, while Western stains are primarily from the United States, Europe, and Australia.18-20 Over the last several years, the discovery of additional cagA alleles led to the addition of the J-Western and Amerindian strain designations. The J-Western, or Japanese subtype of Western CagA, strain was initially believed to be predominantly found in Okinawa, Japan21,22; however, it has recently been shown that J-Western strains have a broader geographic distribution.23 The Amerindian strain is predominately found within the Amerindian populations of South America.10,24,25 Finally, multilocus sequence typing (MLST) of several H. pylori housekeeping genes and the vacA virulence gene has been utilized to develop a classification system that takes advantage of the broader genetic diversity of H. pylori as a means to study its ancestry9,12,13,26; MLST identified seven distinct H. pylori populations, some of which have several sub-populations that are named based upon the geographic location in which the strains are found (Table 1).9,13,14,26-28 This new classification system delineates strains from the classical East Asian, Western, Amerindian or J-Western strain designations, which are based solely on cagA C-terminal polymorphisms.
Table 1. An overview of H. pylori populations*.
| H. pylori population | H. pylori subpopulations | Geographic location or human population | Reference |
|---|---|---|---|
| hpAfrica1 |
hspWAfrica |
W. Africa |
13 |
| |
hspSAfrica |
S. Africa |
13 |
| hpAfrica2 |
|
S. Africa |
13 |
| hpNEAfrica |
|
Ethiopia, N. Nigeria, Somalia, Sudan |
9 |
| hpAsia2 |
|
Bangladesh, N. India, Malaysia, Thailand |
9, 28 |
| hpEastAsia |
hspAmerind |
Native Americans |
13 |
| |
hspEAsia |
East Asians |
13 |
| |
hspMasori |
Taiwanese Aboriginals, Melanesians, Polynesians |
14 |
| hpEurope |
|
Europe, Middle East, India, Iran |
13, 27 |
| hpSahul | Australian Aboriginals, Papua New Guineans | 14 |
In keeping with the idea that H. pylori may be normal flora whose association with the human host is important, recent findings suggest that H. pylori plays a protective role in various aspects of human health (Table 2). Perhaps the strongest evidence for a protective role for H. pylori against disease development are the studies that show an inverse association between H. pylori colonization with occurrence of asthma and allergy, particularly in adolescents.29-32 Interestingly, the presence of CagA seems to increase the protective nature of H. pylori against both asthma and allergy.31 Therefore, the disappearance or eradication of H. pylori may represent another example of the “Hygiene Hypothesis,” which suggests that reduced exposure to pathogens early in life may predispose individuals to diseases such as allergy and asthma later in life.33 A protective role for H. pylori that extends beyond the “Hygiene Hypothesis” has been indicated by studies that show that H. pylori protects against tuberculosis and diabetes. The tuberculosis study utilized human subjects as well as non-human primates to show that H. pylori colonized individuals are less likely to develop active tuberculosis through reactivation.34 Additionally, H. pylori infected individuals have significantly increased Th1 responses and IFN-γ, which is required for TB latency.35 In relation to diabetes, recent work indicates that the human gastric hormones that control energy homeostasis, leptin and ghrenlin, along with body mass index (BMI) are increased in patients following H. pylori eradication.36,37 Furthermore, a cross-sectional study identified a positive association between glycated hemoglobin, a marker for long-term blood glucose levels, and H. pylori in individuals without diabetes; this association was increased in patients with a higher BMI.38 Therefore, data suggest that eradication of H. pylori may play a role in development of metabolic disorders in individuals with an increased BMI.
Table 2. Role of H. pylori as normal flora and pathogen^.
| Potential beneficial effects of H. pylori against: | Known and suspected negative effects of H. pylori: |
|---|---|
| Asthma29-32,183,184 |
Gastritis1,185 |
| Allergy31,184 |
Peptic and duodenal ulcers185,186 |
| Esophageal reflux and Barrett’s Esophagus41,42,45 |
MALT lymphoma2 |
| Esophageal adenocarcinoma39,40 |
Gastric adenocarcinoma2,4,5 |
| Reactivation of Tuberculosis*34,35 |
Idiopathic thrombocytic purpura (reviewed in ref. 37) |
| Inflammatory bowel diseases*187,188 |
Iron deficiency anemia (reviewed in ref. 37) |
| Weight gain (due to effects on leptin and ghrelin)*36,38 |
Atherosclerosis/coronary artery disease*189,190 |
| Graves’s Disease*191 |
Further analysis is required to substantiate these findings. ^Due to space limitations only a few key references are provided.
Finally, as the prevalence of H. pylori has decreased over the last century, a rise in esophageal reflux disease and esophageal adenocarcinoma has been observed. Several studies suggest a protective role for H. pylori in preventing both diseases.39-44 A meta-analysis of 49 studies, as well as two prospective studies, indicate that H. pylori colonization is associated with a reduced risk for Barrett’s Esophagus, which often occurs following esophageal exposure to acid through reflux.43-45 It is believed that the decrease in stomach acid, which occurs due to the loss of parietal cells in H. pylori infected individuals, protects the esophagus from cellular damage that initiates esophageal disease.45 A potential role for CagA in protection against esophageal disease has been suggested by studies that indicate individuals infected with cagA positive H. pylori strains are less likely to develop esophagitis or Barrett’s Esophagus than those infected with cagA negative strains.42,43 These results in turn suggest that there may be a consequence to outright eradication of H. pylori from the human microbiome.
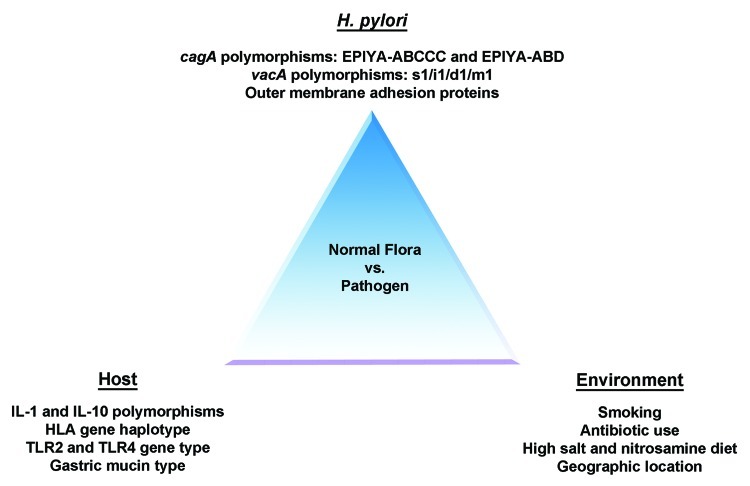
Given the clear evidence that H. pylori is associated with development of gastric disease, along with the growing evidence that suggests that H. pylori may play a beneficial role in some individuals, the question arises as to what factor/factors are responsible for determining the ultimate outcome of H. pylori colonization? Unfortunately, there is likely no single answer to this question. Instead, as previously stated by Merrell and Falkow, “the balance shift that causes colonization to go awry, leading to a deadly disease, is a combination of physiological and genetic factors for both participants of the host-pathogen interaction”46. Clearly, in the case of H. pylori-induced gastric disease, the etiology involves numerous host genetics/responses, environmental factors, and bacterial virulence factors (Table 3). Accordingly, previous reviews have focused on these topics.47,48 As such, herein, we will only briefly mention some discoveries that affect our understanding of host and environmental factors, and then focus the remainder of our discussion on bacterial factors that are clearly important for disease.
Table 3. An overview of factors influencing H. pylori disease development.
| Factor or mechanism | References^ | |
|---|---|---|
| Host: |
Age |
175, 192 |
| |
Male sex |
148, 190 |
| |
IL-1 gene cluster polymorphisms |
193–195 |
| |
IL-10 gene polymorphisms |
195, 196 |
| |
HLA gene haplotype |
197 |
| |
TLR2 and TLR4 gene types |
198 |
| |
Atg16L1 Autophagy gene polymorphism |
199 |
| |
Nod1/Nod2 polymorphism (Chinese population) |
200 |
| |
PTPN11 gene polymorphism |
201, 202 |
| |
Gastric mucins |
203 |
| Bacterial: |
cag pathogenicity island |
15 |
| |
CagA EPIYA polymorphism |
109–113 |
| |
vacA polymorphism |
138–140, 142, 144, 147 |
| |
Outer membrane proteins (HomB, BabA, SabA/B, IceA, OipA, DupA) |
175 |
| Environmental: |
Diet: Red meat/processed meat consumption and production of nitrosamines* |
63 |
| |
Asian Populations: High salt consumption* |
204 |
| |
Mexican Populations: Capsaicin consumption |
61 |
| |
Antibiotic use |
205 |
| |
Smoking: |
204, 206, 207 |
| |
CagA positive H. pylori |
208 |
| |
Geographical location: East Asia vs. North America/Europe/Africa vs. South America: |
10, 18–22, 24, 25 |
Association with disease was found only in individuals with concurrent H. pylori infection. ^Due to space limitations, only a few key references are provided
Host and Environment
From the immunological perspective, it is known that how the immune system responds to H. pylori dictates the ultimate outcome of infection and that the immune response to H. pylori is clearly complicated (refer to reviews49-51). For example, specific classes of T-cells have several possible effects. The presence of T-cells promotes H. pylori-induced chronic-active gastritis.52 As such, patients with peptic ulcer disease show an increase in Th1 and Th2 cellular responses, which likely then contribute to the pathology associated with gastric disease.53 In contrast, the role of T-regulatory (Treg) cells in H. pylori infection appears protective (reviewed in49). Tregs control effector T-cell responses to H. pylori infection in a neonatal mouse model by inducing tolerance and protecting against development of preneoplastic lesions.54 Furthermore, an increased number of CD4+CD25hi Treg cells that secrete the immune suppressive cytokine IL-10 is inversely correlated with IL-8 expression and NF-κB activation in asymptomatic carriers.53 In keeping with this, mild gastritis observed in H. pylori infected children is also linked to a Treg cell response along with increased levels of IL-10 and TGF-β 55. Since H. pylori is known to induce IL-8 expression upon host cell contact, which in turn induces proinflammatory conditions in the stomach,56,57 it is possible that the presence of a Treg cell response regulates IL-8 expression and NF-κB activation by suppression of the proinflammatory cytokine Il-17 from the gastric mucosa as a means to prevent overt disease and bacterial clearance (refer to review by51). This notion is consistent with the finding that the presence of a Treg cell population results in increased H. pylori colonization.53
Supporting the idea that specific T-cell immune responses determine the outcome of H. pylori infection, recent studies indicate that Th17 cells promote H. pylori associated pathology. In the absence of the Th17 polarizing cytokine IL-23, IL-23p19-/- mice exhibit significantly less gastritis and precancerous lesions, while IL-23-/- H. pylori infected mice show decreased gastritis, decreases in the cytokines IL-17 and IFN-γ, and concomitant increases in bacterial burden.58,59 IL-23 therefore appears to contribute to gastric pathology through activation of Th1/Th17 immune responses that control H. pylori infection by initiating inflammation and controlling the degree of gastritis.58,59 The results from these animal experiments have also been extrapolated to human samples. Analysis of patients with a past, but not current, H. pylori infection shows persistent H. pylori specific Th17 responses. Among these individuals, those with gastric pre-cancerous lesions have increased levels of IL-17A compared with those without lesions.60 In summary, the type of immune response mounted by the host influences the outcome of H. pylori infection.
