Abstract
Eyes absent (Eya) is an evolutionarily conserved transcriptional coactivator and protein phosphatase that regulates multiple developmental processes throughout the metazoans. Drosophila eya is necessary for survival as well as for the formation of the adult eye. Eya contains a tyrosine phosphatase domain, and mutations altering presumptive active-site residues lead to strongly reduced activities in ectopic eye induction, in vivo genetic rescue using the Gal4-UAS system, and in vitro phosphatase assays. However, these mutations have not been analyzed during normal development with the correct levels, timing, and patterns of endogenous eya expression. To investigate whether the tyrosine phosphatase activity of Eya plays a role in Drosophila survival or normal eye formation, we generated three eya genomic rescue (eyaGR) constructs that alter key active-site residues and tested them in vivo. In striking contrast to previous studies, all eyaGR constructs fully restore eye formation as well as viability in an eya null mutant background. We conclude that the tyrosine phosphatase activity of Eya is not required for normal eye development or survival in Drosophila. Our study suggests the need for a re-evaluation of the mechanism of Eya action and underscores the importance of studying genes in their native context.
Introduction
Drosophila eye development relies on a network of retinal determination (RD) genes, which encode highly conserved transcription factors and cofactors [1]. A key member of the RD gene network is eyes absent (eya), which is necessary for survival and normal eye formation, as well as sufficient for ectopic eye induction when overexpressed [2], [3]. The four mammalian Eya homologues (Eya1-4) regulate the development of the kidneys, ears, craniofacial and skeletal structures, muscle, thymus, parathyroid, and lungs, as well as acting in the innate immune response, DNA damage repair, inhibition of apoptosis, angiogenesis, and photoperiodism [4]. Notably, Eya1-4 are implicated in several diseases in humans, such as the multi-organ developmental disorderbranchio-oto-renal (BOR) syndrome [5], congenital cataracts [6], and late-onset deafness [7], and are overexpressed in multiple types of cancers [8]–[11].
Eya is known to act as a transcriptional coactivator as well as a protein phosphatase. The highly conserved C-terminal region of Eya, referred to as the Eya domain (ED) [12], contains tyrosine phosphatase activity of the haloacid dehalogenase family [13]–[16]. In vitro phosphatase assays show that Eya proteins derived from plant, mouse, and fly exhibit tyrosine phosphatase activity, although that of Drosophila Eya is very low and is difficult to detect [13], [14]. Multiple lines of evidence suggest that Eya tyrosine phosphatase regulates development in mammals. In humans, loss of tyrosine phosphatase activity is observed in BOR-associated mutations in EYA1 [17], , implying that loss of tyrosine phosphatase function contributes to this disease. Over-expression of murine Eya1, Eya2, or Eya3 results in increased proliferation, migration, invasion, and transformation of breast cancer cells. Interestingly, Eya1/2/3 phosphatase-dead mutations attenuate induction of migration, invasion, and transformation, suggesting that the tyrosine phosphatase activity promotes tumor cell invasiveness [9]. Finally, tyrosine phosphatase activity is necessary for Eya1/2/3 to activate reporter gene expression in mammalian cell culture [15].
In Drosophila, cDNA-based eya mutant transgenes that disrupt the predicted tyrosine phosphatase active site (D493N and E728Q) have drastically decreased ability to induce ectopic eye formation and to rescue eye development in the eya2 eye-specific loss-of-function mutant [13], [14]. In addition, a study based on overexpression of wild type and phosphatase-inactive eya transgenes in the developing eye suggests that Eya tyrosine phosphatase regulates photoreceptor axon targeting [19]. Although these findings suggest that Drosophila Eya tyrosine phosphatase activity may play a role during normal development, this hypothesis has not been tested using a system that accurately reproduces endogenous levels, timing, and patterns of eya expression.
We recently reported a study that used a genomic DNA-based rescue system to evaluate the in vivo significance of two predicted MAPK target sites in the Eya protein. Previous experiments using the Gal4-UAS system had suggested that MAPK-mediated phosphorylation activates Eya during ectopic eye development. In contrast, our genomic rescue-based study found that the two MAPK target residues of Eya are not required for normal eye development or survival in Drosophila [20]. In the current study, we have used the same genomic rescue strategy to gain a more accurate understanding of the role of Eya tyrosine phosphatase activity during normal Drosophila development. Surprisingly, we find that tyrosine phosphatase activity is not required for any known function of the Eya protein during normal Drosophila development.
