Abstract
The homeobox transcription factor Prox1 is highly expressed in adult hepatocytes and is involved in the regulation of bile acid synthesis and gluconeogenesis in the liver by interacting with other transcriptional activators or repressors. Recent studies showed that Prox1 could inhibit proliferation of hepatocellular carcinoma (HCC) cells and reduced Prox1 expression was associated with poor prognosis of HCC patients. However, the underlying mechanism by which Prox1 attenuates HCC growth is still unclear. In this study, we demonstrated that Prox1 induced senescence-like phenotype of HCC cells to reduce cell proliferation. Our results indicated that the tumor suppressor p53 is a key mediator of Prox1-induced growth suppression because Prox1 only induced senescence-like phenotype in HCC cells harboring wild type p53. In addition, knockdown of p53 by shRNA reversed the effect of Prox1. However, chromatin immunoprecipitation assay did not demonstrate the direct binding of Prox1 to proximal promoter of human p53 gene suggesting Prox1 might not directly activate p53 transcription. We found that Prox1 suppressed Twist expression in HCC cells and subsequently relieved its inhibition on p53 gene transcription. The involvement of Twist in the regulation of p53 by Prox1 was supported by the following evidence: (1) Prox1 inhibited Twist expression and promoter activity; (2) knockdown of Twist in SK-HEP-1 cells upregulated p53 expression and (3) ectopic expression of Twist counteracted Prox1-induced p53 transcription and senescence-like phenotype. We also indentified an E-box located at p53 promoter which is required for Twist to inhibit p53 expression. Finally, our animal experiment confirmed that Prox1 suppressed HCC growth in vivo. Collectively, we conclude that Prox1 suppresses proliferation of HCC cells via inhibiting Twist to trigger p53-dependent senescence-like phenotype.
Keywords: Prox1, Twist, p53, senescence, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide. Development of HCC is frequently induced by chronic liver injury which is caused by various etiological factors like hepatitis infection, alcoholism and aflatoxin contamination. HCC patients with large and unresectable tumors as a result of disease progression usually have poor treatment response and prognosis.1,2 Because the therapeutic effect of current anti-cancer drugs on HCC is not satisfied, study of the molecular mechanisms that control proliferation and progression of HCC cells are important for the development of new therapeutic strategies.
Senescence, an irreversible cell cycle arrest response, is a cellular program that can be activated by oncogenic signaling or replicative stress and is an important barrier to suppress tumor progression.3 Several senescence-associated characteristics have been used for the detection of this cellular process. First, senescent cells display senescence-associated β-galactosidase activity due to an increase of lysosome number in these cells.4 Second, senescent cells exhibit a significant change of chromatin remodeling which causes the formation of senescence-associated heterochromatin foci.5 Third, secretion of extracellular matrix proteins and soluble factors are also altered in senescent cells.6 Because senescence blocks continuous proliferation, cancer cells need to develop different strategies to escape from this process. For example, HCC cells may activate Twist, an inducer of tumor metastasis and epithelial-mesenchymal transition (EMT), to override premature senescence by abolishing p53-dependent pathway.7 Recent studies demonstrate that p53 re-activation, c-Myc inhibition or treatment of cyclin-dependent kinase inhibitors (CDKIs) could effectively evoke a senescence response to repress tumor growth.8-10 These results suggest that induction of senescence is a new strategy for cancer treatment.
Transcription factor Prospero-related homeobox 1 (Prox1) was originally cloned as the homolog of the Drosophila prospero gene and its expression was found in lens, heart, liver, kidney, skeletal muscle, pancreas, and central nervous system at different developmental stages.11,12 Although the role of Prox1 in development has been understood gradually by using different genetic animal studies, its function in tumorigenesis is largely unclear. Altered Prox1 expression has been found in a variety of human cancers. Overexpression of Prox1 is frequently detected in colon cancer, brain tumor and Kaposi sarcoma.13-15 Animal studies also demonstrate that Prox1 acts as a tumor promoter in these cancers. Our recent study showed that Prox1 promotes EMT by downregulating E-cadherin expression in colon cancer cells.16 We also find that Prox1 increases cell invasiveness by regulating integrins and matrix metalloproteinases. Conversely, defective Prox1 function caused by genomic deletion, mutation, and promoter methylation is generally found in breast cancer, hematological malignancies, pancreatic cancer, and liver cancer indicating a tumor suppressor role of this transcription factor.17-20 These studies suggest that Prox1 may function as an oncogene or a tumor suppressor gene in a cancer type-specific manner.