From the environmental perspective, diet and geographical location appear to have the largest influence on disease. Several studies have indicated that consumption of nitrosamines in processed meats and vegetables, as well as a high salt diet both increase the risk for H. pylori induced disease.61-64 Due to variations in diet around the world, the role of diet in the incidence of gastric carcinoma also varies. In East Asia, where the incidence of gastric carcinoma is high, diets are often high in salt content and nitrosamines (reviewed in64 and discussed in63). Similarly, in Europe, consumption of nitrosamines is associated with gastric carcinoma.63 Studies of Mexican populations, find that consumption of capsaicin, a component of chili peppers, may increase the risk of gastric carcinoma, particularly in individuals who carry an IL1B-31C allele.61 While the role of nitrosamines, and other dietary components, have been studied in human populations, direct causation between diet, H. pylori, and disease is difficult to prove due to the inability to directly regulate patient’s diets. As such, researchers are now using long-term colonization of dietary controlled non-human primates to address these questions. Indeed, gastric neoplasia is induced in rhesus macaques only when nitrosamine compounds are administered in the presence of H. pylori colonization; thus supporting the synergistic role of these compounds in gastric disease.63
Overall, the data suggest that as humans and H. pylori have co-evolved over the last 58,000 y, H. pylori has developed mechanisms to survive the harsh conditions found in the gastric environment, as well as mechanisms by which to evade and utilize the human immune response for survival. Conversely, humans appear to utilize H. pylori colonization as a means to protect against several severe disease states. However, despite the evidence that supports the protective role and generally asymptomatic colonization of H. pylori in the vast majority of individuals, some individuals are clearly predisposed to severe disease as a result of colonization. Thus far, host polymorphisms, bacterial polymorphisms, and environmental conditions (Table 3) have all been implicated in conversion of H. pylori from a commensal to a pathogen, but the precise mechanism underlying this conversion remains unclear. What is clear is that overall disease severity is attributed to the production of several H. pylori virulence factors, each of which show variability across strains: such as the toxins, CagA and VacA, and the outer membrane proteins, HomB, BabA, IceA, SabA/B, DupA, and OipA. The remainder of this review will focus on the role of CagA polymorphism in cancer development, as it is the most severe disease pathology associated with H. pylori infection, as well as how polymorphism in VacA may contribute to disease.
CagA
CagA is one of the best-studied virulence factors of H. pylori; this toxin is 120–145 kDa in size, and is encoded by cagA, which is carried on the cag-PAI.15 H. pylori strains that carry the PAI are known to be more virulent than those that do not.15 Following translocation into the host cell by the type IV secretion system (T4SS), which is encoded for by the other genes contained within the PAI, CagA exerts its effects directly on eukaryotic cells.16,65,66
The exact role of CagA appears to be multifactorial. For instance, effective H. pylori colonization of the apical surface of polarized monolayers requires CagA mediated acquisition of nutrients.67,68 CagA also regulates the host’s immune response to H. pylori as a means to aid in persistence of the bacteria. In the presence of CagA, bone marrow derived dendritic cells (BMDC) from wild type C57BL/6 mice are unable to mature and clear H. pylori.69 Furthermore, in BMDC’s infected with wild type H. pylori, mRNA expression of the proinflammatory cytokines TNF-α and IL-12p40 are decreased, while mRNA expression of IL-10 is increased; this response is CagA dependent.69 In CagA-transgenic mice (CagA-Tg), expression of CagA inhibits the ability of BMDCs to mediate the differentiation of CD4+ T-cells to Th1 cells and the ability to respond to LPS.69 From these studies, CagA appears to negatively regulate dendritic cells as a means to promote persistence of H. pylori.69
Following translocation into eukaryotic cells by the T4SS, CagA binds to phosphatidylserine in the plasma membrane of host cells and is phosphorylated on tyrosine residues found within conserved C-terminal Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs70,71; the host cell kinases c-Src and c-Abl, are responsible for phosphorylation.72-75 Phospho-CagA forms a complex with SHP-2, a protein tyrosine phosphatase.16,66,76 Formation of this complex results in recruitment to, and constitutive activation of, SHP-2 at the host cell membrane.71 The many downstream signaling events resulting from CagA-induced SHP-2 activation have been extensively reviewed elsewhere.77-79 However, for the purpose of our discussion, we point out that the deregulation of SHP-2 by CagA results in cell scattering and elongation. This CagA induced cell elongation is termed the “hummingbird phenotype” due to the long extensions observed on infected cells and depends on Ras independent modification of Erk as well as the interaction of CagA with partitioning-defective 1 (PAR1)/microtubule affinity-regulating kinase (MARK).16,80-82 The interaction of CagA with PAR1 is also important for stabilizing CagA within the eukaryotic cell following translocation; deletion of the C-terminal EPIYA motifs results in a significant decrease in the half-life of CagA.83 Not only can CagA induce cell scattering and elongation in a phosphorylation-dependent manner, CagA can also initiate this process in a phosphorylation independent manner through mediation of NF-κB and β-catenin activity.84 Regardless of the mechanism underlying initiation of cell scattering and elongation, following CagA injection, changes in expression of genes involved in epithelial-mesenchymal transition, a process known to be involved in the promotion of cancer dissemination in metastatic cancers and tissue fibrosis, occurs.85 As such, during H. pylori infection of AGS cells, epithelial cell markers are found to be downregulated while markers of mesenchymal cells are upregulated.85
CagA domains
Recent studies of CagA have investigated how the N- and C-terminal domains of the protein are involved in targeting CagA within the eukaryotic cell as well as modulating changes in host cell signaling. Studies using MDCK cells expressing various domains of CagA found that the N-terminus of the protein targets CagA to the cell-cell junctions and promotes cell migration. Conversely, the C-terminus targets CagA to the cytoplasm and is required for pseudopodial formation.86 Conflictingly, independent studies using AGS cells and transfection of various CagA EPIYA constructs indicate that the C-terminal EPIYA motif is required for CagA membrane localization and that this occurs independent of phosphorylation.87 A third study, which also used transfection of CagA but in MDCK cells, aimed to clarify the role of the N- and C-termini in CagA membrane localization. This work showed that CagA contains two independent domains that target CagA to the membrane; one at the N-terminus and one at the C-terminus.88 Thus, the conflicting results of the two previous studies were due to the particular region found in the truncated CagA products, which were different in each study.88 Based on these results, Steininger, et al.89 have suggested the following model: the first 200 amino acids of CagA may serve as a membrane binding domain that is required to mediate proper CagA translocation into the host cell, targets CagA to cell-cell adhesions, and serves to inhibit EPIYA-mediated cell signaling. Conversely, a second membrane targeting domain found in the C-terminus functions to tether CagA to the plasma membrane for EPIYA-mediated signaling events; both the N- and C-terminal regions of CagA interact with each other to accomplish membrane tethering.88,89 Interestingly, the N-terminal targeting domain appears to inhibit apical surface constriction and host cell elongation by reducing CagA effects on β-catenin, which is believed to be an important component that contributes to carcinogenesis.88 Consequently, activity of the N-terminal domain on β-catenin signaling may contribute to inhibition of carcinogenesis.88 It is worth noting that since these studies used truncated versions of CagA, it is unknown precisely how the domains of intact CagA interact inside the host cell.
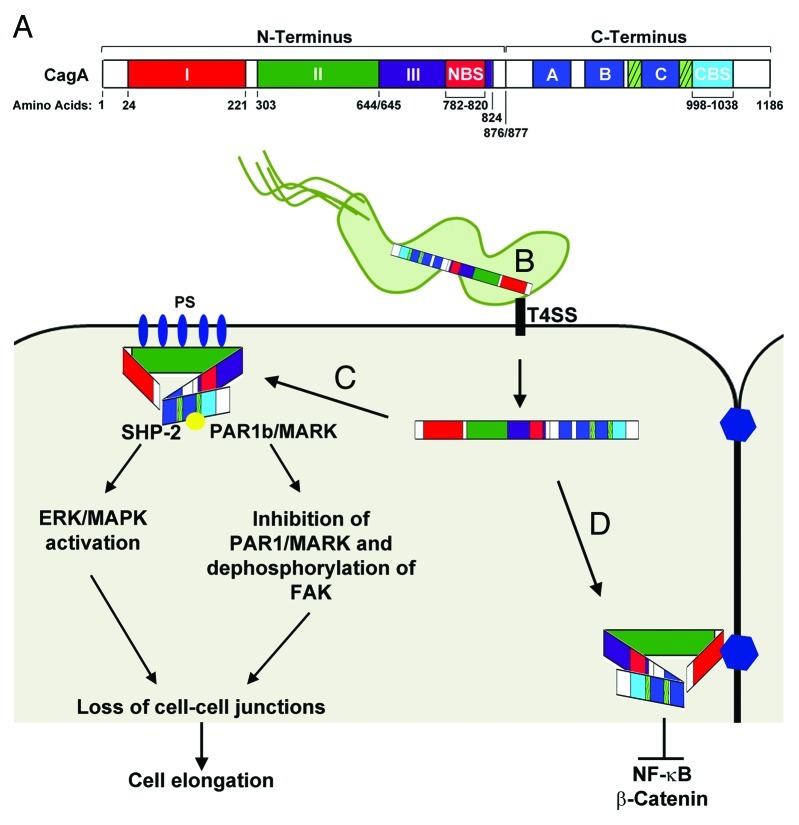
More recently, another study has further fine-tuned our understanding of the different CagA domains.90 Examination of CagA by X-ray crystallography shows an ordered N-terminus that is comprised of three domains (I, II, and III) (Fig. 1A); Domain I corresponds to residues 24–221, which overlaps with residues 1–200 in the previously described model.88,90 Basic residues located in Domain II are responsible for CagA’s interaction with phosphatidylserine located within the host cell membrane (Fig. 1B). Domain III was shown to contain a N-terminal binding sequence (NBS) that interacts with a C-terminal binding sequence (CBS) (Fig. 1C).90 Consistent with a previous study that showed that CagA lacks a defined secondary structure,91 NMR and X-ray crystallography revealed that tertiary structure of the C-terminus also lacks an ordered and solid structure.90 It has been suggested that during interactions between the NBS and CBS, the NBS serves as a regulatory sequence that facilitates binding of CagA to its target proteins (Fig. 1C).90 In this regard, interaction between the NBS and CBS may alter the C-terminal secondary structure whereby folding surrounding or within the Cag multimerization (CM) sequence may decrease turnover of the complex between CagA and PAR1/MARK; thus facilitating binding of other proteins.90,92
Figure 1. CagA Functional Domains. (A) Linear representation of CagA domains. The CagA N-terminus (1–876) contains three domains (I, II, and III). Domain I spans residues 24–221, Domain II spans residues 303–644, and Domain III spans residues 654–824. A N-terminal binding sequence (NBS) is located within Domain III from residues 782–820. The CagA C-terminus (877–1186) contains the EPIYA-ABC motifs (dark blue boxes), the Cag multimerization sequences (hatched boxes), and a C-terminal binding sequence (CBS) located from residues 998–1038.90 (B) The first 200 amino acids of the N-terminus (Domain I) target CagA inside H. pylori to the type IV secretion system (T4SS). The T4SS interacts with β-1 integrin (black rectangle) and is responsible for translocation of CagA into the epithelial cell. Residues 800–1216 alone target CagA to the cytoplasm.88 (C) CagA interacts with phosphatidylserine (PS; blue ovals) on the inner surface of the plasma membrane via basic residues located on Domain II. Interaction of the NBS and CBS condenses the structure of CagA allowing the unstructured C-terminus to remain accessible to Src-kinases as well as SHP-2. Phosphorylation of CagA (yellow circle) by Src-kinases results in activation of SHP-2 and the ERK/MAPK pathways leading to cell elongation. (D) Targeting of CagA to cell-cell junctions by Domain I results in inhibition of NF-κB and β-catenin and inhibition of EPIYA mediated cell signaling pathways. For more detailed information concerning the signaling pathways affected by CagA see78. Figure was adapted from.89,90,92
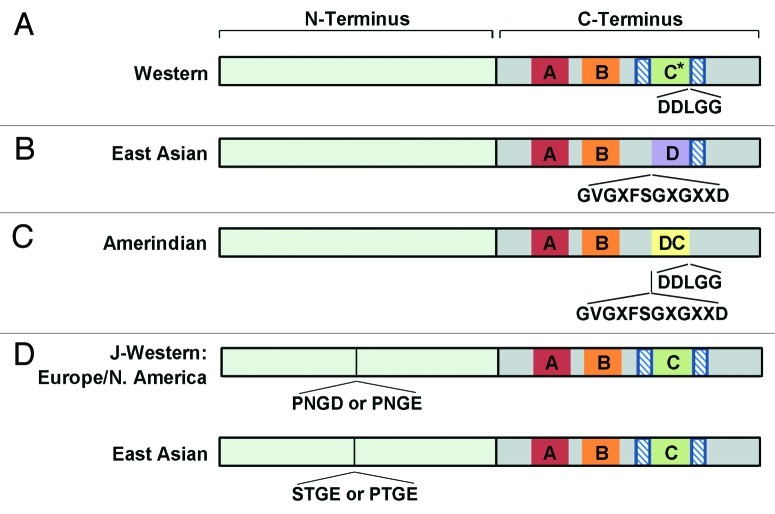
As alluded to in the previous paragraph, another component of CagA is a conserved 16 amino acid motif, FPLXRXXVXDLSKVG, found within the C-terminus. This motif mediates CagA multimerization and CagA’s interaction with SHP-2; this segment is termed the Cag multimerization or CM sequence.93,94 Furthermore, the presence of the 14 amino acid sequence, FPLKRHDKVDDLSK, within this region led to identification of this region as the MARK2 inhibitory sequence (MKI) due to its role as the binding site for PAR1/MARK kinase, which functions in CagA-mediated disruption of epithelial cell polarity.82,91 The C-termini of both East Asian and Western H. pylori isolates contain CM sequences; in East Asian strains, this sequence is found immediately downstream of the EPIYA-D motif, and in Western strains this sequence is found upstream of each EPIYA-C motif and downstream of the last EPIYA-C motif (Fig. 2A and 2B).94 This same region of the C-terminus has also been termed the conserved repeat responsible for phosphorylation independent activity (CRPIA) due to its role in promoting CagA phosphorylation independent activation of β-catenin and NF-κB signaling.84
Figure 2. CagA variations by geographic region. (A) Western, (B) East Asian CagA, and D) J-Western CagA contain a Cag multimerization (CM)/CRPIA/MKI sequence (FPLXRXXVXDLSKVG) for CagA dimerization and binding to PAR1/MARK (hatched boxes). *Western CagA can have up to five EPIYA-C repeats. (C) The last EPIYA motif of Amerindian CagA contains a motif that is a chimera of the East Asian EPIYA-D and Western EPIYA-C motifs as determined by the presence of a five amino acid sequence found in Western CagA (DDLGG) and a 12 amino acid sequence found in East Asian CagA (GVGXFSGXGXXD). (D) J-Western strains of either European or North American origin contain either a PNGD or PNGE N-terminal amino acid insertion, while strains of East Asian origin contain a STGE or a PTGE N-terminal amino acid insertion starting at residue 207 when compared with H. pylori strain 26695. Figure generated from studies described in.23
En masse, these studies show that the various CagA domains contribute to targeting within the host cell, which ultimately affects how CagA alters host cell signaling pathways. While all of these CagA domain studies are intriguing, interpretation of the data are complicated due to use of transfection models, differences in the cell types used, and differences in the polarization state of the various cell lines.86-88 Differences also occur between normal and cancer cell lines as well as between polarized epithelial monolayers and sub-confluent cells in terms of signaling and junctional complex formation95-97. Thus, it will be interesting to see if these results can be recapitulated in an infection model, especially since the levels of CagA are clearly different between these model systems.77,89 One way to address these differences in model systems would be the creation of isogenic strains that could be used in a rodent model. Use of these same strains could then be expanded into a primate model as it is the closest model to replicate human disease.