Materials and Methods
Recombineering-induced Point Mutagenesis and Fly Transgenesis
We used a two-step recombineering method to create the D493N (GAT->AAT) and E728Q (GAG->CAG) point mutations in the eya+GR construct as described previously [21]. The recombineering products were sequenced and subjected to restriction enzyme fingerprint digest prior to transgenesis. We then employed φC31 to integrate constructs into attP2 (68A) on the third chromosome [22]. Site-specific integration into attP2 was confirmed by genomic PCR with attP and attB primers [23], and transgenic flies carrying the three different eya transgenes were confirmed by genomic DNA PCR sequencing. Primer sequences are available on request.
Fly Work
Flies were raised on standard media at 25°C. Fertility of rescued flies was tested as follows: five males or females of a given genotype were mated to 10 w1118 females or five males in each vial, respectively, and allowed to lay eggs for 48 hours before being discarded. Triplicate vials were set up for eyacli/CyO, Df/CyO and eyacli/Df; eya*GR/+. All resulting F1 progeny were counted during the first three days of eclosion.
Histology and Immunohistochemistry
Tangential sections of three-day-old adult eyes were performed as described [24]. Images of eye sections and whole adult eyes were taken with a Zeiss AxioPlan 2 microscope and AxioVision software. Images of whole-mount adult eyes were processed with CZ Focus software.
Larval imaginal discs [25] and eye-brain complexes [26] were dissected and stained as previously described. Primary antibodies used were 1∶300 mouse anti-Eya (10H6, Developmental Studies Hybridoma Bank), 1∶400 rat anti-Elav (7E8A10, Developmental Studies Hybridoma Bank), 1∶100 mouse anti-Chaoptin (24B10, Developmental Studies Hybridoma Bank), and 1∶800 rabbit anti-β-galactosidase (ab9361, Abcam). Secondary antibodies used (1∶500 for discs, 1∶200 for brains) were Alexa Fluor 488 goat anti-mouse and anti-rat (Molecular Probes) and Cy3 goat anti-rabbit (Jackson ImmunoResearch). Photography was carried out using a Zeiss LSM 510 confocal microscope and processed with Image J software.
Electroretinograms
Six three-day-old adults were assayed for each genotype. ERGs were performed as described previously [27]. Recordings were processed with AxoGraph X software.
Statistical Analysis
For viability tests, goodness of fit was evaluated using the chi-square test. For fertility tests and scoring overshooting of axon bundles, data are presented as the mean ± standard deviation (s.d.). Statistical significance (p values) of each data set was tested using ANOVA.
Results
Tyrosine Phosphatase-dead Genomic Rescue Constructs
In this study, we utilized eya genomic rescue constructs (eyaGR, Fig. 1) and site-specific transgenesis [22], [28] to investigate whether tyrosine phosphatase activity of Eya is required for Drosophila eye development or survival. The wild type eya genomic rescue construct (eya+GR) fully rescues viability and eye formation in an eya mutant background, and is used as a positive control throughout our study. We generated three genomic rescue constructs encoding tyrosine phosphatase-dead Eya proteins. The residues Asp 493 and Glu 728 we chose to mutate have been reported previously to be required for Eya tyrosine phosphatase activity in vitro [13], [14]. Two of the point-mutant constructs, eyaD493NGR and eyaE728QGR, encode a protein with a single amino-acid substitution: D493N and E728Q, respectively. In the third construct, eyaNQGR, we engineered both D493N and E728Q mutations in eya+GR. Hereafter eyaD493NGR, eyaE728QGR and eyaNQGR are collectively described as eya*GR.
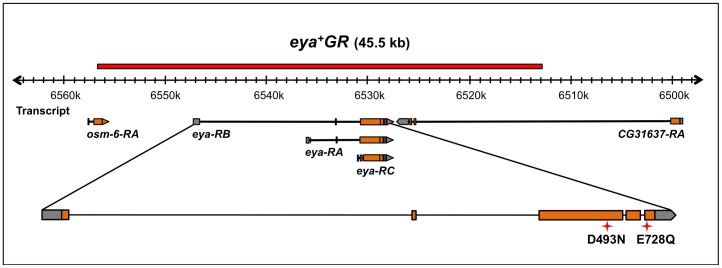
Figure 1. Schematic of the eya+GR transgene and eya locus with the position of mutations indicated.
