Abstract
The intimate interplay between the gut microbiota and the host may contribute to health and disease in the host. Experiments using conventionalized and conventionally raised animal models have illustrated the role of the intestinal microbiota in shaping and maintaining the host immune system. However, it is still unclear whether colonization at birth or at adulthood induces different host responses. Here, we perform comparative transcriptome analyses to elucidate the impact of the gut microbiota on the development and maintenance of the immune system in adult conventionalized (after 16 and 30 days of colonization) and conventionally raised mice, which were obtained in two independent laboratories. Transcriptional profiles of jejunum, ileum and colon were compared between germfree, conventionally raised mice and conventionalized mice. Germfree mice from the two different facilities clustered together, establishing the validity of the comparative analysis. Nevertheless, significant spatial differences were detected along the gut; the jejunum and colon exhibited a transient response (conventionalized mice) that eventually returned to a homeostatic level (conventionally raised). In contrast, the ileal response to microbiota was similar in conventionalized and conventionally raised mice. Overall, this comparative analysis supports the hypothesis that co-development of the gut microbiota and its host initiates at early stage of development and indicates that despite the achieved homeostasis, immune development is substantially different in mice conventionalized in adulthood. These findings imply that colonization during development is required to meet the window of opportunity where the gut microbiota can imprint the host’s mucosal immune-homeostasis in a way that cannot be achieved at later stages in life.
Keywords: germfree adult mice, microbiota, conventionalized mice, conventionally raised mice, transcriptome, comparative analysis
Introduction
Mammals become colonized by microbes during and after birth, although emerging evidence suggests that the in utero environment may not be strictly sterile, as originally thought.1 Post-natal colonization is followed by the dynamic process of (sequential) colonization of the gut by members of the microbiota, which is driven by complex interactions, including life-style, diet, host genotype, use of antibiotics and disease.2 This dynamic process results eventually in vastly diverse bacterial populations that establish a symbiotic relationship with the host.3 The gut is in continuous contact with trillions of microbes, but nevertheless succeeds to maintain a state of homeostasis that depends on tightly controlled immune responses. The gut microbiota is proposed to shape the host immunity and maintain homeostasis, through complex microbial cross-talk with the mucosal immune system that involves a variety of highly integrated signaling pathways and gene regulatory networks.4,5 These local interactions also elicit systemic effects on the host through the microbial regulation of metabolism of dietary components in the liver, blood and urine in addition to its impact in brain and behavior.2,6
Interactions between the host immune system and the colonizing gut microbiota initiate at birth.7 In mammals, primary colonization has been shown to contribute to developmental programming of gut homeostasis as well as the host immune system.8,9 The substantial post-natal development of the immune-system is accompanied with prominent changes in mucosal and systemic metabolism as well as the development of the hypothalamic-pituitary-adrenal (HPA) axis, which impacts on the gastrointestinal (GI) tract through its action on the enteric nervous system.10 This integrated development that occurs during the first years of life, suggest that the normal development of microbiota during this stage profoundly impacts on the host’s development and state of homeostasis, which could have lifelong consequences.11,12 This hypothesis inspired us to carry out a comparative transcriptome analysis using existing data sets (see refs. 5 and13) to investigate whether the immune homeostasis that is achieved in germfree mice exposed to gut microbiota during adulthood display similar characteristics as conventionally raised mice.
Results and Discussion
Comparative analysis of the immune features of conventionalized and conventionally raised adult mice
We recently published a study where we examined time- and region-transcriptional responses that are elicited in the mucosa of jejunum, ileum and colon during microbial colonization of adult germfree mice.5 These responses included tolerant responses, which ensured that a novel state of microbiota-accommodating homeostasis was reached in a region-specific timeframe, taking 16 to 30 d post-conventionalization in the small intestine, whereas colonic homeostasis was reached earlier, within eight to 16 d.5 To evaluate whether the impact of conventionalization of adult germfree mice leads to a mucosal immune-status resembling that of the gut mucosa in normal mice, comparative transcriptome analyses were performed using data sets obtained from conventionally raised mice that are exposed to the gut microbiota from birth onwards. To this end, the transcriptional profiles from conventionalized adult germfree mice after 16 and 30 d post-conventionalization5 were compared with the data from host transcriptional profiles of germfree and conventionally raised mice.13 We focused on transcriptional signatures reflecting the interaction between the host immune responses and microbial composition throughout the gut.