CagA EPIYA motifs
While there has been some previous debate as to the role of the various domains of CagA, effects on host cell signaling are primarily attributed to the EPIYA motif region that is contained within the C-terminus of the protein (reviewed in77). The EPIYA motifs are comprised of Glu-Pro-Ile-Tyr-Ala residues, where the Tyr residues serve as the site of phosphorylation.70 Differences exist in the number of EPIYA repeats found across strains, as well as in the amino acid sequences that flank the EPIYA repeats. This variation has been used as a means to classify the EPIYA containing regions into motifs termed EPIYA-A, -B, -C, and –D. Furthermore, since geographic variability exists in grouping of these motifs, this has been used to classify CagA as either Western (contains EPIYA-A, -B, and -C, where the C motif may be duplicated multiple times) or East Asian (contains EPIYA-A, -B, and –D)70,76,93,98. Recently the sequence and EPIYA motifs of two additional CagA motifs, J-Western and Amerindian, have also been characterized10,20-22,24,25,99 and compared with the Western and East Asian CagA EPIYA motifs (Fig. 2).23,100 Analysis of both nucleotide and amino acid sequences across H. pylori strains suggests that all CagA variants can be derived by homologous recombination at the CM sequence, recombination surrounding the EPIYA sequence, or by illegitimate recombination between short similar DNA sequences (1–12 base pairs) that appear to be AT rich.101 Moreover, as discussed below, additional studies have investigated evolution of the J-Western and Amerindian motifs.
Analysis of Amerindian CagA EPIYA motifs show that the last EPIYA motif appears to be a hybrid between Western EPIYA-C and East Asian EPIYA-D motifs and thus, has been designated EPIYA-DC (Fig. 2C).23 As such, the Amerindian CagA EPIYA-DC sequence contains a five amino acid sequence (DDLGG) found in the EPIYA-C motifs, and a 12 amino acid sequence (GVGXFSGXGXXD) that is similar to a segment found in EPIYA-D motifs.23 Regardless of how Amerindian CagA evolved, the result is that Amerindian CagA appears less virulent than both Western and East Asian CagA; the level of IL-8 induction is decreased and it appears that Amerindian CagA is capable of serving as a dominant negative inhibitor of CagA.25
In comparison, initial analysis suggested that the EPIYA motifs of J-Western CagA were indistinguishable from Western CagA. However, further analysis of strains from Okinawa that also contain the vacA m2 allele (discussed below) and analysis of indels or hypervariable regions in the CagA proteins, reveal a four amino acid insertion in the N-terminus of J-Western CagA that is not present in any of the other forms of CagA (Fig. 2D).23,100 In conjunction with the discovery that J-Western CagA has geographical origins other than Okinawa, Japan, it was found that this four amino acid sequence also varies based on H. pylori origin.23 In this regard, a PNGD or PNGE insertion is present in J-Western CagA’s that have a European or North American geographical origin, while a STGE or PTGE insertion is present in strains with an East Asian origin.23 Furthermore, while 45 amino acid differences exist between J-Western and Western CagA, 112 differences exist between J-Western and East Asian CagA.23
Similar to variations in the C-terminal EPIYA motifs, analysis of the CagA N-terminus suggests that N-terminal recombination events may also have occurred; CagA from strains isolated in Japan show similarity to CagA from strains isolated from various locations around the world as well as similarity to East Asian CagA.23 Therefore, Duncan et al. suggest that since the diversity is specific to parts of cagA, and is not present throughout the gene, these recombination events are coupled to selection.23 Thus, the derivation of the various CagA alleles appears to involve a series of rearrangements/recombinations that occurred during the course of bacterial evolution within the host.
An immediate result of the variations in the EPIYA motifs can be seen by differences in tyrosine phosphorylation of the CagA EPIYA-ABC and EPIYA-ABD motifs. Elegant studies by Mueller et al. investigated the process of tyrosine phosphorylation of both the EPIYA-ABC and EPIYA-ABD motifs by Src family kinases.102 These studies revealed that c-Src can phosphorylate both the EPIYA-C and –D motifs and that this phosphorylation occurs within 90 min of CagA translocation. Conversely, c-Abl can phosphorylate all four varieties of EPIYA motifs and this phosphorylation occurs within 180 min of translocation. Interestingly, the “hummingbird phenotype” cannot be induced by phosphorylation of any single motif in Western CagA, indicating that phosphorylation of at least two motifs is required for cell elongation.102 Finally, the study demonstrated that single CagA molecules are simultaneously phosphorylated at only one or two EPIYA motifs, and never at all three motifs.102 Since CagA is known to dimerize,93 the dual phosphorylation may occur either on one CagA molecule or such that each monomer contains one phosphorylated EPIYA motif.102 En masse, the model that arises from these studies is that CagA is first phosphorylated by c-Src at the EPIYA-C or –D motif, followed by phosphorylation of the EPIYA-A or –B motif by c-Abl.102 The fact that CagA can only be simultaneously phosphorylated at a maximum of two motifs, begs the question of why a substantial increase in apparent levels of phosphorylation are observed when cells are infected with an EPIYA-ABD motif containing strain as compared with an EPIYA-ABC motif containing strain.93,103,104 One possibility could be that numerous components interplay to facilitate this finding. This notion is supported by previous studies showing that East Asian CagA forms a stronger interaction with SHP-280,104,105 due to the presence of a perfect SHP-2 consensus binding motif located around the EPIYA-D motif; in Western EPIYA-C motifs, the sequence differs at the pY+5 amino acid.106 Once this high affinity interaction occurs, CagA dimerization mediated by the CM sequence through interaction with PAR1 dimers within the cell could facilitate a stronger interaction of CagA with SHP-2, resulting in increased SHP-2 deregulation and cell elongation.107 In turn, phosphorylation of the EPIYA-A and –B motifs results in interaction of CagA with Csk, and Csk serves to inhibit Src functioning in a negative feedback loop.105,108 In this model, Western CagA with an EPIYA-ABC motif binds more efficiently to Csk than an East Asian EPIYA-ABD motif, despite the ability of EPIYA-A and EPIYA-B motifs from both Western and East Asian CagA to bind Csk.105 Therefore, it appears that this feedback loop is enhanced in Western strains.105 Thus it is possible that the increase in binding affinity of EPIYA-D to SHP-2, CagA multimerization, and inhibition of Src activity by increased EPIYA-A and –B motifs in Western strains may be the reason underlying the apparent increase in phosphorylation observed with EPIYA-D motifs.
Role of the CagA EPIYA motifs in disease
Given the high degree of variability of the CagA EPIYA region, the major question that arises is whether this variability and subsequent differences in phosphorylation affect H. pylori mediated disease? This seems to be the case since studies of H. pylori strains isolated from patients in the United States, Italy, Brazil, and Portugal have shown that CagA containing higher numbers of EPIYA-C motifs are associated with the development of gastric carcinoma109-112 or more severe precancerous lesions.113 However, in direct contrast, study of H. pylori strains isolated from patients in Columbia, Venezuela, Mexico, and another Italian population showed no correlation between increasing EPIYA-C motifs and disease.109,114-116 These differences in the association of the CagA EPIYA-C motifs and disease severity may be attributed to geographical differences in the EPIYA motifs themselves.111,116 Conversely, the conflicting data may suggest that other factors must affect the ultimate role of CagA polymorphism on disease.