A 45.5 kb region of the genomic DNA surrounding the eya locus (shown as a red bar) was recombineered into attB-P[acman]-ApR. eya has three alternative transcripts (eya-RA, -RB, and -RC). Red 4-point stars indicate the tyrosine phosphatase active site mutations; D493N and E728Q are in the 3rd and 5th exons of eya, respectively.
eya*GR Transgenes Fully Rescue Eye Development as well as Viability in an eya Mutant Background
Previous data show that the D493N mutation in Drosophila Eya leads to tyrosine phosphatase inactivation, a low frequency of ectopic eye induction, and incomplete genetic rescue using the Gal4-UAS system [13], [14]. Likewise, murine Eya3 harboring the E478Q mutation, which is homologous to Drosophila E728Q, has severely decreased in vitro phosphatase activity [13]. Therefore, we predicted that eyaD493NGR and eyaE728QGR would either rescue eya mutants poorly or not at all. In order to investigate the effect of mutating both catalytic residues at once, we also generated the double-mutant genomic rescue construct eyaNQGR. Hereafter we only show data for eyaNQGR since all three eya*GR constructs behaved identically. We first tested the ability of eya*GR to rescue eya2 homozygous flies, which are viable and fertile but completely lack eyes due to a deletion of an enhancer required for eya expression during eye development [2]. We compared the external and internal eye morphology relative to that of flies rescued with eya+GR or wild type flies (Fig. 2). In contrast to our prediction, eya*GR fully rescues the eye-specific eya2 mutant. Eyes of eya2 adults rescued with a single copy of eya*GR reveal indistinguishable external and internal morphology from wild type.
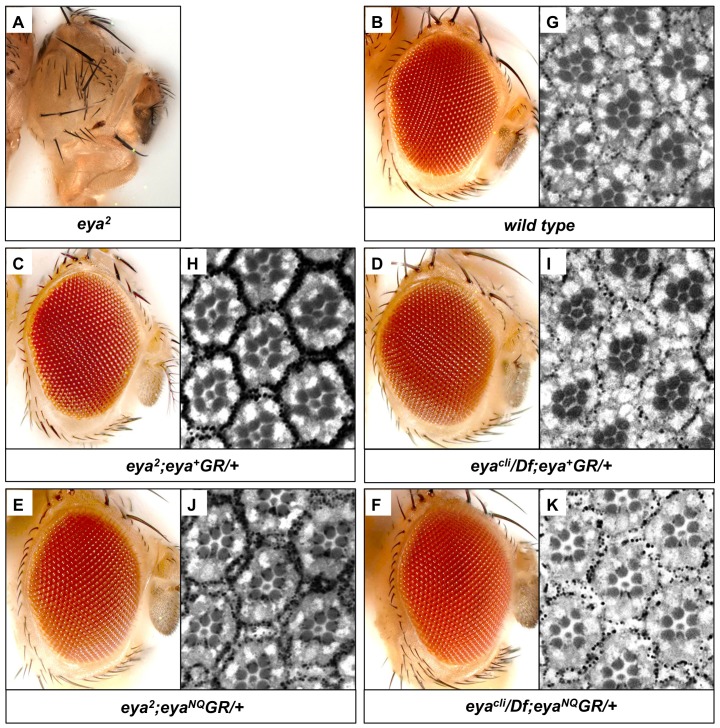
Figure 2. eya*GR constructs restore eye development and viability in eya mutants.
(A) eya2 homozygous adults completely lack eyes. eya2 adults rescued with one copy of eya+GR (C,H) or eyaNQGR (E,J) show similar external and internal eye morphology compared with wild-type Canton S (B,G). A single copy of eya+GR (D,I) or eyaNQGR (F,K) can fully rescue eye morphology and survival in eyacli/Df flies, which have no endogenous eya function.
To test whether eya*GR can restore viability, we performed rescue assays in a null background. The null allele eyaclillD (eyacli) harbors a nonsense mutation and is embryonic lethal [29]. Df(2L)BSC354 (hereafter referred to as Df) deletes the entire eya gene and fails to complement eyacli [30]. We crossed eya*GR into the background of eyacli/Df and found rescued adults are viable and fertile, both in males and females. Since Eya regulates muscle development [31], we also tested whether the rescued flies have normal muscle function. The eyacli/Df; eya*GR/+ adults appear to move and fly normally, suggesting that they have no gross defects in muscle development (data not shown). To investigate the viability of rescued flies, we crossed w; eyacli/CyO; eya*GR males to w; Df/CyO virgin females. Progeny are present at Mendelian ratios (Table 1), suggesting that the rescued flies survive as well as their heterozygous siblings. Hence, a single copy of eya*GR is functionally equivalent to a single copy of endogenous eya in promoting survival. Because eya is also required for fertility [32], we next tested whether the rescued null flies are fertile and found both males and females produce the same number of offspring as positive controls when crossed to w1118 females and males (Fig. 3). Moreover, eya null flies carrying one copy of eya*GR have eyes indistinguishable from those of flies rescued with eya+GR or wild type, both in external and internal morphology (Fig. 2). In summary, eya*GR transgenes fully restore viability, fertility, and eye development in eya null mutants.