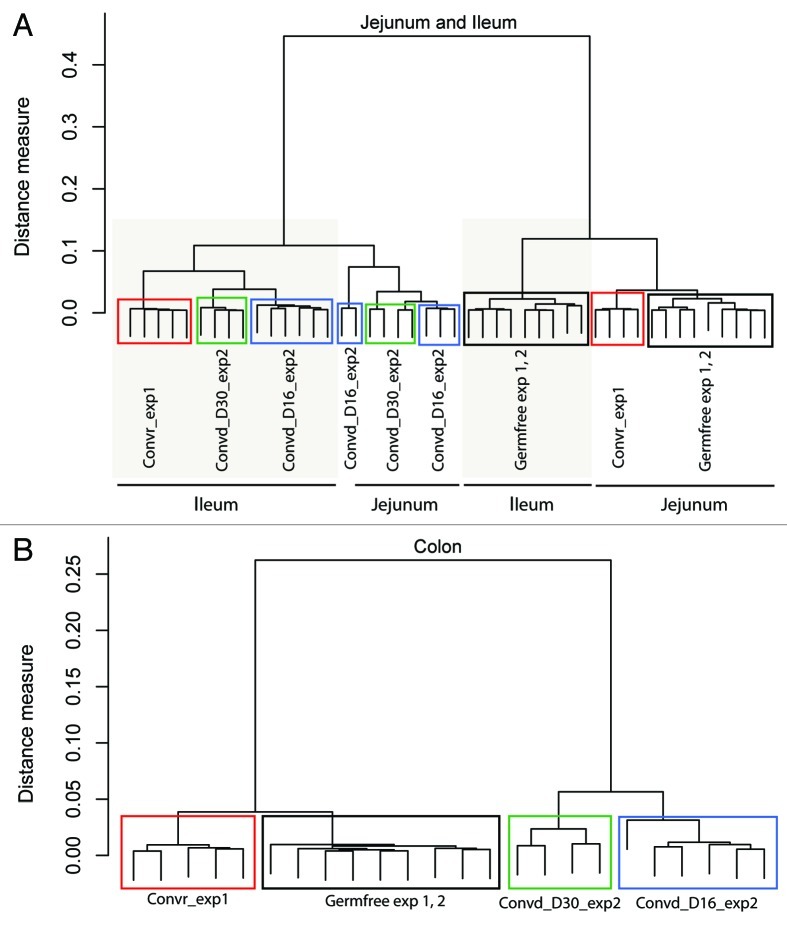
To check for the validity of the comparative analysis, Pearson correlation clustering analysis was performed on the data obtained from the 23 normalized arrays per tissue (see Materials and Methods for detailed description). Our combined data analysis, which represent different regions of the intestine of the two germfree mice control groups, conventionally raised and conventionalized mice (16 and 30 d post-conventionalization), clearly established that the germfree animals of the two experiments had very similar transcriptome patterns that were clearly and consistently distinct from those obtained after conventionalization or from conventionally raised mice (Fig. 1). Pearson correlation analysis of the transcriptome data showed that the data sets did not cluster on basis of the experimental set-up, including variation of the diet (autoclaved chow diet13 and commercial laboratory chow diet5) or the age of the germfree mice (8, 10 and 12 weeks old) (see refs. 5 and 13 for details), enabling us to eliminate the consequence of the aging in the germfree state. The ileum transcriptomes of conventionalized and conventionally raised mice clustered close to each other, whereas both the jejunum and colon transcriptomes of conventionally raised mice clustered closer to the germfree mice and were quite distinct from the transcriptomes of the same intestinal regions in conventionalized mice. Overall, the clustering of the transcriptome data sets strictly followed the intervention, confirming the validity of their comparative analysis.
Figure 1. Region-specific clustering distinguishes conventionalized and conventionally raised adult mice. Dendogram using Ward clustering on the Pearson distance measure of the transcriptome data obtained from the two animal experiments under comparison; exp1; (see ref. 13) and exp2; (see ref. 5). The dendogram shows that the germfree animals grouped together and did not cluster based on the experiment, diet used or animal age, establishing the validity of the comparative analysis performed here. Convr = conventionally raised mice, Convd-D16 = day 16 of conventionalization, Convd-D30 = day 30 of conventionalization.