Prox1 is essential for liver development and is highly expressed in both embryonic hepatoblasts and adult hepatocytes. Livers from Prox1-null mice are much smaller than that of wild type mice because of reduced proliferation of hepatoblasts.21,22 In addition, inactivation of Prox1 results in abnormal cellular proliferation, downregulation of the cell cycle inhibitors p27 and p57, and inappropriate apoptosis.23 Therefore, Prox1 is an important regulator in the control of cell growth and may play a role in liver tumorigenesis. Recent studies demonstrated that reduction of Prox1 was found in HCC tissues and low expression of Prox1 was associated with poor prognosis and un-differentiation status.24,25 These data implicate a potential tumor suppressor role of Prox1 in HCC. However, the underlying mechanism by which Prox1 inhibits the development of HCC is unknown. In this study, we provide the first evidence that Prox1 inhibits Twist expression to upregulate p53-dependent senescence-like phenotype in HCC cells.
Results
Prox1 inhibits proliferation of HCC cells by inducing p53-dependent senescence-like phenotype
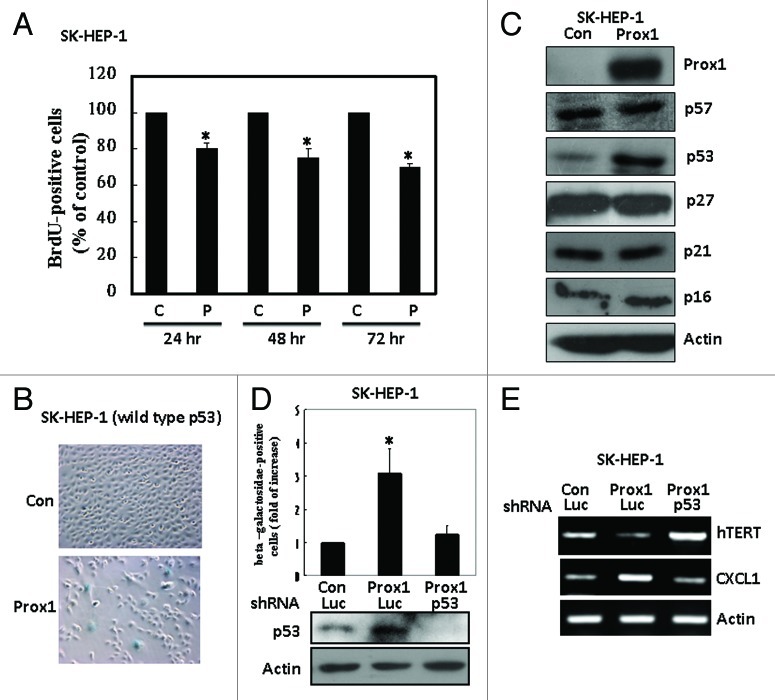
Screening of various HCC cell lines showed that SK-HEP-1 and Mahlavu cells are Prox1-negative while HepG2 and Hep3B cells are Prox1-positive (Fig. S1). We used SK-HEP-1 cells for overexpression study and found that Prox1 reduced proliferation (as indicated by BrdU incorporation) in a time-dependent manner with a 30% of inhibition was found at 72 h after Prox1 expression in these cells (Fig. 1A). However, no significant alteration of cell cycle distribution was detected (Fig. S2). In addition, no significant apoptotic death was found in Prox1-overexpressing cells by using caspase-3 activation as an indication (Fig. S3). Interestingly, we found that percentage of β-galactosidase-positive cells was significantly increased after Prox1 overexpression indicating an induction of senescence-like phenotype (Fig. 1B). Because senescence phenotype is strongly associated with increase of CDKIs, we examined the expression of CDKI proteins in control and Prox1-overexpressing cells. Among these inhibitory proteins, only p53 was dramatically increased (Fig. 1C). We used different experimental approaches to verify the importance of p53 in Prox1-induced senescence-like phenotype. We first demonstrated that increase of β-galactosidase-positive cells by Prox1 expression was totally abolished by knockdown of p53 by shRNA (Fig. 1D). In addition to β-galactosidase staining, we examined the expression of p53-regulated senescence-associated genes. Human telomerase reverse transcriptase (hTERT) and chemokine C-X-C motif ligand 1 (CXCL1) have been shown to be downregulated and upregulated separately by p53 during p53-mediated senescence. As shown in Figure 1E, we found that Prox1 significantly repressed hTERT expression and this effect was abolished when p53 was inhibited by shRNA. Our data also demonstrated that CXCL1 was increased by Prox1 and this effect was attenuated by p53 knockdown. Finally, we checked the effect of Prox1 on Hep3B (p53 deletion) and Mahlava (p53 mutation) cells and found that Prox1 could not trigger senescence-like phenotype in these cells (Fig. 2). Collectively, our results suggested that p53 is a key mediator of senescence in HCC cells induced by Prox1.