Table 1. Flies carrying one copy of eya*GR show normal viability.
| Genotype | Non-Cy | Cy | Total | χ2 | ||
| Observed | Expected | Observed | Expected | |||
| eya+GR | 120 | 128 | 264 | 256 | 384 | 0.75 |
| eyaNQGR | 129 | 135 | 274 | 268 | 403 | 0.40 |
| eyaD493NGR | 110 | 122 | 256 | 244 | 366 | 1.77 |
| eyaE728QGR | 111 | 114 | 230 | 227 | 341 | 0.11 |
Progeny from w; eyacli/CyO; eya*GR×w; Df/CyO cross are present at Mendelian ratios, indicating that rescued files carrying a single copy of eya*GR do not have a survival disadvantage compared with their heterozygous siblings. χ2 critical (1 d.f. p 0.05) = 3.84.
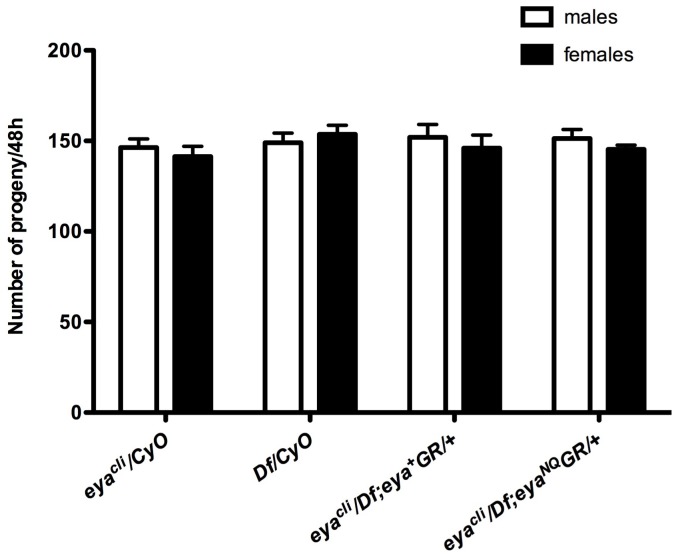
Figure 3. Adults rescued with eya*GR show normal fertility.
Fertility assays of animals of the indicated sex and genotypes show both males and females carrying one copy of eya*GR have normal fertility. No statistically significant difference was observed compared to control eyacli/CyO and Df/CyO flies using ANOVA. Error bars indicate standard deviation.
Flies Rescued with Phosphatase-inactive Eya Show Normal Development of Larval Eye Imaginal Discs
The adult Drosophila eye arises from a larval precursor structure called the eye imaginal disc. In normal eye formation, Eya expression begins in the second instar eye disc, prior to the formation of the morphogenetic furrow (MF), which marks the onset of differentiation in the eye disc. During the third instar larval stage, the MF progresses across the eye disc,with Eya expressed in a zone of undifferentiated cells anterior to the MF as well as in progressively differentiating cells posterior to the MF. In contrast, eya2 mutants have smaller eye discs due to massive apoptosis, loss of differentiation and reduced or no detectable Eya expression [2]. Given our in vivo rescue results, we predicted the eyacli/Df; eya*GR/+ larval eye discs would develop normally. As predicted, the level and pattern of Eya expression appear indistinguishable in the eye discs of positive control and rescued larvae (Fig. 4). In addition, the eye discs of rescued flies are normal in size. We also visualized Elav, a neuron-specific antigen [33], to investigate photoreceptor differentiation. Third instar eye imaginal discs from Df heterozygotes and rescued flies all yield similar Elav expression patterns (Fig. 4), implying that the tyrosine phosphatase-inactive mutations do not adversely affect differentiation of photoreceptors. To summarize, one copy of eya*GR is sufficient to support normal larval eye disc development.