The idea that geographical diversity affects disease outcome is supported by molecular epidemiological studies of H. pylori strains that carry East Asian CagA. Early evaluation of Korean H. pylori strains did not find an association between disease and higher numbers of EPIYA-C motifs.117 It should be noted that the findings of these studies are likely affected by sample size and the fact that East Asian strains typically carry the EPIYA-ABD motif. Indeed, work by Argent et al. found that 88.3% of East Asian strains harbored the EPIYA-ABD allele and these authors suggested that Western strains may in fact increase the number of EPIYA-C motifs as a means to increase virulence.98 As such, co-culture studies of strains carrying various EPIYA motifs showed that East Asian strains expressing the EPIYA-ABD motif induced significantly more IL-8 secretion than Western strains with a single EPIYA-C motif; yet as the number of EPIYA-C motifs increased, the amount of IL-8 secreted also significantly increased.98 Analysis of H. pylori strains from Japanese cancer patients provided the first evidence for the role of the EPIYA-ABD allele in H. pylori CagA associated carcinoma; all of the isolated strains carried the EPIYA-D motif.104 Additionally, sera from Japanese patients was highly reactive to a 100 kDa fragment of CagA that was isolated from Japanese clinical isolates using a high-molecular weight cell-associated proteins (HM-CAP) assay.118 In contrast, when this assay was performed using isolates from the United States, a larger CagA fragment that was not as reactive as the Japanese HM-CAP fragment was generated.118 These different sized cleavage products suggest that there may also be structural differences in the various CagA proteins.118 Of note, these proposed structural differences do not appear to alter the stability of CagA within the host cell since CagA EPIYA-ABD and CagA EPIYA-ABCCC do not exhibit a statistically significant difference in their half-lives.83 Finally, the first definitive evidence for the role of the EPIYA-ABD motif in cancer development was obtained from a large scale molecular epidemiological study of South Korean H. pylori isolates; this work was the first to show that the presence of an EPIYA-ABD motif was statistically associated with the development of gastric cancer.119 Taken together, current studies suggest that gastric cancer in East Asian countries is most often associated with the EPIYA-ABD motif119 and more severe disease in Western countries is associated with multiple EPIYA-C motifs.109-112
CagA was first classified as an oncoprotein following studies using a C57BL/6J transgenic mouse model in which CagA containing an EPIYA-ABDD was expressed in the stomach of these animals.120,121 This resulted in the formation of gastric polyps and adenocarcinoma of the stomach and small intestine; this was dependent on tyrosine phosphorylation of CagA and deregulation of SHP-2 120. Notably, due to transgenic expression in this model, CagA EPIYA-ABDD also became systemically expressed and caused hematological abnormalities in some animals . Development of these conditions is attributed to the deregulation of SHP-2 by CagA EPIYA-ABDD,120 which is known to avidly bind and activate SHP-2 105. Interestingly, gastric tumor formation in the CagA EPIYA-ABDD transgenic mice occurs in the absence of overt mucosal inflammation and metaplastic change, suggesting that the formation of gastric carcinoma may not require chronic inflammation.120 This is in contrast to the current belief that inflammation is crucial for the formation of precancerous lesions, which are believed to result from pathogenic T-cell mediated immune responses. Indeed, the absence of lymphocytes results in resistance to gastritis.53,58 To study the role of the Western EPIYA motif in CagA induced carcinogenesis, a second transgenic mouse model was also developed.121 Expression of CagA containing the EPIYA-ABCCC motif in these animals induced thickening of the gastric mucosa comparable to that seen in the EPIYA-ABDD transgenic model.121 Similar to the earlier study,120 Western CagA induced tumor formation in the absence of chronic inflammation and intestinal metaplasia; however, the incidence of tumor formation was less than that induced by CagA EPIYA-ABDD and no hematological abnormalities developed.121 Interestingly, both studies showed that once the CagA-induced transformed phenotype was established, CagA was no longer required for disease progression indicating that CagA plays a role in the early steps of gastric carcinoma.120,121 The discrepancy that tumor formation was induced in the absence of inflammation, a condition previously suggested to be required for carcinogenesis in humans,121 may be a side effect of the use of a transgene model. Together, these findings suggest that while Western CagA can induce tumor formation, it appears less potent than East Asian CagA. Given the differential effects on SHP-2, the authors suggest that tumor formation requires a threshold level of SHP-2 activation; the increase in SHP-2 activity induced by East Asian CagA may be the contributing factor to the increase in gastric carcinoma in East Asian countries.121
At the same time that the transgenic mouse model was developed, a CagA transgenic Drosophila model also emerged.122 This model showed that expression of CagA results in apical targeting and epithelial disorganization and that CagA mimics the eukaryotic Grb2-associated binder (Gab) adaptor protein that can activate SHP-2 122. However, later work showed that in this model, targeting of CagA to the apical region of the epithelium depends on EPIYA motif activation of myosin light chain (MLC) and not SHP-2 123. Furthermore, work using a S2 cell culture model showed that CagA activates MLC in a Rho and Rho-Kinase (ROCK) dependent manner.123 The role of Rho/ROCK in epithelial disruption is consistent with an earlier study conducted in AGS cells that shows that the characteristic “hummingbird phenotype” induced by CagA resulted from a failure of the trailing edge to retract, a process regulated by Rho and ROCK.124 Those studies suggest that Rho and MLC, rather than SHP-2, may be the critical host cell proteins required for epithelial disruption.123 Overall, the use of different cell culture and transgenic models has led to a dispute concerning exactly how CagA affects host cell signaling pathways. As such, it is clear that a common robust infection model is needed to tweeze out the mechanism underlying CagA carcinogenesis.
VacA
A second important toxin in H. pylori’s arsenal of virulence factors is VacA. vacA was discovered in 1988 and unlike cagA, which is not found in all strains, is present in virtually all H. pylori strains (refer to reviews125,126). VacA is a pore forming toxin that is initially expressed as a 140 kDa protoxin and, following secretion via an autotransporter mechanism, is cleaved into a mature 88 kDa secreted toxin.127 This secreted form is further cleaved to form 33 kDa and 55 kDa fragments termed p33 and p55, respectively (reviewed in126,128,129). Though clearly not prototypical, VacA has been suggested to have structural similarity to AB toxins.130 Moreover, recent discussion suggests that VacA could serve as a model for a new subtype of AB toxins, whereby the A subunit (comprised of p33) is responsible for pore formation rather than enzymatic activity, and the B subunit (composed of p55) is responsible for interaction with the host cell membrane.128 In order for VacA to induce its toxic effects, it binds to sphingomyelin or the receptor type protein-tyrosine phosphatases RPTPα and RPTPβ, both of which are contained within eukaryotic cell membrane lipid rafts.131-133 Following Cdc42 dependent pinocytic uptake, VacA is trafficked to the mitochondria where it induces apoptosis and formation of large intracellular vacuoles .127,134 While the mechanism underlying VacA targeting to the mitochondria remains unclear, it is clear that VacA induced pore formation in the mitochondrial membrane is responsible for induction of apoptosis (reviewed in126,129,135).
As a vast majority of information is known about how VacA functions and signals in host cells (refer to reviews78,126,129,135,136), herein, we will only briefly mention some discoveries that affect our understanding of how VacA polymorphisms influence disease state. Many molecular epidemiological studies suggest that VacA is important for disease (discussed below). Furthermore, analysis of sera from patients with intestinal metaplasia, gastric carcinoma, duodenal ulcers, and non-atrophic gastritis suggests an association between neutralizing VacA antibodies and gastric carcinoma; the amount of neutralizing antibodies tended to increase as patients progressed from pre-neoplastic lesions to those with cancer.137 Additionally, serum-neutralizing antibodies appear to depend on vacA polymorphism (discussed below), and antibodies to VacA also appear to persist longer than antibodies to H. pylori itself; in patients who tested negative for H. pylori but positive for VacA, the risk for gastric carcinoma was increased compared with those who tested positive for both.137
VacA polymorphisms
Similar to cagA, vacA also contains allelic diversity that varies across H. pylori strains.138 VacA contains four identified variable regions, each of which are subdivided into types (Fig. 3). The best characterized regions are the signal (s) region, which encompasses the N-terminus and a portion of the signal sequence and is typed as s1 or s2; and the mid (m) region of the gene, which is typed as m1 or m2.138 Since they were only recently discovered, much less is known about the role of the intermediate (i) region or the deletion (d) region in disease progression.139,140 The i-region lies between the s- and m-regions and is typed as i1, i2, or i3 (Fig. 3).139,141 The d-region, located between the i- and m-regions is typed as d1 if there is no deletion (Fig. 3A), or d2 if a 69 to 89 base pair deletion is present (Fig. 3B).140
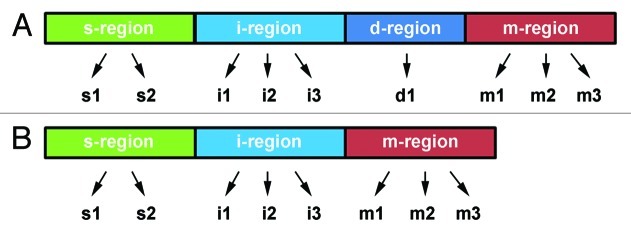
Figure 3.vacA polymorphism. Like cagA, vacA contains polymorphism that results in VacA toxins with varying degrees of virulence. Each region of the gene has multiple alleles (indicated by arrows), which can combine in any combination. (A) While the s-, i-, and m-regions are always present, the presence of the d-region is termed d1. (B) VacA that contains a 69–89 base pair deletion between the i- and m-regions is termed d2. Figure was modified from.135
vacA can recombine to form a wide variety of combinations, some of which are less common than others. Of the possible allelic combinations, the vacA s1/m1 alleles are the most virulent combination, while the s1/m2 and s2/m2 genotypes display virtually no cytotoxicity.138,142-147 Subsequent identification of the i-region showed that the i1 polymorphism is a significant, independent risk factor for gastric carcinoma139 and is strongly associated with the s1 polymorphism, both of which are associated with peptic ulcers.148 As such, it has been suggested that typing the i-region of VacA may suffice to determine the pathogenic potential of the toxin.139,148 While only one study has investigated the role of the d-region in disease, this study showed that the s1, m1, i1, and d1 polymorphisms significantly increase the risk for gastric cancer in Western countries.140 Based on the results of that and a previous study by the same group, it was suggested that the d-region polymorphism could be used as a better predictor for disease.140 Together, while each of the vacA polymorphisms has been used as a predictor for VacA-induced disease severity, it is clear that other factors, such as the presence of CagA (discussed below), contribute to disease. For this reason individually typing vacA alone may not provide sufficient information to understand the virulence potential of a strain and typing of multiple virulence factors appears to be required to understand strain dependent disease contributions.
Epidemiological Connection Between CagA and VacA
Since the mid-1990s, studies have attempted to correlate the independent presence of CagA, VacA, and several other virulence factors with disease state, oftentimes producing mixed results (discussed below). Studies conducted in Senegal, Taiwan, South Korea, Greece, Turkey, Alaska, and Europe identified a correlation between CagA status and more severe disease states (duodenal ulcers, peptic ulcer disease, progression of preneoplastic lesions, and gastric cancer).42,144,149-156 Additionally, patients identified as VacA seropositive were shown to have an increased risk of gastric carcinoma that occurred in conjunction with the s1/m1 polymorphism.157-159 It has become clear that looking at these toxins independently is insufficient. Thus, examination of the combined cagA and vacA status of a strain has shown a significant association between the presence of both genes and disease state.144,148,151-154,160 These studies show that most cagA positive H. pylori strains are associated with the vacA s1 and/or m1 genotypes.42,99,144,150-155,161-164 Together these studies support the notion that individuals who carry cagA positive, vacA s1/m1 H. pylori isolates may be candidates for eradication therapy as a means to prevent severe disease (duodenal ulcers, peptic ulcer disease, progression of preneoplastic lesions, and gastric cancer).