Figure 1. Prox1 induces cell senescence-like phenotype via p53 induction. (A) SK-HEP-1 cells were transfected with pcDNA (C) or Prox1 (P) expression vector. After different times, DNA synthesis was assessed by using BrdU incorporation assay and the number of cells with positive staining of the control group was defined as 100%. *p < 0.05 (B) Senescence-like phenotype was assessed by β-galactosidase staining at 72 h after transfection and typical microscope figures were shown. (C) Expression of various CDKIs in control and Prox1-overexpressing cells was studied by western blotting. (D) SK-HEP-1 cells were co-transfected with pcDNA (Con) or Prox1 with luciferase (Luc) or p53 shRNA for 48 h. β-galactosidase-positive cell number of the group transfected with pcDNA and luciferase shRNA was defined as 1. *p < 0.05. Protein level of p53 was studied by western blotting. (E) SK-HEP-1 cells were co-transfected with pcDNA (Con), Prox1 expression vector, luciferase (Luc) or p53 shRNA for 48 h. Expression of hTERT and CXCL1 was examined by RT-PCR.
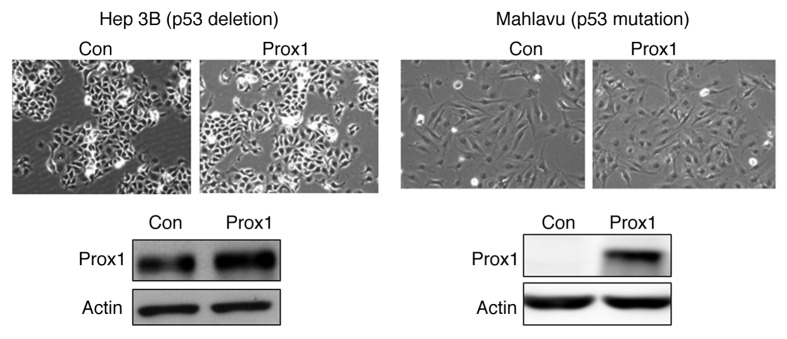
Figure 2. Prox1 did not increase β-galactosidase staining in HCC cells with p53 deficiency. p53-deleted Hep3B cells or p53-mutated Mahlavu cells were transfected with pcDNA (Con) or Prox1 expression vector. After 72 h, cells were subjected to β-galactosidase staining and Prox1 protein level was studied by western blotting.