Temporal and spatial immune features of the conventionalized vs. conventionally raised adult mice
The gut-mucosa gene expression patterns of jejunum, ileum and colon from germfree, conventionalized and conventionally raised mice, were compared across time of exposure to the gut microbiota using the time series analysis software, Short Time-series Expression Miner (STEM) (see the Material and Methods section for detailed description). Clustering of genes was determined separately for each segment of the GI tract (Fig. S1). Only the statistically significant temporal expression profiles identified by STEM to include immune-related GO categories (p < 0.001), were used to search for signaling pathways and potential transcriptional signature networks using ingenuity pathway analysis (IPA) (Fig. S1).
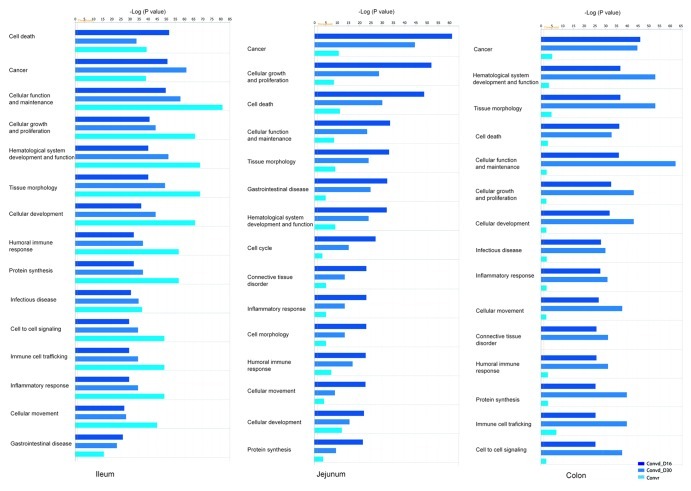
IPA comparative analysis highlighted the temporal and region-specific differences in the signaling pathways that characterized the conventionalized adult mice at a stage where a microbiota-accommodating homeostasis was achieved, in contrast to the conventionally raised mice, when both groups are compared with the germfree control. In the ileum, which represents the main site for immune activation and regulation in the GI tract, the gut microbiota elicited similar immune induced responses in both conventionalized mice and conventionally raised mice (Fig. 2). In contrast, in both jejunum and colon the immune related processes remained much more strongly stimulated in conventionalized as compared with conventionally raised mice, relative to the germfree control (Fig. 2). Taken together these data imply that the ileal responses to the microbiota are highly persistent throughout life. In contrast, in conventionally raised mice, jejunal and colonic mucosal responses appear to dampen down, while they are sustained in mice that are conventionalized in adulthood.
Figure 2. Temporal and spatial immune features of the conventionalized in contrast to conventionally raised mice. Ingenuity biological processes that were significantly modulated in the gut tissue of conventionalized mice as well as conventionally raised mice compared with the germfree control. Significance was calculated via a one-tailed Fisher’s Exact test in IPA and is represented as –log (p value); -log values exceeding 1.30 were significant p < 0.05, (n = 4−5 mice/time point).
Region-specific transcriptome signatures distinguish conventionalized from conventionally raised adult mice
We tested the hypothesis that there might be a core set of regulatory genes along the gut that could serve as a transcriptional signature to distinguish between the mucosal homeostasis of mice conventionalized during adulthood and conventionally raised animals. To test this hypothesis, we mined the genes identified by STEM (Fig. S1) and performed IPA group-wise analysis by comparing the gene sets that are significantly differentially expressed in conventionalized mice (days 16 and 30) and conventionally raised mice relative to the germfree control mice.
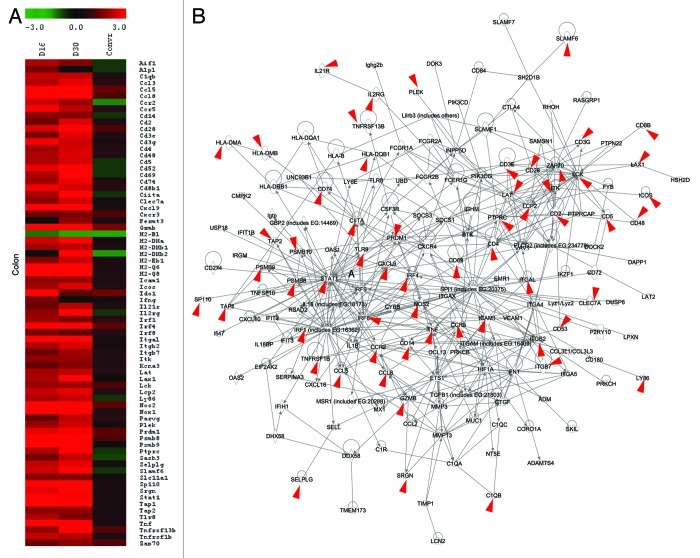
The resulting IPA derived transcriptional signatures distinctly highlighted the differential signatures of colon and jejunum between conventionalized and conventionally raised animals (Fig. 3 and S2), but failed to clearly discriminate the ileum transcriptome patterns (as the STEM identified genes were similarly induced in both conventionalized and conventionally raised mice, Fig. S1). These transcriptional signatures were highly induced only in conventionalized but not in conventionally raised mice, relative to the germfree control. Moreover, the jejunal and colonic signatures displayed high similarity (76% and 89%, respectively) with the transcriptional signatures that were previously identified to govern the dynamic mucosal responses of adult germfree mice upon their conventionalization (for details see ref. 5). These analyses reinforce and detail the conclusion that was reached on basis of the dendogram derived from the Pearson correlation analysis of the arrays (Fig. 1), prior to applying any biological interpretation software.