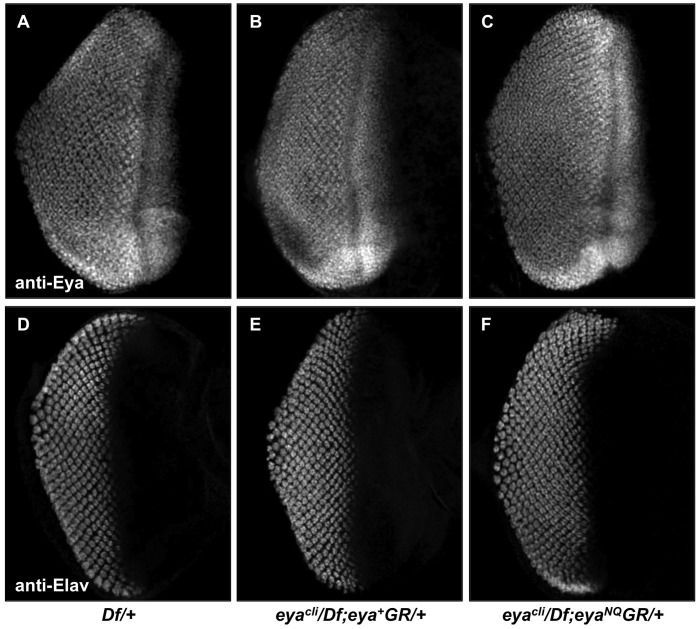
Figure 4. eya*GR constructs rescue larval eye imaginal disc development.
Immunohistochemical detection of Eya (A–C) and the pan-neural marker Elav (D–F) in Df/+ (A,D), eyacli/Df; eya+GR/+ (B,E) and eyacli/Df; eyaNQGR/+ (C,F) third instar larval eye imaginal discs. Expression patterns and levels of Eya and Elav are indistinguishable among eya null eye discs carrying one copy of eya+GR, eyaNQGR and Df heterozygotes.
eya*GR Rescued Flies have a Normal Visual Response
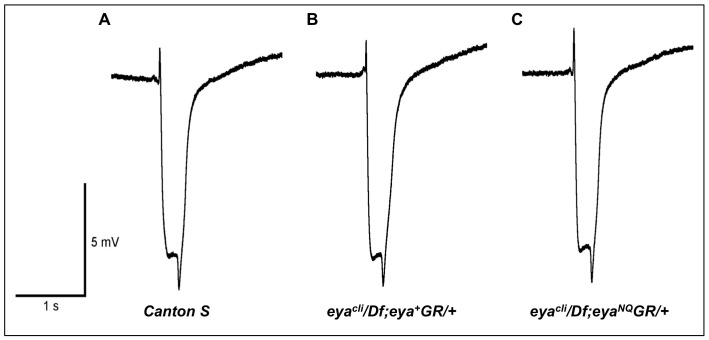
The results described above suggest that the tyrosine phosphatase catalytic residues Asp 493 and Glu 728 are not required for normal eye formation. To test whether these mutations affect eye function in rescued adults, we performed electroretinograms (ERG), a physiological assay of retinal function. Three-day-old wild type, eyacli/Df; eya+GR/+ and eyacli/Df; eya*GR/+ files were assayed, and all showed indistinguishable ERG recordings (Fig. 5), indicating that Eya tyrosine phosphatase activity is dispensable for adult eye function.
Figure 5. Mutations in the tyrosine phosphatase active site of Eya do not affect response to light.
ERG recordings from wild-type Canton S (A), eyacli/Df; eya+GR/+ (B) and eyacli/Df; eyaNQGR/+ (C) 3-day-old retinas show similar curves and suggest rescued animals have a normal light response.