Prox1 increases p53 level via an indirect mechanism
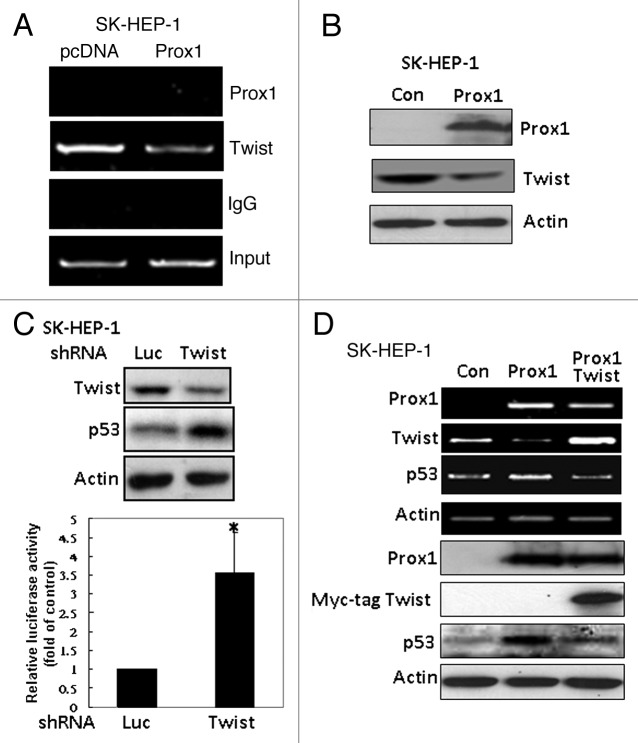
We next studied whether p53 is a direct transcriptional target of Prox1. Unexpectedly, we could not detect the binding of Prox1 to the p53 proximal promoter (Fig. 3A). Therefore, we hypothesized that Prox1 might inhibit a transcriptional repressor of p53 gene to restore p53 expression. Our ChIP data demonstrated that Twist, a negative regulator of p53 which has been reported to override p53-induced senescence,7 was overexpressed in SK-HEP-1 cells and it constitutively bound to p53 promoter (Fig. 3A). Enforced expression of Prox1 reduced Twist protein level (Fig. 3B) and attenuated its binding to p53 promoter (Fig. 3A). Knockdown of Twist expression by shRNA increased p53 promoter activity and protein level (Fig. 3C). More importantly, overexpression of Twist effectively counteracted Prox1-increased p53 expression (Fig. 3D).
Figure 3. Inhibition of Twist by Prox1 is required for the upregulation of p53. (A) SK-HEP-1 cells were trasnfected with pcDNA or Prox1 expression vector. After 48 h, ChIP assay was performed by using anti-Prox1 or anti-Twist antibody to detect the binding of these two proteins to human p53 proximal promoter region. (B) Effect of Prox1 on Twist expression was investigated by western blotting. (C) SK-HEP-1 cells were co-transfected human p53 promoter with luciferase (Luc) or Twist shRNA for 48 h. Protein levels of Twist and p53 (upper panel) and luciferase activity (bottom panel) were determined. Promoter activity of cells co-transfected with luciferase shRNA was defined as 100%. *p < 0.05 when compared with the control group. (D) SK-HEP-1 cells were transfected with pcDNA (Con), Prox1 or Prox+Myc-tag Twist vector. After 48 h, RT-PCR and western blotting were performed to study Prox1, Twist and p53 expression.
To confirm these results, we used HepG2 cells which expressed endogenous Prox1 and wild type p53 for investigation. As shown in Figure 4A, expression of Twist repressed p53 promoter activity as well as protein level in HepG2 cells. Conversely, knockdown of endogenous Prox1 caused upregulation of Twist and downregulation of p53 (Fig. 4B). It has been demonstrated that Twist binds to the consensus E-box sequence 5′-CANNTG-3′ to activate or repress gene transcription.26 By using bioinformatics analysis, we identified an E-box located at -286/-281 region of human p53 promoter. We then tested the effect of Twist on wild type and E-box mutant of human p53 gene promoter. As shown in Figure 4C, ectopic expression of Twist in HepG2 cells inhibited wild-type p53 promoter activity but not the E-box mutant. In SK-HEP-1 cells which expressed high level of Twist, the promoter activity of E-box mutant was higher than that of wild-type promoter due to diminishment of Twist-mediated inhibition (Fig. 4D). These data suggested that Prox1 increases p53 expression by suppressing Twist.