Despite the evidence that cagA positivity, cagA and vacA seropositivity, and/or vacA polymorphism contribute to disease severity, numerous studies have not found this association.163-174 For example, in Tunisia, vacA type is significantly different between patients with peptic ulceration and gastritis, while cagA status is not.175 In China, no association between cagA status and peptic ulceration or chronic gastritis was established, likely due to the high presence of cagA in both patient populations.164 The differences observed between disease severity and toxin type/presence in these epidemiological studies may be due to differences that exist in H. pylori strains well beyond the described cagA and vacA polymorphisms. As noted at the outset of this review, environmental, geographic, and host influences could contribute to the differences observed in disease severity between these studies. As such, while individual evaluation of cagA and vacA genotypes show that both contribute to disease; the lack of evaluation of both genotypes in combination with other factors is problematic for determining how both toxins contribute to disease.149
Functional Interaction Between CagA and VacA
While numerous epidemiological studies have been conducted to determine how VacA and CagA are distributed between populations, the area that clearly remains understudied is the functional connection between the two toxins (this topic has also recently been reviewed in129). The first study investigating the interaction between CagA and VacA found that CagA contains an amino acid sequence that resembles the immune-receptor tyrosine based activation motif (ITAM) that is found on the cytoplasmic region of immune receptors; tyrosine phosphorylated CagA localizes to and clusters in lipid rafts.132 Furthermore, CagA can associate with receptor tyrosine phosphorylated GIT1/Cat1, which is a substrate of the VacA receptor RPTPα/β.132,133 The authors suggested that the interaction between the toxins in lipid rafts results in negative regulation; VacA appears responsible for reduced tyrosine phosphorylation of CagA during early stages of infection. A second study illustrated another CagA and VacA functional interaction. This study showed that CagA activates the PLC-γ-Ca2+-calcineurin pathway, which results in translocation of NFAT to the nucleus and transcription of NFAT regulated genes.176 To counteract CagA-induced effects on host cell signaling, VacA inhibits T-cell Ca2+ uptake thus preventing dephosphorylation of calcineurin and translocation of NFAT to the nucleus.176
While CagA and VacA can prevent host cell morphological changes induced by the other, it remains unclear how this process occurs.177 A study investigating host cell signaling pathways showed that VacA prevents CagA-induced cell elongation by suppressing the EGRF and HER2/Neu receptors, resulting in inhibition of the Erk1/2 kinase pathway.178 To prevent VacA-induced host cell apoptosis, phosphorylated CagA decreases SKF tyrosine activity within the host cell and blocks trafficking of VacA containing compartments the mitochondria; CagA does not appear to inhibit pinocytotic uptake of VacA.179 Therefore, it was suggested that for the small amount of VacA that reaches the mitochondria, non-phosphorylated CagA may induce nuclear translocation of NF-κB and production of IL-8 as a means to block VacA induced apoptosis.179 In direct contrast, a later study showed that in host cells with attached H. pylori, translocated CagA inhibits VacA-induced apoptosis by preventing pinocytotic uptake of VacA, resulting in a decrease in vacuole formation within the host cell.180 Moreover, uninfected cells were susceptible to VacA induced apoptosis.180 Finally in terms of CagA and VacA interaction, a recent study revealed that H. pylori may utilize both CagA and VacA for acquisition of iron from mucosal epithelial cells.181 In this model, CagA increases transferrin uptake from the basolateral surface in an EPIYA dependent manner, delivering it to the apical surface to be released into the lumen, while VacA increases transferrin-receptor uptake from the basolateral surface, delivering it to sites of bacterial attachment.181 Akada et al. suggest that when H. pylori attach to gastric epithelial cells, CagA and VacA may interact in a manner that facilitates persistent colonization since CagA prevents VacA-mediated apoptosis. In contrast, when H. pylori are planktonic, VacA induces cellular apoptosis in both infiltrating immune cells and epithelial cells to acquire nutrients required for H. pylori survival.180
While these studies opened the door to understanding how CagA and VacA functionally interact and outline a direction for future studies, currently few studies have actually investigated in detail the combined role of CagA and VacA polymorphism in relation to disease state. One such study performed sequence analysis of the VacA i-region of H. pylori isolated from South Korean patients.182 In this study, polymorphism at amino acid 196 is associated with more severe disease, and polymorphism at amino acid 231 is linked to strains that do not carry the CagA EPIYA-ABD allele. Together, the data suggest that polymorphisms within each toxin may affect the functional interaction between the two toxins.182 Clearly, more intense study is required in this area.
Conclusion
Current evidence supports the hypothesis that H. pylori was acquired by humans prior to the great human migration out of Africa. As such, H. pylori has evolved mechanisms to persist within the harsh human gastric environment and may even serve to protect humans from diseases such as asthma, allergy, and esophageal cancer. While half of the world’s population is infected with H. pylori, only a subset of individuals actually develop H. pylori induced disease. As such, a debate exists as to whether H. pylori should be eradicated from the human microbiome. Since recent studies suggest that numerous factors (Fig. 4) contribute to the establishment of disease, it perhaps seems more appropriate to individually assess each infected individual for eradication therapy. As the evidence is starting to suggest that H. pylori protects us against several disease states, overtly eradicating H. pylori from the human microbiome may open “Pandora’s Box” and be more detrimental than helpful to overall human health. However, if we can understand the mechanisms underlying the conversion of H. pylori from the commensal found in most of the world’s population, to the pathogen only found in a subset of individuals, we would have the means to selectively treat those who are at the greatest risk for H. pylori induced gastric carcinoma. As such, there are several questions regarding the pathogenic mechanism of H. pylori that require further study: (1) How do the different CagA EPIYA motifs affect host cell signaling pathways; (2) Does CagA inhibit VacA induced apoptosis at the step of pinocytosis or does CagA target a protein required for vesicular transport within the host to inhibit VacA trafficking; (3) How do the polymorphisms in each toxin affect the ability of CagA and VacA to functionally interact; and (4) How does this interaction ultimately contribute to disease severity? Future studies will aim to answer these questions in order to provide a better understanding of H. pylori –induced gastric disease.
Figure 4. Balance between H. pylori as normal flora vs. a pathogen. The balance between H. pylori’s role in protecting against disease vs. causing gastric pathologies depends on bacterial, host and environmental factors. As such, based on the combination of these factors in any one individual, it may be detrimental to eradicate H. pylori from the human stomach.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
Research in the Merrell laboratory is made possible by grants from NIH, DOD, USMCI and USUHS. The contents of this manuscript represent the views of the authors and not those of the NIH or DOD. We would like to thank Dr. Beth Carpenter for critical review of the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/23797
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–5. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127–31. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J, Hansen S, Rodriguez L, Gelb AB, Warnke RA, Jellum E, et al. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–71. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard DM, Crabtree JE. Helicobacter pylori and gastric cancer. Curr Opin Gastroenterol. 2006;22:620–5. doi: 10.1097/01.mog.0000245539.50765.f6. [DOI] [PubMed] [Google Scholar]
- 5.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 6.Infection with Helicobacter pylori. IARC monographs on the evaluation of carcinogenic risks to humans / World Health Organization. International Agency for Research on Cancer. 1994;61:177–240. [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Perez GI, Salomaa A, Kosunen TU, Daverman B, Rautelin H, Aromaa A, et al. Evidence that cagA(+) Helicobacter pylori strains are disappearing more rapidly than cagA(-) strains. Gut. 2002;50:295–8. doi: 10.1136/gut.50.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–33. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 9.Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–8. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersulyte D, Kalia A, Gilman RH, Mendez M, Herrera P, Cabrera L, et al. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5:e15076. doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirth T, Wang X, Linz B, Novick RP, Lum JK, Blaser M, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc Natl Acad Sci U S A. 2004;101:4746–51. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moodley Y, Linz B. Helicobacter pylori Sequences Reflect Past Human Migrations. Genome Dyn. 2009;6:62–74. doi: 10.1159/000235763. [DOI] [PubMed] [Google Scholar]
- 13.Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–5. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 14.Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, Wu JY, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–30. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648–53. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci U S A. 1999;96:14559–64. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Z, Censini S, Bayeli PF, Telford JL, Figura N, Rappuoli R, et al. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect Immun. 1995;63:94–8. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaoka Y, Osato MS, Sepulveda AR, Gutierrez O, Figura N, Kim JG, et al. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91–6. doi: 10.1017/S0950268899003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 20.Kersulyte D, Mukhopadhyay AK, Velapatiño B, Su W, Pan Z, Garcia C, et al. Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol. 2000;182:3210–8. doi: 10.1128/JB.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43:3906–16. doi: 10.1128/JCM.43.8.3906-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong BX, Mai VT, Tanaka H, Ly T, Thong TM, Hai HH, et al. Diverse characteristics of the CagA gene of Helicobacter pylori strains collected from patients from southern vietnam with gastric cancer and peptic ulcer. J Clin Microbiol. 2009;47:4021–8. doi: 10.1128/JCM.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duncan SS, Valk PL, Shaffer CL, Bordenstein SR, Cover TL. J-Western forms of Helicobacter pylori cagA constitute a distinct phylogenetic group with a widespread geographic distribution. J Bacteriol. 2012;194:1593–604. doi: 10.1128/JB.06340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mane SP, Dominguez-Bello MG, Blaser MJ, Sobral BW, Hontecillas R, Skoneczka J, et al. Host-interactive genes in Amerindian Helicobacter pylori diverge from their Old World homologs and mediate inflammatory responses. J Bacteriol. 2010;192:3078–92. doi: 10.1128/JB.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki M, Kiga K, Kersulyte D, Cok J, Hooper CC, Mimuro H, et al. Attenuated CagA oncoprotein in Helicobacter pylori from Amerindians in Peruvian Amazon. J Biol Chem. 2011;286:29964–72. doi: 10.1074/jbc.M111.263715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correa P, Piazuelo MB. Evolutionary History of the Helicobacter pylori Genome: Implications for Gastric Carcinogenesis. Gut Liver. 2012;6:21–8. doi: 10.5009/gnl.2012.6.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devi SM, Ahmed I, Francalacci P, Hussain MA, Akhter Y, Alvi A, et al. Ancestral European roots of Helicobacter pylori in India. BMC Genomics. 2007;8:184. doi: 10.1186/1471-2164-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tay CY, Mitchell H, Dong Q, Goh KL, Dawes IW, Lan R. Population structure of Helicobacter pylori among ethnic groups in Malaysia: recent acquisition of the bacterium by the Malay population. BMC Microbiol. 2009;9:126. doi: 10.1186/1471-2180-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zevit N, Balicer RD, Cohen HA, Karsh D, Niv Y, Shamir R. Inverse association between Helicobacter pylori and pediatric asthma in a high-prevalence population. Helicobacter. 2012;17:30–5. doi: 10.1111/j.1523-5378.2011.00895.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198:553–60. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167:821–7. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 32.Reibman J, Marmor M, Filner J, Fernandez-Beros ME, Rogers L, Perez-Perez GI, et al. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS One. 2008;3:e4060. doi: 10.1371/journal.pone.0004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–60. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry S, de Jong BC, Solnick JV, de la Luz Sanchez M, Yang S, Lin PL, et al. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5:e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry S, Chang AH, Sanchez L, Yang S, Haggerty TD, Parsonnet J. The immune response to tuberculosis infection in the setting of Helicobacter pylori and helminth infections. Epidemiol Infect. 2012;•••:1–12. doi: 10.1017/S0950268812001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francois F, Roper J, Joseph N, Pei Z, Chhada A, Shak JR, et al. The effect of H. pylori eradication on meal-associated changes in plasma ghrelin and leptin. BMC Gastroenterol. 2011;11:37. doi: 10.1186/1471-230X-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malfertheiner P, Selgrad M. Helicobacter pylori infection and current clinical areas of contention. Curr Opin Gastroenterol. 2010;26:618–23. doi: 10.1097/MOG.0b013e32833efede. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Blaser MJ. Association between gastric Helicobacter pylori colonization and glycated hemoglobin levels. J Infect Dis. 2012;205:1195–202. doi: 10.1093/infdis/jis106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rokkas T, Pistiolas D, Sechopoulos P, Robotis I, Margantinis G. Relationship between Helicobacter pylori infection and esophageal neoplasia: a meta-analysis. Clin Gastroenterol Hepatol. 2007;5:1413–7, 1417, e1-2. doi: 10.1016/j.cgh.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 40.de Martel C, Llosa AE, Farr SM, Friedman GD, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of development of esophageal adenocarcinoma. J Infect Dis. 2005;191:761–7. doi: 10.1086/427659. [DOI] [PubMed] [Google Scholar]
- 41.Vaezi MF, Falk GW, Peek RM, Vicari JJ, Goldblum JR, Perez-Perez GI, et al. CagA-positive strains of Helicobacter pylori may protect against Barrett’s esophagus. Am J Gastroenterol. 2000;95:2206–11. doi: 10.1111/j.1572-0241.2000.02305.x. [DOI] [PubMed] [Google Scholar]
- 42.Miernyk K, Morris J, Bruden D, McMahon B, Hurlburt D, Sacco F, et al. Characterization of Helicobacter pylori cagA and vacA genotypes among Alaskans and their correlation with clinical disease. J Clin Microbiol. 2011;49:3114–21. doi: 10.1128/JCM.00469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corley DA, Kubo A, Levin TR, Block G, Habel L, Zhao W, et al. Helicobacter pylori infection and the risk of Barrett’s oesophagus: a community-based study. Gut. 2008;57:727–33. doi: 10.1136/gut.2007.132068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corley DA, Kubo A, Levin TR, Block G, Habel L, Rumore G, et al. Helicobacter pylori and gastroesophageal reflux disease: a case-control study. Helicobacter. 2008;13:352–60. doi: 10.1111/j.1523-5378.2008.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischbach LA, Nordenstedt H, Kramer JR, Gandhi S, Dick-Onuoha S, Lewis A, et al. The association between Barrett’s esophagus and Helicobacter pylori infection: a meta-analysis. Helicobacter. 2012;17:163–75. doi: 10.1111/j.1523-5378.2011.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrell DS, Falkow S. Frontal and stealth attack strategies in microbial pathogenesis. Nature. 2004;430:250–6. doi: 10.1038/nature02760. [DOI] [PubMed] [Google Scholar]
- 47.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–73. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorer MS, Talarico S, Salama NR. Helicobacter pylori’s unconventional role in health and disease. PLoS Pathog. 2009;5:e1000544. doi: 10.1371/journal.ppat.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237–59. doi: 10.1016/j.bpg.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Arnold IC, Hitzler I, Müller A. The Immunomodulatory Properties of Helicobacter pylori Confer Protection Against Allergic and Chronic Inflammatory Disorders. Front Cell Infect Microbiol. 2012;2:10. doi: 10.3389/fcimb.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabir S. The role of interleukin-17 in the Helicobacter pylori induced infection and immunity. Helicobacter. 2011;16:1–8. doi: 10.1111/j.1523-5378.2010.00812.x. [DOI] [PubMed] [Google Scholar]
- 52.Eaton KA, Ringler SR, Danon SJ. Murine splenocytes induce severe gastritis and delayed-type hypersensitivity and suppress bacterial colonization in Helicobacter pylori-infected SCID mice. Infect Immun. 1999;67:4594–602. doi: 10.1128/iai.67.9.4594-4602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson K, Kenefeck R, Pidgeon EL, Shakib S, Patel S, Polson RJ, et al. Helicobacter pylori-induced peptic ulcer disease is associated with inadequate regulatory T cell responses. Gut. 2008;57:1375–85. doi: 10.1136/gut.2007.137539. [DOI] [PubMed] [Google Scholar]
- 54.Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, et al. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology. 2011;140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris PR, Wright SW, Serrano C, Riera F, Duarte I, Torres J, et al. Helicobacter pylori gastritis in children is associated with a regulatory T-cell response. Gastroenterology. 2008;134:491–9. doi: 10.1053/j.gastro.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Sharma SA, Tummuru MK, Blaser MJ, Kerr LD. Activation of IL-8 gene expression by Helicobacter pylori is regulated by transcription factor nuclear factor-kappa B in gastric epithelial cells. J Immunol. 1998;160:2401–7. [PubMed] [Google Scholar]
- 57.Crabtree JE, Xiang Z, Lindley IJ, Tompkins DS, Rappuoli R, Covacci A. Induction of interleukin-8 secretion from gastric epithelial cells by a cagA negative isogenic mutant of Helicobacter pylori. J Clin Pathol. 1995;48:967–9. doi: 10.1136/jcp.48.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hitzler I, Kohler E, Engler DB, Yazgan AS, Müller A. The role of Th cell subsets in the control of Helicobacter infections and in T cell-driven gastric immunopathology. Front Immunol. 2012;3:142. doi: 10.3389/fimmu.2012.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horvath DJ, Jr., Washington MK, Cope VA, Algood HM. IL-23 Contributes to Control of Chronic Helicobacter Pylori Infection and the Development of T Helper Responses in a Mouse Model. Front Immunol. 2012;3:56. doi: 10.3389/fimmu.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serelli-Lee V, Ling KL, Ho C, Yeong LH, Lim GK, Ho B, et al. Persistent Helicobacter pylori specific Th17 responses in patients with past H. pylori infection are associated with elevated gastric mucosal IL-1β. PLoS One. 2012;7:e39199. doi: 10.1371/journal.pone.0039199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.López-Carrillo L, Camargo MC, Schneider BG, Sicinschi LA, Hernández-Ramírez RU, Correa P, et al. Capsaicin consumption, Helicobacter pylori CagA status and IL1B-31C>T genotypes: a host and environment interaction in gastric cancer. Food Chem Toxicol. 2012;50:2118–22. doi: 10.1016/j.fct.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.López-Carrillo L, López-Cervantes M, Robles-Díaz G, Ramírez-Espitia A, Mohar-Betancourt A, Meneses-García A, et al. Capsaicin consumption, Helicobacter pylori positivity and gastric cancer in Mexico. Int J Cancer. 2003;106:277–82. doi: 10.1002/ijc.11195. [DOI] [PubMed] [Google Scholar]
- 63.Liu H, Merrell DS, Semino-Mora C, Goldman M, Rahman A, Mog S, et al. Diet synergistically affects helicobacter pylori-induced gastric carcinogenesis in nonhuman primates. Gastroenterology. 2009;137:1367–79, e1-6. doi: 10.1053/j.gastro.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.González CA, López-Carrillo L. Helicobacter pylori, nutrition and smoking interactions: their impact in gastric carcinogenesis. Scand J Gastroenterol. 2010;45:6–14. doi: 10.3109/00365520903401959. [DOI] [PubMed] [Google Scholar]
- 65.Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 66.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci U S A. 2000;97:1263–8. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–4. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tanaka H, Yoshida M, Nishiumi S, Ohnishi N, Kobayashi K, Yamamoto K, et al. The CagA protein of Helicobacter pylori suppresses the functions of dendritic cell in mice. Arch Biochem Biophys. 2010;498:35–42. doi: 10.1016/j.abb.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 70.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A. 1993;90:5791–5. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe. 2010;7:399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 72.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277:6775–8. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 73.Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43:971–80. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 74.Poppe M, Feller SM, Römer G, Wessler S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene. 2007;26:3462–72. doi: 10.1038/sj.onc.1210139. [DOI] [PubMed] [Google Scholar]
- 75.Tammer I, Brandt S, Hartig R, König W, Backert S. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132:1309–19. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 76.Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori Cag. Protein Sci. 2002;295:683–6. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 77.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–76. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 78.Jones KR, Whitmire JM, Merrell DS. A Tale of Two Toxins: Helicobacter Pylori CagA and VacA Modulate Host Pathways that Impact Disease. Front Microbiol. 2010;1:115. doi: 10.3389/fmicb.2010.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hatakeyama M. Helicobacter pylori and gastric carcinogenesis. J Gastroenterol. 2009;44:239–48. doi: 10.1007/s00535-009-0014-1. [DOI] [PubMed] [Google Scholar]
- 80.Higashi H, Nakaya A, Tsutsumi R, Yokoyama K, Fujii Y, Ishikawa S, et al. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J Biol Chem. 2004;279:17205–16. doi: 10.1074/jbc.M309964200. [DOI] [PubMed] [Google Scholar]
- 81.Kikuchi K, Murata-Kamiya N, Kondo S, Hatakeyama M. Helicobacter pylori stimulates epithelial cell migration via CagA-mediated perturbation of host cell signaling. Microbes Infect. 2012;14:470–6. doi: 10.1016/j.micinf.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 82.Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, Saito Y, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447:330–3. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 83.Ishikawa S, Ohta T, Hatakeyama M. Stability of Helicobacter pylori CagA oncoprotein in human gastric epithelial cells. FEBS Lett. 2009;583:2414–8. doi: 10.1016/j.febslet.2009.06.043. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 85.Sohn SH, Lee YC. The genome-wide expression profile of gastric epithelial cells infected by naturally occurring cagA isogenic strains of Helicobacter pylori. Environ Toxicol Pharmacol. 2011;32:382–9. doi: 10.1016/j.etap.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 86.Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci U S A. 2005;102:16339–44. doi: 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Higashi H, Yokoyama K, Fujii Y, Ren S, Yuasa H, Saadat I, et al. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J Biol Chem. 2005;280:23130–7. doi: 10.1074/jbc.M503583200. [DOI] [PubMed] [Google Scholar]
- 88.Pelz C, Steininger S, Weiss C, Coscia F, Vogelmann R. A novel inhibitory domain of Helicobacter pylori protein CagA reduces CagA effects on host cell biology. J Biol Chem. 2011;286:8999–9008. doi: 10.1074/jbc.M110.166504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steininger S, Pelz C, Vogelmann R. Purpose of recently detected inhibitory domain of the Helicobacter pylori protein CagA. Gut Microbes. 2011;2:167–72. doi: 10.4161/gmic.2.3.15872. [DOI] [PubMed] [Google Scholar]
- 90.Hayashi T, Senda M, Morohashi H, Higashi H, Horio M, Kashiba Y, et al. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe. 2012;12:20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 91.Nesić D, Miller MC, Quinkert ZT, Stein M, Chait BT, Stebbins CE. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17:130–2. doi: 10.1038/nsmb.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Backert S, Tegtmeyer N. Helicobacter pylori CagA tertiary structure reveals functional insights. Cell Host Microbe. 2012;12:3–5. doi: 10.1016/j.chom.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 93.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, et al. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci U S A. 2002;99:14428–33. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ren S, Higashi H, Lu H, Azuma T, Hatakeyama M. Structural basis and functional consequence of Helicobacter pylori CagA multimerization in cells. J Biol Chem. 2006;281:32344–52. doi: 10.1074/jbc.M606172200. [DOI] [PubMed] [Google Scholar]
- 95.van der Wouden JM, Maier O, van IJzendoorn SC, Hoekstra D. Membrane dynamics and the regulation of epithelial cell polarity. Int Rev Cytol. 2003;226:127–64. doi: 10.1016/S0074-7696(03)01003-9. [DOI] [PubMed] [Google Scholar]
- 96.Wang Q, Margolis B. Apical junctional complexes and cell polarity. Kidney Int. 2007;72:1448–58. doi: 10.1038/sj.ki.5002579. [DOI] [PubMed] [Google Scholar]