Figure 3. Transcriptome signature for the colonic responses of conventionalized and conventionally raised adult mice to the gut microbiota. (A) Heat map of the genes that constitute the core regulatory protein-protein interaction network (B) derived by plotting STEM output genes (see Fig. S1) of the temporal expression profiles involved in immune response. Transcriptional data was projected onto the interaction map. Red arrows refer to genes associated with inflammatory bowel disease (n = 4−6/ group).
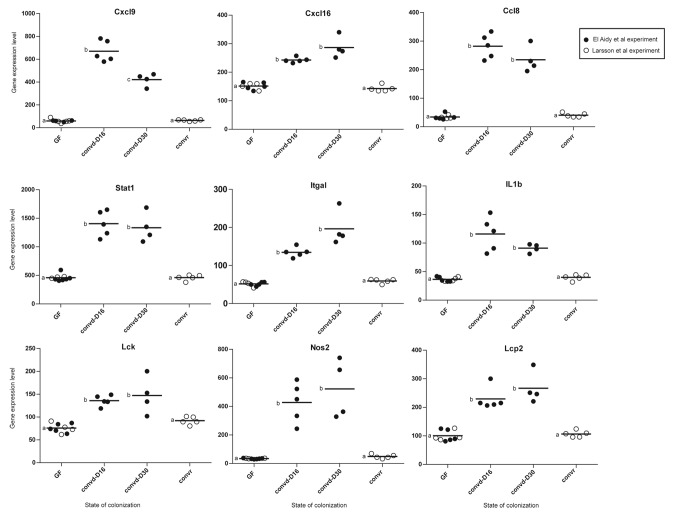
Particularly in the colon, 77% of the identified transcriptional signatures that distinguish conventionalized from conventionally raised mice orthologs to Crohn disease-associated genes that have been discovered in genome-wide association studies (GWAS) (Fig. 3B). This observation supports the notion that the gut microbiota can also impact on the responsiveness of “risk genes” and signaling pathways that are dysregulated in patients with inflammatory gastrointestinal diseases. Interestingly, the transcriptional signature identified above to distinguish between the colon of conventionalized and conventionally raised mice, belong to the same signaling pathways as the ileal transcripts, which remained highly induced in both groups. Presumably, this region-specific gene signaling depends on whether continual induction of these genes at that particular intestinal region would be advantageous or detrimental to the host according to the function of the encoded products. Interestingly, the majority of the transcription signatures identified in the colon of adult conventionalized but not conventionally raised mice are proinflammatory genes. Among the identified genes, is the chemokine ligend Cxcl16, which was highly induced only in the colon of conventionalized mice (Fig. 4). Cxcl16 is a potent proinflammatory neutrophil chemoattractant and activator.14 Recently, Olszak and colleagues showed that Cxcl16 is an age and organ dependent factor that is regulated by the exposure to the gut microbiota only early in life where it is essential for establishing colonic tolerance to environmental exposures.12 The findings of Olszak et al. demonstrates that the age-sensitive contact with the microbiota is crucial for regulating susceptibility to tissue inflammation.12 Similarly, Stat1 (Signal Transducers and Activators of Transcription family, 1) , Ccl8 (chemokine ligand 8), Cxcl9 (chemokine ligand 9), Itgal (Integrin α legand), Il1b (interleukin 1 β) and Lck (lymphocyte-specific protein tyrosine kinase) genes were among the identified transcriptional signature (Fig. 4). These genes encoding proinflammatory mediators were previously shown to be highly induced in mice and zebrafish upon early exposure to microbial factors but become “tolerizeable” by silencing during repetitive exposure later in life. The transient silencing of these genes were shown to occur during development and involved epigenetic mechanisms via chromatin modification to prevent unwarranted inflammation particularly in the densely colonized colon.15,16 The antimicrobial effector Nos2, which was slightly induced in the colon of conventionally raised mice in comparison with their germfree control,13 was strongly more stimulated in the conventionalized mice (Fig. 4). Collectively, these findings support the biological relevance of the identified transcriptional signatures to distinguish between the microbiota-accommodating homeostasis reached in adult-conventionalized jejunum and colon from that in conventionally raised mice. This argument implies that exposure to microbial factors in early life and young adulthood, elicits long-lasting effects on the identified gene signatures, and in their absence, later life exposure to factors that stimulate these signatures may induce an inflammatory response.