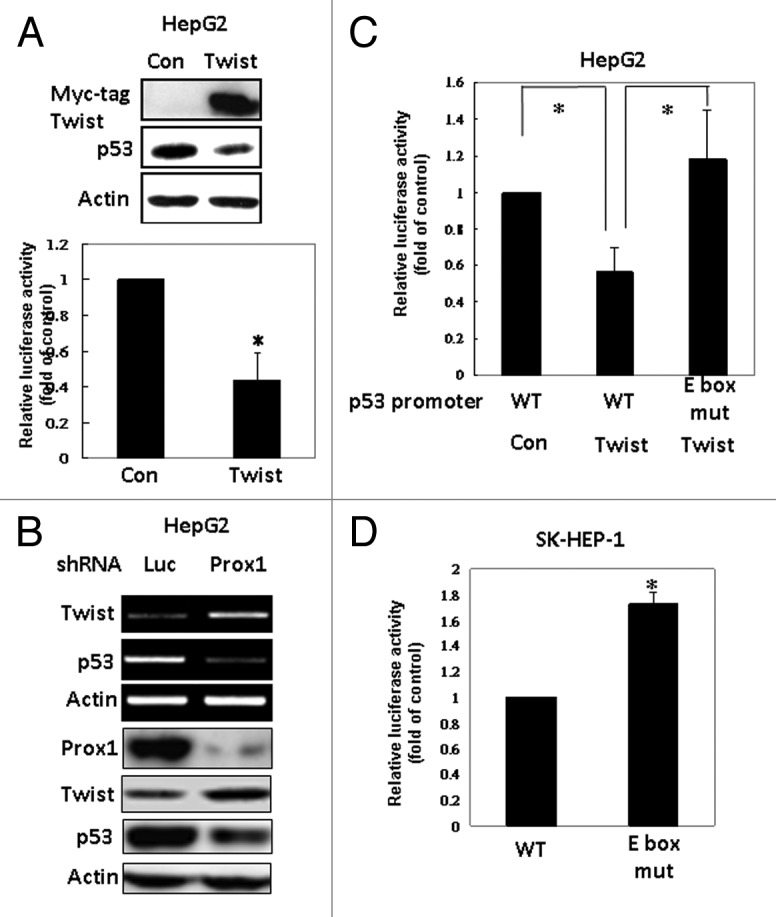
Figure 4. Twist inhibits p53 expression via E-box in the promoter. (A) HepG2 cells were co-transfected p53 promoter with pcDNA (Con) or Myc-tag Twist vector. After 24 h, p53 promoter activity and protein level were determined. (B) HepG2 cells were transfected with luciferase (as a control) or Prox1 shRNA. Protein and mRNA levels of Prox1, Twist and p53 were studied. (C) HepG2 cells were co-transfected human wild type (WT) or E-box mutant of human p53 promoter with pcDNA (Con) or Myc-tagged Twist for 48 h and luciferase activity was determined. Promoter activity of cells co-transfected with pcDNA was defined as 100%. *p < 0.05 when compared with the control group. (D) SK-HEP-1 cells were transfected with wild-type or E-box mutant p53 promoter and promoter activity was determined.
Twist counteracts Prox1-induced senescence-like phenotype and inhibition of anchorage-independent growth
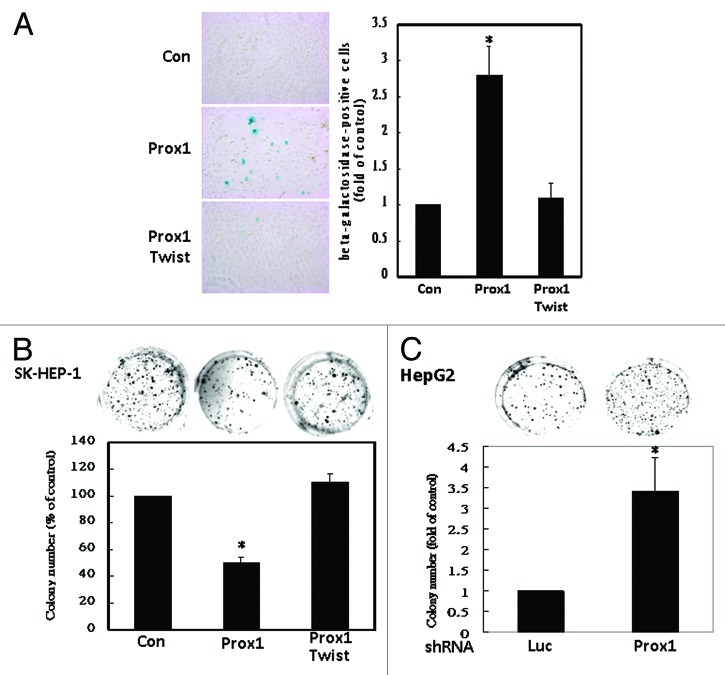
We next studied whether Twist could antagonize anti-proliferative function of Prox1. As shown in Figure 5A, Twist reversed the increase of β-galactosidase-positive cells induced by Prox1. Induction of senescence-like phenotype by Prox1 also caused reduction of anchorage-independent growth (colony formation) which could be counteracted by co-expression of Twist (Fig. 5B). On the contrary, knockdown of endogenous Prox1 increased colony formation of HepG2 cells (Fig. 5C). These results suggested that Twist could overcome Prox1-induced senescence-like phenotype and inhibition of anchorage-independent growth.