- 97.van Staveren WC, Solís DY, Hébrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 98.Argent RH, Hale JL, El-Omar EM, Atherton JC. Differences in Helicobacter pylori CagA tyrosine phosphorylation motif patterns between western and East Asian strains, and influences on interleukin-8 secretion. J Med Microbiol. 2008;57:1062–7. doi: 10.1099/jmm.0.2008/001818-0. [DOI] [PubMed] [Google Scholar]
- 99.Camorlinga-Ponce M, Perez-Perez G, Gonzalez-Valencia G, Mendoza I, Peñaloza-Espinosa R, Ramos I, et al. Helicobacter pylori genotyping from American indigenous groups shows novel Amerindian vacA and cagA alleles and Asian, African and European admixture. PLoS One. 2011;6:e27212. doi: 10.1371/journal.pone.0027212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shiota S, Matsunari O, Yamaoka Y. Relationship between J-Western CagA subtype and the vacA m2 region of Helicobacter pylori. J Clin Microbiol. 2010;48:3033–4. doi: 10.1128/JCM.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Furuta Y, Yahara K, Hatakeyama M, Kobayashi I. Evolution of cagA oncogene of Helicobacter pylori through recombination. PLoS One. 2011;6:e23499. doi: 10.1371/journal.pone.0023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, et al. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest. 2012;122:1553–66. doi: 10.1172/JCI61143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hatakeyama M, Higashi H. Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis. Cancer Sci. 2005;96:835–43. doi: 10.1111/j.1349-7006.2005.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Azuma T, Yamazaki S, Yamakawa A, Ohtani M, Muramatsu A, Suto H, et al. Association between diversity in the Src homology 2 domain--containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J Infect Dis. 2004;189:820–7. doi: 10.1086/381782. [DOI] [PubMed] [Google Scholar]
- 105.Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S, et al. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology. 2006;130:1181–90. doi: 10.1053/j.gastro.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 106.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–94. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 107.Nagase L, Murata-Kamiya N, Hatakeyama M. Potentiation of Helicobacter pylori CagA protein virulence through homodimerization. J Biol Chem. 2011;286:33622–31. doi: 10.1074/jbc.M111.258673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsutsumi R, Higashi H, Higuchi M, Okada M, Hatakeyama M. Attenuation of Helicobacter pylori CagA x SHP-2 signaling by interaction between CagA and C-terminal Src kinase. J Biol Chem. 2003;278:3664–70. doi: 10.1074/jbc.M208155200. [DOI] [PubMed] [Google Scholar]
- 109.Yamaoka Y, El-Zimaity HM, Gutierrez O, Figura N, Kim JG, Kodama T, et al. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999;117:342–9. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 110.Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, et al. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91–9. doi: 10.1053/j.gastro.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 111.Batista SA, Rocha GA, Rocha AM, Saraiva IE, Cabral MM, Oliveira RC, et al. Higher number of Helicobacter pylori CagA EPIYA C phosphorylation sites increases the risk of gastric cancer, but not duodenal ulcer. BMC Microbiol. 2011;11:61. doi: 10.1186/1471-2180-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ferreira RM, Machado JC, Leite M, Carneiro F, Figueiredo C. The number of Helicobacter pylori CagA EPIYA C tyrosine phosphorylation motifs influences the pattern of gastritis and the development of gastric carcinoma. Histopathology. 2012;60:992–8. doi: 10.1111/j.1365-2559.2012.04190.x. [DOI] [PubMed] [Google Scholar]
- 113.Sicinschi LA, Correa P, Peek RM, Camargo MC, Piazuelo MB, Romero-Gallo J, et al. CagA C-terminal variations in Helicobacter pylori strains from Colombian patients with gastric precancerous lesions. Clin Microbiol Infect. 2010;16:369–78. doi: 10.1111/j.1469-0691.2009.02811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Acosta N, Quiroga A, Delgado P, Bravo MM, Jaramillo C. Helicobacter pylori CagA protein polymorphisms and their lack of association with pathogenesis. World J Gastroenterol. 2010;16:3936–43. doi: 10.3748/wjg.v16.i31.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rizzato C, Torres J, Plummer M, Muñoz N, Franceschi S, Camorlinga-Ponce M, et al. Variations in Helicobacter pylori cytotoxin-associated genes and their influence in progression to gastric cancer: implications for prevention. PLoS One. 2012;7:e29605. doi: 10.1371/journal.pone.0029605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Figura N, Moretti E, Roviello F, Papini F, Marrelli D. cagA structural types of Helicobacter pylori strains isolated from patients with gastric carcinoma and chronic gastritis only. Intern Emerg Med. 2012;7(Suppl 2):S103–5. doi: 10.1007/s11739-012-0759-z. [DOI] [PubMed] [Google Scholar]
- 117.Choi KD, Kim N, Lee DH, Kim JM, Kim JS, Jung HC, et al. Analysis of the 3′ variable region of the cagA gene of Helicobacter pylori isolated in Koreans. Dig Dis Sci. 2007;52:960–6. doi: 10.1007/s10620-005-9030-z. [DOI] [PubMed] [Google Scholar]
- 118.Yamada K, Sugiyama T, Mihara H, Kajiura S, Saito S, Itaya Y, et al. Fragmented CagA protein is highly immunoreactive in Japanese patients. Helicobacter. 2012;17:187–92. doi: 10.1111/j.1523-5378.2011.00930.x. [DOI] [PubMed] [Google Scholar]
- 119.Jones KR, Joo YM, Jang S, Yoo YJ, Lee HS, Chung IS, et al. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J Clin Microbiol. 2009;47:959–68. doi: 10.1128/JCM.02330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105:1003–8. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miura M, Ohnishi N, Tanaka S, Yanagiya K, Hatakeyama M. Differential oncogenic potential of geographically distinct Helicobacter pylori CagA isoforms in mice. Int J Cancer. 2009;125:2497–504. doi: 10.1002/ijc.24740. [DOI] [PubMed] [Google Scholar]
- 122.Botham CM, Wandler AM, Guillemin K. A transgenic Drosophila model demonstrates that the Helicobacter pylori CagA protein functions as a eukaryotic Gab adaptor. PLoS Pathog. 2008;4:e1000064. doi: 10.1371/journal.ppat.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muyskens JB, Guillemin K. Helicobacter pylori CagA disrupts epithelial patterning by activating myosin light chain. PLoS One. 2011;6:e17856. doi: 10.1371/journal.pone.0017856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bourzac KM, Botham CM, Guillemin K. Helicobacter pylori CagA induces AGS cell elongation through a cell retraction defect that is independent of Cdc42, Rac1, and Arp2/3. Infect Immun. 2007;75:1203–13. doi: 10.1128/IAI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113:321–33. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rassow J, Meinecke M. Helicobacter pylori VacA: a new perspective on an invasive chloride channel. Microbes Infect. 2012;14:1026–33. doi: 10.1016/j.micinf.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–9. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 128.Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol. 2012;20:165–74. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 129.Kim IJ, Blanke SR. Remodeling the host environment: modulation of the gastric epithelium by the Helicobacter pylori vacuolating toxin (VacA) Front Cell Infect Microbiol. 2012;2:37. doi: 10.3389/fcimb.2012.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, et al. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801–7. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. PLoS Pathog. 2008;4:e1000073. doi: 10.1371/journal.ppat.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Asahi M, Tanaka Y, Izumi T, Ito Y, Naiki H, Kersulyte D, et al. Helicobacter pylori CagA containing ITAM-like sequences localized to lipid rafts negatively regulates VacA-induced signaling in vivo. Helicobacter. 2003;8:1–14. doi: 10.1046/j.1523-5378.2003.00118.x. [DOI] [PubMed] [Google Scholar]
- 133.Schraw W, Li Y, McClain MS, van der Goot FG, Cover TL. Association of Helicobacter pylori vacuolating toxin (VacA) with lipid rafts. J Biol Chem. 2002;277:34642–50. doi: 10.1074/jbc.M203466200. [DOI] [PubMed] [Google Scholar]
- 134.Gupta VR, Wilson BA, Blanke SR. Sphingomyelin is important for the cellular entry and intracellular localization of Helicobacter pylori VacA. Cell Microbiol. 2010;12:1517–33. doi: 10.1111/j.1462-5822.2010.01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Backert S, Tegtmeyer N. the versatility of the Helicobacter pylori vacuolating cytotoxin vacA in signal transduction and molecular crosstalk. Toxins (Basel) 2010;2:69–92. doi: 10.3390/toxins2010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ayala G, Galván-Portillo M, Chihu L, Fierros G, Sánchez A, Carrillo B, et al. Study Group Resistance to antibiotics and characterization of Helicobacter pylori strains isolated from antrum and body from adults in Mexico. Microb Drug Resist. 2011;17:149–55. doi: 10.1089/mdr.2010.0154. [DOI] [PubMed] [Google Scholar]
- 138.Atherton JC, Cao P, Peek RM, Jr., Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–7. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 139.Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, et al. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology. 2007;133:926–36. doi: 10.1053/j.gastro.2007.06.056. [DOI] [PubMed] [Google Scholar]
- 140.Ogiwara H, Sugimoto M, Ohno T, Vilaichone RK, Mahachai V, Graham DY, et al. Role of deletion located between the intermediate and middle regions of the Helicobacter pylori vacA gene in cases of gastroduodenal diseases. J Clin Microbiol. 2009;47:3493–500. doi: 10.1128/JCM.00887-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chung C, Olivares A, Torres E, Yilmaz O, Cohen H, Perez-Perez G. Diversity of VacA intermediate region among Helicobacter pylori strains from several regions of the world. J Clin Microbiol. 2010;48:690–6. doi: 10.1128/JCM.01815-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meining A, Stolte M, Hatz R, Lehn N, Miehlke S, Morgner A, et al. Differing degree and distribution of gastritis in Helicobacter pylori-associated diseases. Virchows Arch. 1997;431:11–5. doi: 10.1007/s004280050063. [DOI] [PubMed] [Google Scholar]
- 143.Miehlke S, Hackelsberger A, Meining A, Hatz R, Lehn N, Malfertheiner P, et al. Severe expression of corpus gastritis is characteristic in gastric cancer patients infected with Helicobacter pylori. Br J Cancer. 1998;78:263–6. doi: 10.1038/bjc.1998.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Miehlke S, Kirsch C, Agha-Amiri K, Günther T, Lehn N, Malfertheiner P, et al. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int J Cancer. 2000;87:322–7. doi: 10.1002/1097-0215(20000801)87:3<322::AID-IJC3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 145.Letley DP, Rhead JL, Twells RJ, Dove B, Atherton JC. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. J Biol Chem. 2003;278:26734–41. doi: 10.1074/jbc.M304071200. [DOI] [PubMed] [Google Scholar]
- 146.McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, et al. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J Bacteriol. 2001;183:6499–508. doi: 10.1128/JB.183.22.6499-6508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Letley DP, Atherton JC. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J Bacteriol. 2000;182:3278–80. doi: 10.1128/JB.182.11.3278-3280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yordanov D, Boyanova L, Markovska R, Gergova G, Mitov I. Significance of Helicobacter pylori vacA intermediate region genotyping-a Bulgarian study. Diagn Microbiol Infect Dis. 2012;74:253–7. doi: 10.1016/j.diagmicrobio.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 149.Breurec S, Michel R, Seck A, Brisse S, Come D, Dieye FB, et al. Clinical relevance of cagA and vacA gene polymorphisms in Helicobacter pylori isolates from Senegalese patients. 2012;18:Clinical microbiology and infection. 153–9. doi: 10.1111/j.1469-0691.2011.03524.x. [DOI] [PubMed] [Google Scholar]
- 150.Jang S, Jones KR, Olsen CH, Joo YM, Yoo YJ, Chung IS, et al. Epidemiological link between gastric disease and polymorphisms in VacA and CagA. J Clin Microbiol. 2010;48:559–67. doi: 10.1128/JCM.01501-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Panayotopoulou EG, Sgouras DN, Papadakos KS, Petraki K, Breurec S, Michopoulos S, et al. CagA and VacA polymorphisms are associated with distinct pathological features in Helicobacter pylori-infected adults with peptic ulcer and non-peptic ulcer disease. J Clin Microbiol. 2010;48:2237–9. doi: 10.1128/JCM.00662-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Erzin Y, Koksal V, Altun S, Dobrucali A, Aslan M, Erdamar S, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter. 2006;11:574–80. doi: 10.1111/j.1523-5378.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- 153.van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/S0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 154.Warburton VJ, Everett S, Mapstone NP, Axon AT, Hawkey P, Dixon MF. Clinical and histological associations of cagA and vacA genotypes in Helicobacter pylori gastritis. J Clin Pathol. 1998;51:55–61. doi: 10.1136/jcp.51.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.González CA, Figueiredo C, Lic CB, Ferreira RM, Pardo ML, Ruiz Liso JM, et al. Helicobacter pylori cagA and vacA genotypes as predictors of progression of gastric preneoplastic lesions: a long-term follow-up in a high-risk area in Spain. Am J Gastroenterol. 2011;106:867–74. doi: 10.1038/ajg.2011.1. [DOI] [PubMed] [Google Scholar]