Figure 4. Dot plots represent the expression levels of transcriptional signatures identified for colonic tissues of conventionalized mice and were previously shown to be age-sensitive and regulated through epigenetic mechanisms.12,15,16 Significant differences between time points are indicated by distinctive characters above the measurement groups (p < 0.05; n = 4−6 / group).
Material and Methods
Animal experimental design, sampling and microarray processing
Intestinal mucosa transcriptomes of male, germfree, conventionalized and conventionally raised C57BL/6J mice (8−12 weeks old) were previously obtained in two independent animal experiments and using animals of different sources (for detailed description of the two experiments see refs. 5 and 13). In the two experiments, intestinal tissue samples were obtained as previously described.5,13 RNA was isolated from the gut tissues that were snap frozen immediately after harvest and were stored at -80ᵒ C until RNA isolation using the RNeasyMini Kit (Qiagen). For RNA labeling in the experiment of Larsson et al., microarray hybridization and scanning were performed at the Uppsala array-platform core facility at Uppsala University using MoGene 1.0 ST chips (Affymetrix), according to the manufacturer’s instructions.13 In the experiment of El Aidy et al., samples were hybridized on Affymetrix GeneChip Mouse Gene 1.1 ST arrays, according to the manufacturer’s instructions.5
Comparative transcriptome analysis and biological interpretation of expression data sets
Quality control of the data sets obtained from the two independent experiments5,13 was performed using Bioconductor packages integrated in an online pipeline.17 The complete data sets encompassed 23 Affymetrix arrays; germfree (n = 4) and conventionally raised mice (n = 5) from the experiment of Larsson et al. and germfree (n = 5) mice as well as conventionalized mice at days 16 (n = 5) and 30 (n = 4) post conventionalization from the experiment of El Aidy et al. Various advanced quality metrics, diagnostic plots, pseudo-images and classification methods were applied to ascertain only excellent quality arrays were used in the subsequent analyses.18 Normalized expression estimates were obtained from the raw intensity values using the robust multiarray analysis (RMA) pre-processing algorithm available in the Bioconductor library AffyPLM using default settings.19 Next an empirical Bayes method, called ComBat, was used to correct for the systematic errors (batch effects).20,21
Complementary methods were applied to relate changes in gene expression to functional changes. Hierarchical clustering of modulated genes in the mucosa that were unique for each mouse group (or several groups) was performed by MeV (Multi-experiment Viewer).22 Comparison of time series gene expression data using STEM was used to identify transcriptome signature of genes. The time series was selected based on the duration of intestinal exposure to the gut microbiota [germfree as time (t)1, day 16 of conventionalization as t2, day 30 of conventionalization as t3 and finally the conventionally raised group as t4]. STEM allows to identify significant temporal expression profiles and the genes associated with these profiles and to compare the behavior of these genes across multiple time-points. STEM supports gene ontology (GO) term category gene enrichment analyses for sets of genes having the same temporal expression pattern.23,24 The statistical significance of the profiles generated by STEM was calculated via a permutation test (n = 1000) corrected using a false discovery rate (FDR < 0.001).24 Biological interpretations of the significantly altered signaling pathways and functional processes, in response to conventionalization were identified using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems) (www.ingenuity.com). Our IPA analyses included comparison of differentially regulated genes in the jejunum, ileum and colon on days 16 and 30 post-conventionalization and in conventionally raised mice, in each case relative to expression observed in the control group (germfree). The input was all differentially regulated genes (p value ≤ 0.001, FC ≥ 1.2 and intensity ≥ 20) of the intestinal segments.