Figure 5. Prox1-induced senescence-like phenotype and inhibition of anchorage- independent growth were abolished by Twist. (A) SK-HEP-1 cells were transfected with pcDNA (Con), Prox1 or Myc-tagged Twist expression vectors. β-galactosidase staining was performed and the number of positive signal of the control group was defined as 1. *p < 0.05. (B) SK-HEP-1 cells were trasnfected with control, Prox1 or Twist expression vectors. Colonies were counted and the number of the control group was defined as 100%.*p < 0.05. (C) HepG2 cells were trasnfected with luciferase (Luc) or Prox1 shRNA. Colonies were counted and the number of control group was defined as 1. *p < 0.05.
Inhibition of tumor growth by Prox1 in vivo
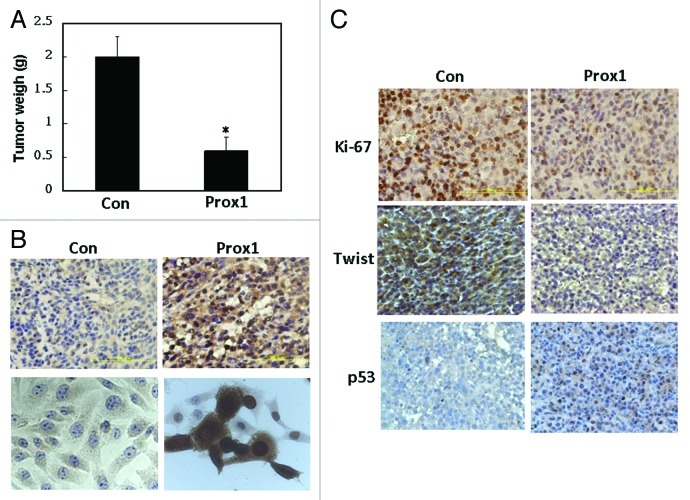
To test the anti-proliferative effect of Prox1 in vivo, subcutaneous injection model was used. As shown in Figure 6A, Prox1 inhibited tumor growth of SK-HEP-1 cells by 65–70%. Our immunohistochemical staining (by using the antibody from Abnova) showed that Prox1 was expressed both in the nucleus and cytoplasm (Fig. 6B, upper panel). We also did immuncytochemical staining of control and Prox1-expressing SK-HEP-1 cells to confirm the specificity of Prox1 staining. No staining was detected in control cells and strong Prox1 signal was found both in the nucleus and cytoplasm (Fig. 6B, bottom panel). To further confirm the results, we used another Prox1 antibody (obtained from AngioBio) to do immunocytochemical and immunohistochemical staining. Similar nuclear and cytoplasmic staining pattern was found (Fig. S4). Reduction of tumor growth was also confirmed by a significant reduction of cell proliferation marker Ki-67 in Prox1-expressing tumor tissues (Fig. 6C). In consistent with the results of cell-based experiments, tumors of the control group showed high level of Twist protein (Fig. 6C). Conversely, it was dramatically reduced in Prox1 group. In addition, few p53-positive cells were detected in control group while it was significantly increased in Prox1 group (Fig. 6C). These data suggested Prox1 exhibits potent anti-proliferative activity by suppressing Twist and inducing p53 in vivo.
Figure 6. Prox1 inhibits tumor growth in vivo. (A) SK-HEP-1 cells were transfected with pcDNA or Prox1 expression vector and subjected to subcutaneous injection as described in Materials and Methods. Tumor growth was continuously monitored and mice were sacrificed at 14 d after injection. Tumors were removed and weighted. *p < 0.05 when compared with the control group. (B) Immunohistochemical staining of tumor tissues (upper panel) and immunocytochemical staining of control and Prox1- expressing SK-HEP-1 cells (bottom panel) were conducted. Representative images were shown. (C) Tumor tissue sections were stained with Ki-67, Twist and p53 antibodies and representative images were shown.