Photoreceptor Axon Projections are Normal in Adults Rescued with eya*GR
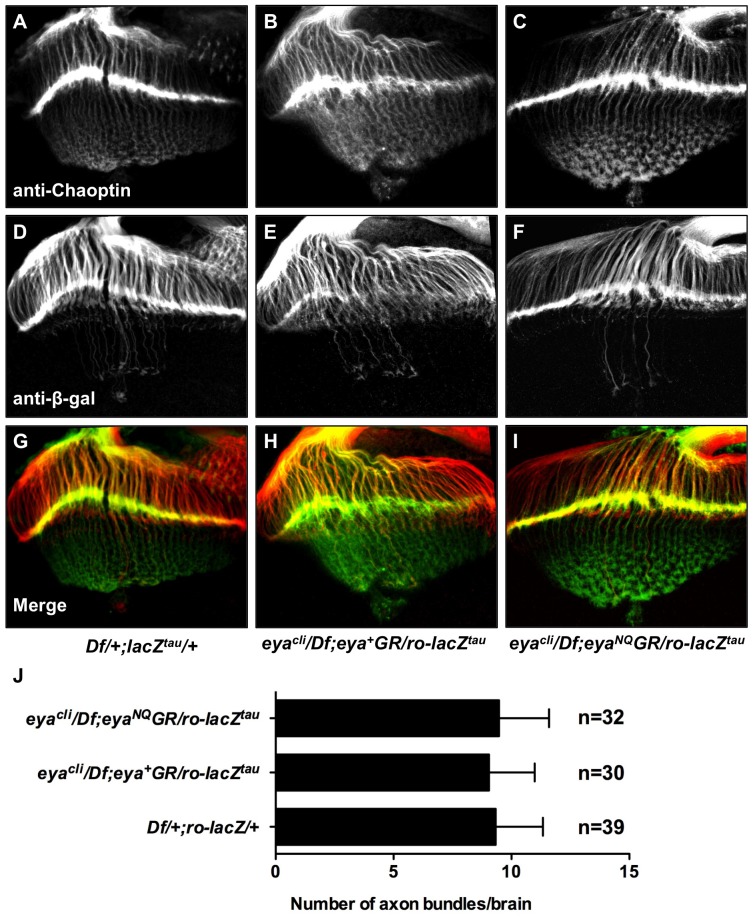
Finally, previous studies have also shown that protein tyrosine phosphatase activity contributes to the regulation of photoreceptor axon targeting [19], [34]. We therefore asked if Eya phosphatase function mediated by D493 and E728 is required for normal axon targeting. We used the reporter line ro-lacZtau, which is driven by the R2–R5 specific rough enhancer and encodes a Tau-β-galactosidase fusion protein that marks R2–R5 photoreceptor axons [34]. Immunohistochemistry shows eya*GR rescued animals have normal photoreceptor axon projections (Fig. 6). Moreover, the average number of overshooting axon bundles per brain is the same for each genotype (Fig. 6). Based on these axon projection and the ERG results, we conclude that eye function is not affected by loss of Eya tyrosine phosphatase activity.
Figure 6. Animals rescued with eya*GR have normal photoreceptor axon projections.
Third instar eye-brain complexes from Df/+; ro-lacZtau/+ (A,D,G), eyacli/Df; eya+GR/ro-lacZtau (B,E,H), and eyacli/Df; eyaNQGR/ro-lacZtau (C,F,I) stained with anti-Chaoptin and anti-β-galactosidase. (A–C) Projections from photoreceptors R1–8 are visualized by using mAb24B10 (anti-Chaoptin). (D–F) Projections from photoreceptors R2–R5, which terminate in the lamina, are revealed using ro-lacZtau (anti-β-galactosidase). (G-I) Merge of channels. eyaNQGR rescued animals show normal photoreceptor axon projections, similar to flies rescued with eya+GR and Df/+; ro-lacZtau/+. (J) 30–40 eye-brain complexes were scored for overshooting axon bundles by investigators who were blind to the genotype. The average number of overshooting axon bundles per brain is the same for Df/+; ro-lacZtau/+, eyacli/Df; eya+GR/ro-lacZtau and eyacli/Df; eya*GR/ro-lacZtau. n, number of brains scored. Error bars indicate standard deviation.
Discussion
As a dual-function protein, Eya has been best characterized as a transcriptional coactivator [35]–[37]. The second function – phosphatase activity – needs further investigation. In this study, we show that mutations in the tyrosine phosphatase active site of Eya do not affect Drosophila survival or normal eye development. However, previous studies suggest that the Eya domain (ED), in which the tyrosine phosphatase active site is located, is required for other functions during development. First, lethal missense and nonsense mutations within the ED of Drosophila Eya have been reported [38], indicating that this domain is indeed required for normal Eya function. Second, cell culture and GST pull-down studies have shown that the Drosophila ED binds to the transcription factor Sine oculis (So/Six), perhaps serving to recruit Eya to target genes, where it can act as a transcriptional co-activator within an Eya/So complex [18], [38]. Similarly, the ED of mammalian EYA can also bind to the SIX proteins [35], [39]. Taken together, these results suggest that while the tyrosine phosphatase activity of the ED is no longer required in Drosophila, it appears that some other function of the ED has either been acquired or retained since the divergence of insects and vertebrates.
cDNA-based studies have also suggested that an internal proline-serine-threonine-rich (PST) domain in Eya is required for transcriptional activation in cell culture reporter assays and for efficient induction of ectopic eyes in vivo [36]. Consistent with these results, our genomic rescue assays also show that the PST domain is required for Drosophila survival as well as normal eye development (data not shown; to be reported elsewhere). These results validate our genomic rescue system and indicate that, unlike the transactivation function, Eya tyrosine phosphatase activity is not necessary for Drosophila normal development.