Conclusion
From the comparative analysis reported herein, it is tempting to speculate that the spatial differences observed between adult conventionally raised and conventionalized mice in terms of the molecular characteristics of the immune homeostasis elicited by the gut microbiota involves specific gene signaling and epigenetic mechanisms that are initiated soon after birth and during early adulthood, by exposure of the gut-mucosa of newborn and young mice to the gut bacteria and their metabolites. This concept is in good agreement with several recent reports that suggest the presence of a window of opportunity in early life during which the gut microbiota shapes the host resilience.10,12,25-27
Our comparative study suggests that the identified gene signature that distinguishes conventionally raised from adult-conventionalized mice are region-specific and the ileum, unlike the jejunum and colon, seems to be tuned to “remain alerted to the gut microbiota,” supporting its prominent role in the maintenance of immune homeostasis throughout life. The identified transcriptional signatures appear to be age-dependant and to involve microbially regulated molecules that consequently regulate the susceptibility to tissue inflammation. Furthermore, these observations suggest that the timing of the microbial regulation of epigenetic differentiation and maturation is of great relevance to the developmental programming of the immune response, which is the concept behind the microbiota deficiency hypothesis. This hypothesis postulates that colonization with a “healthy” microbiota during the vulnerable developmental period exerts effects that may decrease susceptibility to diseases including allergy and inflammatory bowel disease, whereas its absence or dysbiosis, as in antibiotic treatment in childhood, may have reverse effects.25,28 Overall, our comparative analysis increases urgency to improve our understanding of the intricate network between the maternal gut microbiota transfer, infant nutrition, use of antibiotics and genome variation and the development of the gut microbiota during the early stage of life. Especially during this developmental stage, such incredibly complex interactions appear to be involved in shaping the host immune homeostasis and consequently in regulating later unwarranted inflammation.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/23362
References
- 1.Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159:187–93. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 3.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–70. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–73. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El Aidy S, van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, et al. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 2012;5:567–79. doi: 10.1038/mi.2012.32. [DOI] [PubMed] [Google Scholar]
- 6.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 7.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 8.Lundin A, Bok CM, Aronsson L, Björkholm B, Gustafsson JA, Pott S, et al. Gut flora, Toll-like receptors and nuclear receptors: a tripartite communication that tunes innate immunity in large intestine. Cell Microbiol. 2008;10:1093–103. doi: 10.1111/j.1462-5822.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 9.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seckl JR, Meaney MJ. Glucocorticoid programming. Ann N Y Acad Sci. 2004;1032:63–84. doi: 10.1196/annals.1314.006. [DOI] [PubMed] [Google Scholar]
- 12.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–31. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittal P, Romero R, Kusanovic JP, Edwin SS, Gotsch F, Mazaki-Tovi S, et al. CXCL6 (granulocyte chemotactic protein-2): a novel chemokine involved in the innate immune response of the amniotic cavity. Am J Reprod Immunol. 2008;60:246–57. doi: 10.1111/j.1600-0897.2008.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–8. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 16.Galindo-Villegas J, García-Moreno D, de Oliveira S, Meseguer J, Mulero V. Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. Proc Natl Acad Sci U S A. 2012;109:E2605–14. doi: 10.1073/pnas.1209920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin K, Kools H, de Groot PJ, Gavai AK, Basnet RK, Cheng F, et al. MADMAX - Management and analysis database for multiple ~omics experiments. J Integr Bioinform. 2011;8:160. doi: 10.2390/biecoll-jib-2011-160. [DOI] [PubMed] [Google Scholar]
- 19.Heber S, Sick B. Quality assessment of Affymetrix GeneChip data. OMICS. 2006;10:358–68. doi: 10.1089/omi.2006.10.358. [DOI] [PubMed] [Google Scholar]
- 20.Bolstad BM, Collin F, Simpson KM, Irizarry RA, Speed TP. Experimental design and low-level analysis of microarray data. Int Rev Neurobiol. 2004;60:25–58. doi: 10.1016/S0074-7742(04)60002-X. [DOI] [PubMed] [Google Scholar]
- 21.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 22.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M, et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One. 2011;6:e28279. doi: 10.1371/journal.pone.0028279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shreiner A, Huffnagle GB, Noverr MC. The “Microflora Hypothesis” of allergic disease. In: Huffnagle GB, Noverr MC, ed. GI microbiota and regulation of the immune system. Landes Bioscience and Springer Science + Business Media, 2008:113-34. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.