- 156.Markovska R, Boyanova L, Yordanov D, Gergova G, Mitov I. Helicobacter pylori oipA genetic diversity and its associations with both disease and cagA, vacA s, m, and i alleles among Bulgarian patients. Diagn Microbiol Infect Dis. 2011;71:335–40. doi: 10.1016/j.diagmicrobio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 157.Figueroa G, Troncoso M, Toledo MS, Faúndez G, Acuña R. Prevalence of serum antibodies to Helicobacter pylori VacA and CagA and gastric diseases in Chile. J Med Microbiol. 2002;51:300–4. doi: 10.1099/0022-1317-51-4-300. [DOI] [PubMed] [Google Scholar]
- 158.Rudi J, Kolb C, Maiwald M, Zuna I, von Herbay A, Galle PR, et al. Serum antibodies against Helicobacter pylori proteins VacA and CagA are associated with increased risk for gastric adenocarcinoma. Dig Dis Sci. 1997;42:1652–9. doi: 10.1023/A:1018849112533. [DOI] [PubMed] [Google Scholar]
- 159.de Figueiredo Soares T, de Magalhães Queiroz DM, Mendes EN, Rocha GA, Rocha Oliveira AM, Alvares Cabral MM, et al. The interrelationship between Helicobacter pylori vacuolating cytotoxin and gastric carcinoma. Am J Gastroenterol. 1998;93:1841–7. doi: 10.1016/S0002-9270(98)00414-6. [DOI] [PubMed] [Google Scholar]
- 160.Boukhris SA, Benajah DA, El Rhazi K, Ibrahimi SA, Nejjari C, Amarti A, et al. Prevalence and distribution of Helicobacter pylori cagA and vacA genotypes in the Moroccan population with gastric disease. European journal of clinical microbiology & infectious diseases. 2011 doi: 10.1007/s10096-011-1501-x. [DOI] [PubMed] [Google Scholar]
- 161.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter. 1998;3:241–53. doi: 10.1046/j.1523-5378.1998.08056.x. [DOI] [PubMed] [Google Scholar]
- 162.Ortiz-Princz D, Guariglia-Oropeza V, Avila M, Correnti M, Perrone M, Gutierrez B, et al. Helicobacter pylori cagA and vacA genotypes in Cuban and Venezuelan populations. Mem Inst Oswaldo Cruz. 2010;105:331–5. doi: 10.1590/S0074-02762010000300016. [DOI] [PubMed] [Google Scholar]
- 163.Perng CL, Lin HJ, Sun IC, Tseng GY, Facg Helicobacter pylori cagA, iceA and vacA status in Taiwanese patients with peptic ulcer and gastritis. J Gastroenterol Hepatol. 2003;18:1244–9. doi: 10.1046/j.1440-1746.2003.03214.x. [DOI] [PubMed] [Google Scholar]
- 164.Chen XJ, Yan J, Shen YF. Dominant cagA/vacA genotypes and coinfection frequency of H. pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases. Chin Med J (Engl) 2005;118:460–7. [PubMed] [Google Scholar]
- 165.Khodaii Z, Ghaderian SM, Akbarzadeh Najar R, Nejati H, Tabatabaei Panah AS. cagA and vacA status and influence of Helicobacter pylori infection on serum oxidative DNA damage in Iranian patients with peptic ulcer disease. Ir J Med Sci. 2011;180:155–61. doi: 10.1007/s11845-010-0548-5. [DOI] [PubMed] [Google Scholar]
- 166.Jafari F, Shokrzadeh L, Dabiri H, Baghaei K, Yamaoka Y, Zojaji H, et al. vacA genotypes of Helicobacter pylori in relation to cagA status and clinical outcomes in Iranian populations. Jpn J Infect Dis. 2008;61:290–3. [PMC free article] [PubMed] [Google Scholar]
- 167.Tuncel IE, Hussein NR, Bolek BK, Arikan S, Salih BA. Helicobacter pylori virulence factors and their role in peptic ulcer diseases in Turkey. Acta Gastroenterol Belg. 2010;73:235–8. [PubMed] [Google Scholar]
- 168.Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–9. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Andreson H, Lõivukene K, Sillakivi T, Maaroos HI, Ustav M, Peetsalu A, et al. Association of cagA and vacA genotypes of Helicobacter pylori with gastric diseases in Estonia. J Clin Microbiol. 2002;40:298–300. doi: 10.1128/JCM.40.1.298-300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, et al. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–4. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tanih NF, McMillan M, Naidoo N, Ndip LM, Weaver LT, Ndip RN. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in South African patients with upper gastrointestinal diseases. Acta Trop. 2010;116:68–73. doi: 10.1016/j.actatropica.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 172.Yamaoka Y, Kodama T, Kashima K, Graham DY. Antibody against Helicobacter pylori CagA and VacA and the risk for gastric cancer. J Clin Pathol. 1999;52:215–8. doi: 10.1136/jcp.52.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shimoyama T, Neelam B, Fukuda S, Tanaka M, Munakata A, Crabtree JE. VacA seropositivity is not associated with the development of gastric cancer in a Japanese population. Eur J Gastroenterol Hepatol. 1999;11:887–90. doi: 10.1097/00042737-199908000-00013. [DOI] [PubMed] [Google Scholar]
- 174.Suriani R, Colozza M, Cardesi E, Mazzucco D, Marino M, Grosso S, et al. CagA and VacA Helicobacter pylori antibodies in gastric cancer. Can J Gastroenterol. 2008;22:255–8. doi: 10.1155/2008/521724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ben Mansour K, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, et al. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;9:10. doi: 10.1186/1476-0711-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yokoyama K, Higashi H, Ishikawa S, Fujii Y, Kondo S, Kato H, et al. Functional antagonism between Helicobacter pylori CagA and vacuolating toxin VacA in control of the NFAT signaling pathway in gastric epithelial cells. Proc Natl Acad Sci U S A. 2005;102:9661–6. doi: 10.1073/pnas.0502529102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Argent RH, Thomas RJ, Letley DP, Rittig MG, Hardie KR, Atherton JC. Functional association between the Helicobacter pylori virulence factors VacA and CagA. J Med Microbiol. 2008;57:145–50. doi: 10.1099/jmm.0.47465-0. [DOI] [PubMed] [Google Scholar]
- 178.Tegtmeyer N, Zabler D, Schmidt D, Hartig R, Brandt S, Backert S. Importance of EGF receptor, HER2/Neu and Erk1/2 kinase signalling for host cell elongation and scattering induced by the Helicobacter pylori CagA protein: antagonistic effects of the vacuolating cytotoxin VacA. Cell Microbiol. 2009;11:488–505. doi: 10.1111/j.1462-5822.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 179.Oldani A, Cormont M, Hofman V, Chiozzi V, Oregioni O, Canonici A, et al. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5:e1000603. doi: 10.1371/journal.ppat.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Akada JK, Aoki H, Torigoe Y, Kitagawa T, Kurazono H, Hoshida H, et al. Helicobacter pylori CagA inhibits endocytosis of cytotoxin VacA in host cells. Dis Model Mech. 2010;3:605–17. doi: 10.1242/dmm.004879. [DOI] [PubMed] [Google Scholar]
- 181.Tan S, Noto JM, Romero-Gallo J, Peek RM, Jr., Amieva MR. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7:e1002050. doi: 10.1371/journal.ppat.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jones KR, Jang S, Chang JY, Kim J, Chung IS, Olsen CH, et al. Polymorphisms in the intermediate region of VacA impact Helicobacter pylori-induced disease development. J Clin Microbiol. 2011;49:101–10. doi: 10.1128/JCM.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122:1082–96. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Blaser MJ, Chen Y, Reibman J. Does Helicobacter pylori protect against asthma and allergy? Gut. 2008;57:561–7. doi: 10.1136/gut.2007.133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dooley CP, Cohen H, Fitzgibbons PL, Bauer M, Appleman MD, Perez-Perez GI, et al. Prevalence of Helicobacter pylori infection and histologic gastritis in asymptomatic persons. N Engl J Med. 1989;321:1562–6. doi: 10.1056/NEJM198912073212302. [DOI] [PubMed] [Google Scholar]
- 186.Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–40. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 187.Luther J, Dave M, Higgins PD, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis. 2010;16:1077–84. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Luther J, Owyang SY, Takeuchi T, Cole TS, Zhang M, Liu M, et al. Helicobacter pylori DNA decreases pro-inflammatory cytokine production by dendritic cells and attenuates dextran sodium sulphate-induced colitis. Gut. 2011;60:1479–86. doi: 10.1136/gut.2010.220087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rožanković PB, Huzjan AL, Cupić H, Benčić IJ, Bašić S, Demarin V. Influence of CagA-positive Helicobacter pylori strains on atherosclerotic carotid disease. J Neurol. 2011;258:753–61. doi: 10.1007/s00415-010-5824-9. [DOI] [PubMed] [Google Scholar]
- 190.Longo-Mbenza B, Nsenga JN, Mokondjimobe E, Gombet T, Assori IN, Ibara JR, et al. Helicobacter pylori infection is identified as a cardiovascular risk factor in Central Africans. Vasc Health Risk Manag. 2012;6:455–61. doi: 10.2147/VHRM.S28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bassi V, Marino G, Iengo A, Fattoruso O, Santinelli C. Autoimmune thyroid diseases and Helicobacter pylori: the correlation is present only in Graves’s disease. World J Gastroenterol. 2012;18:1093–7. doi: 10.3748/wjg.v18.i10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Boyanova L, Yordanov D, Gergova G, Markovska R, Mitov I. Benefits of Helicobacter pylori cagE genotyping in addition to cagA genotyping: a Bulgarian study. Antonie Van Leeuwenhoek. 2011;100:529–35. doi: 10.1007/s10482-011-9608-8. [DOI] [PubMed] [Google Scholar]
- 193.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 194.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, et al. The role of interleukin-1 polymorphisms in the pathogenesis of gastric cancer. Nature. 2001;412:99. doi: 10.1038/35083631. [DOI] [PubMed] [Google Scholar]
- 195.Sicinschi LA, Lopez-Carrillo L, Camargo MC, Correa P, Sierra RA, Henry RR, et al. Gastric cancer risk in a Mexican population: role of Helicobacter pylori CagA positive infection and polymorphisms in interleukin-1 and -10 genes. Int J Cancer. 2006;118:649–57. doi: 10.1002/ijc.21364. [DOI] [PubMed] [Google Scholar]
- 196.Chen W, Shu D, Chadwick VS. Helicobacter pylori infection: mechanism of colonization and functional dyspepsia Reduced colonization of gastric mucosa by Helicobacter pylori in mice deficient in interleukin-10. J Gastroenterol Hepatol. 2001;16:377–83. doi: 10.1046/j.1440-1746.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- 197.Azuma T, Ito S, Sato F, Yamazaki Y, Miyaji H, Ito Y, et al. The role of the HLA-DQA1 gene in resistance to atrophic gastritis and gastric adenocarcinoma induced by Helicobacter pylori infection. Cancer. 1998;82:1013–8. doi: 10.1002/(SICI)1097-0142(19980315)82:6<1013::AID-CNCR2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 198.de Oliveira JG, Silva AE. Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol. 2012;18:1235–42. doi: 10.3748/wjg.v18.i11.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote Helicobacter pylori infection in humans. Gastroenterology. 2012;142:1160–71. doi: 10.1053/j.gastro.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang P, Zhang L, Jiang JM, Ma D, Tao HX, Yuan SL, et al. Association of NOD1 and NOD2 genes polymorphisms with Helicobacter pylori related gastric cancer in a Chinese population. World J Gastroenterol. 2012;18:2112–20. doi: 10.3748/wjg.v18.i17.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Goto Y, Ando T, Yamamoto K, Tamakoshi A, El-Omar E, Goto H, et al. Association between serum pepsinogens and polymorphismof PTPN11 encoding SHP-2 among Helicobacter pylori seropositive Japanese. Int J Cancer. 2006;118:203–8. doi: 10.1002/ijc.21338. [DOI] [PubMed] [Google Scholar]
- 202.Jiang J, Jia ZF, Kong F, Jin MS, Wang YP, Tian S, et al. Association of polymorphism of PTPN 11 encoding SHP-2 with gastric atrophy but not gastric cancer in Helicobacter pylori seropositive Chinese population. BMC Gastroenterol. 2012;12:89. doi: 10.1186/1471-230X-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Skoog EC, Sjöling A, Navabi N, Holgersson J, Lundin SB, Lindén SK. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One. 2012;7:e36378. doi: 10.1371/journal.pone.0036378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhong C, Li KN, Bi JW, Wang BC. Sodium intake, salt taste and gastric cancer risk according to helicobacter pylori infection, smoking, histological type and tumor site in china. Asian Pac J Cancer Prev. 2012;13:2481–4. doi: 10.7314/APJCP.2012.13.6.2481. [DOI] [PubMed] [Google Scholar]
- 205.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–94. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Brenner H, Arndt V, Bode G, Stegmaier C, Ziegler H, Stümer T. Risk of gastric cancer among smokers infected with Helicobacter pylori. Int J Cancer. 2002;98:446–9. doi: 10.1002/ijc.10201. [DOI] [PubMed] [Google Scholar]
- 207.Shikata K, Doi Y, Yonemoto K, Arima H, Ninomiya T, Kubo M, et al. Population-based prospective study of the combined influence of cigarette smoking and Helicobacter pylori infection on gastric cancer incidence: the Hisayama Study. Am J Epidemiol. 2008;168:1409–15. doi: 10.1093/aje/kwn276. [DOI] [PubMed] [Google Scholar]
- 208.Wang XQ, Yan H, Terry PD, Wang JS, Cheng L, Wu WA, et al. Interactions between CagA and smoking in gastric cancer. World J Gastroenterol. 2011;17:3330–4. doi: 10.3748/wjg.v17.i28.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]