The differences between our findings based on a genomic rescue strategy and previous assays using ectopic eye induction and cDNA-based Gal4-UAS genetic rescue assays may be observed for several reasons. First, normal eye formation and reprogramming of non-retinal tissue into an ectopic eye involve overlapping but distinct genetic programs, as suggested by previous studies [40]. Second, the Gal4-UAS system does not recapitulate the wild-type pattern or timing of eya expression while our genomic rescue constructs are likely to contain the full complement of regulatory sequences needed, allowing the transgene to be expressed in the same spatiotemporal pattern as the endogenous gene of interest. Evidence supporting the genomic rescue strategy comes from our findings that, unlike ectopic eye formation in Drosophila, normal eye development and survival are unaffected by mutations in two previously studied MAPK target residues of Eya using genomic rescue constructs [20]. Third, in contrast to the random integration of the P-element mediated UAS-eya transgenes used in previous studies, we used site-specific integration, minimizing genomic position effects on different constructs. Previous studies have reported that the extent of rescue is indeed highly variable among four independent insertion lines of a UAS-eyaD493N transgene [14]. Finally, although recent studies have shown that an eyaD493N transgene fails to rescue eya2 using site-specific heat shock-mediated induction [41], this approach has several drawbacks, including inaccurate timing, patterns, and levels of transgene induction [42]. In other words, no heat shock regimen is likely to drive transgene expression timing, patterns, or levels in a manner approaching that of the endogenous eya gene. This report highlights the importance of the genomic rescue system for evaluating the function of specific protein domains, motifs, and even single residues or base pairs in vivo.
Although our studies suggest that Eya tyrosine phosphatase is dispensable for Drosophila development under standard laboratory conditions, it remains possible that Eya tyrosine phosphatase contributes to the animal’s response to environmental stressors. Previous studies reported that murine Eya1 and Eya3 can dephosphorylate H2AX, a histone variant associated with DNA damage, promoting efficient DNA repair [43]. Since Drosophila has an H2AX homologue, it will be interesting to determine if Drosophila Eya is also able to dephosphorylate H2AX. If this is the case, then our eya null mutants rescued with phosphatase-inactive eya*GR may be more sensitive to conditions that cause DNA damage.
To summarize, contrary to current models, the tyrosine phosphatase activity of Eya is dispensable for all known Eya functions in Drosophila. Whether the tyrosine phosphatase activity is required for mammalian Eya function in vivo remains to be tested. Our studies shed new light on a protein that plays a key role in a variety of developmental processes throughout the metazoans.
Acknowledgments
We thank Nele Haelterman for ERG technical assistance, Ilaria Rebay for providing the ro-lacZtau fly stock and Georg Halder for comments on the manuscript. MJ thanks the China Scholarship Council (CSC) for financial support.
Funding Statement
This work was funded by the Retina Research Foundation, National Eye Institute grant R01 EY011232 (GM), the Chinese Scholarship Council (MJ), and the NEI/NIH Core Grant for Vision Research EY-002520. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pappu KS, Mardon G (2004) Genetic control of retinal specification and determination in Drosophila. Int J Dev Biol 48: 913–924. [DOI] [PubMed] [Google Scholar]
- 2. Bonini NM, Leiserson WM, Benzer S (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72: 379–395. [DOI] [PubMed] [Google Scholar]
- 3. Bonini NM, Bui QT, Gray-Board GL, Warrick JM (1997) The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development 124: 4819–4826. [DOI] [PubMed] [Google Scholar]
- 4.Tadjuidje E, Hegde RS (2012) The Eyes Absent proteins in development and disease. Cell Mol Life Sci. [DOI] [PMC free article] [PubMed]
- 5. Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, et al. (1997) A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet 15: 157–164. [DOI] [PubMed] [Google Scholar]
- 6. Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M (2000) Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet 9: 363–366. [DOI] [PubMed] [Google Scholar]
- 7. Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, et al. (2001) Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum Mol Genet 10: 195–200. [DOI] [PubMed] [Google Scholar]
- 8. Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, et al. (2005) Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res 65: 925–932. [PubMed] [Google Scholar]
- 9. Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, et al. (2010) The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene 29: 3715–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller SJ, Lan ZD, Hardiman A, Wu J, Kordich JJ, et al. (2010) Inhibition of Eyes Absent Homolog 4 expression induces malignant peripheral nerve sheath tumor necrosis. Oncogene 29: 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robin TP, Smith A, McKinsey E, Reaves L, Jedlicka P, et al. (2012) EWS/FLI1 Regulates EYA3 in Ewing Sarcoma via Modulation of miRNA-708, Resulting in Increased Cell Survival and Chemoresistance. Mol Cancer Res 10: 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zimmerman JE, Bui QT, Steingrimsson E, Nagle DL, Fu W, et al. (1997) Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res 7: 128–141. [DOI] [PubMed] [Google Scholar]
- 13. Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, et al. (2003) The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature 426: 299–302. [DOI] [PubMed] [Google Scholar]
- 14. Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, et al. (2003) Eyes absent represents a class of protein tyrosine phosphatases. Nature 426: 295–298. [DOI] [PubMed] [Google Scholar]
- 15. Li X, Oghi KA, Zhang J, Krones A, Bush KT, et al. (2003) Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426: 247–254. [DOI] [PubMed] [Google Scholar]
- 16. Okabe Y, Sano T, Nagata S (2009) Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature 460: 520–524. [DOI] [PubMed] [Google Scholar]
- 17. Rayapureddi JP, Hegde RS (2006) Branchio-oto-renal syndrome associated mutations in Eyes Absent 1 result in loss of phosphatase activity. FEBS Lett 580: 3853–3859. [DOI] [PubMed] [Google Scholar]
- 18. Mutsuddi M, Chaffee B, Cassidy J, Silver SJ, Tootle TL, et al. (2005) Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics 170: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiong W, Dabbouseh NM, Rebay I (2009) Interactions with the abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev Cell 16: 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jusiak B, Abulimiti A, Haelterman N, Chen R, Mardon G (2012) MAPK target sites of eyes absent are not required for eye development or survival in Drosophila. PLoS One 7: e50776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, et al.. (2007) Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol Chapter 1: Unit 1 16. [DOI] [PubMed]
- 22. Venken KJ, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- 23. Venken KJ, Kasprowicz J, Kuenen S, Yan J, Hassan BA, et al. (2008) Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res 36: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tomlinson A, Ready DF (1987) Cell fate in the Drosophila ommatidium. Dev Biol 123: 264–275. [DOI] [PubMed] [Google Scholar]
- 25. Pepple KL, Anderson AE, Frankfort BJ, Mardon G (2007) A genetic screen in Drosophila for genes interacting with senseless during neuronal development identifies the importin moleskin. Genetics 175: 125–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu JS, Luo L (2006) A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc 1: 2110–2115. [DOI] [PubMed] [Google Scholar]
- 27. Heisenberg M (1971) Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J Exp Biol 55: 85–100. [DOI] [PubMed] [Google Scholar]
- 28. Groth AC, Fish M, Nusse R, Calos MP (2004) Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyle M, Bonini N, DiNardo S (1997) Expression and function of clift in the development of somatic gonadal precursors within the Drosophila mesoderm. Development 124: 971–982. [DOI] [PubMed] [Google Scholar]
- 30. Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, et al. (2004) Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36: 288–292. [DOI] [PubMed] [Google Scholar]
- 31. Liu YH, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, et al. (2009) A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell 16: 280–291. [DOI] [PubMed] [Google Scholar]
- 32. Bonini NM, Leiserson WM, Benzer S (1998) Multiple roles of the eyes absent gene in Drosophila. Dev Biol 196: 42–57. [DOI] [PubMed] [Google Scholar]
- 33. Koushika SP, Lisbin MJ, White K (1996) ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol 6: 1634–1641. [DOI] [PubMed] [Google Scholar]
- 34. Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, et al. (1999) Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron 22: 707–717. [DOI] [PubMed] [Google Scholar]
- 35. Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, et al. (1999) Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol 19: 6815–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silver SJ, Davies EL, Doyon L, Rebay I (2003) Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol Cell Biol 23: 5989–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu PX, Cheng J, Epstein JA, Maas RL (1997) Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc Natl Acad Sci U S A 94: 11974–11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bui QT, Zimmerman JE, Liu H, Bonini NM (2000) Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikeda K, Watanabe Y, Ohto H, Kawakami K (2002) Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol 22: 6759–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anderson AM, Weasner BM, Weasner BP, Kumar JP (2012) Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139: 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu X, Sano T, Guan Y, Nagata S, Hoffmann JA, et al. (2012) Drosophila EYA Regulates the Immune Response against DNA through an Evolutionarily Conserved Threonine Phosphatase Motif. PLoS One 7: e42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petersen NS (1990) Effects of heat and chemical stress on development. Adv Genet 28: 275–296. [DOI] [PubMed] [Google Scholar]
- 43. Cook PJ, Ju BG, Telese F, Wang X, Glass CK, et al. (2009) Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]