Discussion
Prox1 has been identified to be an important regulator during the development of lymphatic, lens, heart, liver, kidney, skeletal muscle, pancreas, and central nervous system.22,23,27,28 Recent studies showed that Prox1 may be involved in the progression or suppression of malignancies, such as HCC, cholangiocellular carcinoma, hematologic malignancies, breast cancer, and pancreatic cancer.18,19,24,25 How Prox1 acts as an oncogene or tumor suppressor gene in different cancer types is unclear. Petrova et al. demonstrated that Prox1 induced colon cancer progression by promoting the transition from benign to highly dysplastic phenotype indicating a tumor-promoting role.13 Interestingly, they found that Prox1 expression was controlled by the β-catenin/TCF signaling pathway. On the contrary, Prox1 expression was not regulated by β-catenin in HCC cells in which Prox1 functions as a tumor suppressor. Our recent study further elucidated the oncogenic role of Prox1 in colon cancer. We found that Prox1 increased miR-9 expression to inhibit E-cadherin and subsequently enhanced cell invasiveness.16 Importantly, we also found that Prox1 only controlled the expression of miR-9 in colon cancer cells but not HCC cells. One possible explanation is that Prox1 needs to work with other transcription activators or repressors to exert its function. The cancer type-specific action of Prox1 warrants further investigations.
The study of functional role of Prox1 in HCC was first reported by Shimoda et al.24 They showed that Prox1 expression was associated with the differentiation status of HCC tumors. In addition, they also demonstrated that overexpression of Prox1 suppressed proliferation of HCC cells while knockdown of Prox1 by shRNA accelerated cell growth. These data suggest a tumor-suppressive role of Prox1 in HCC cells. However, the underlying mechanism of this anti-proliferative function is largely unknown. In this study, we provide evidence that Prox1 may trigger p53-dependent senescence-like phenotype in HCC cells. Our results also demonstrate that Twist is involved in the upregulation of p53 by Prox1. Indeed, our recent data confirm that Twist is a direct target suppressed by Prox1 via transcription repression.29
Twist is an important oncoprotein and a key regulator of epithelial-mesenchymal transition (EMT). The prominent function of Twist is enhancement of cancer progression and metastasis through EMT induction.30 In addition to induction of EMT, Twist may promote tumorigenesis by inhibiting cancer cell apoptosis. Previous study has demonstrated that Twist may counteract p53-mediated apoptotic effect.31 Ansieau et al. also find that Twist can override oncogene-induced senescence7 implying that Twist can prevent growth inhibition by counteracting senescence. It is well known that p53 can trigger cellular senescence.32,33 In addition, p53-induced senescence is a powerful suppressive mechanism to against tumorigenesis and restoration of p53 expression is known to cause tumor regression.8,34 Our results demonstrate for the first time that Prox1 can inhibit Twist and relieve its inhibition on p53 expression. Upregulation of p53 subsequently causes senescence-like phenotype and inhibits HCC growth in vitro and in vivo. Inhibition of p53 by Twist via multiple mechanisms has been previously demonstrated. Maetro et al. show that Twist may affect p53 protein level indirectly through modulation of the ARF/MDM2/p53 pathway.31 A subsequent study using the Drosophila model to study the functions of Twist orthologs in apoptosis shows that Twist inhibits the p53 pathway independent of ARF and MDM2.35 Recently, Shiota et al. find that Twist directly interacts with the DNA binding domain of p53 to suppress its DNA-binding and transcriptional activity to repress downstream target gene expression.36 Interestingly, we find that Twist reduces p53 mRNA expression and promoter activity. This inhibition required the consensus E-box sequence on p53 promoter suggesting a direct transcriptional repression. Further studies will be needed to clarify how Twist acts via different mechanisms to inhibit p53 function in different types of cancer cells.
Another important finding of this study is that we find Prox1 is located both at nuclei and cytoplasm in our immunocytochemical and immunohistochemical study. This conclusion is supported by the following evidence. First, our immunocytochemical assay clearly demonstrates that parental SK-HEP-1 cells do not have Prox1-positive signal indicating the staining specificity is of no problem. Second, two different antibodies are used for staining and both antibodies show similar nuclear and cytoplamic staining. Prox1 protein has been shown to be predominately distributed into cytoplasmic in the lens placode as well as the lens epithelium and germinative zone throughout development. When fiber cells undergo differentiation, Prox1 protein redistributes into cell nuclei.37 However, the difference of subcellular distribution of Prox1 in cancer cells has little been reported. Whether cytoplasmic Prox1 has any role in HCC development needs further investigation.
In summary, our results provide a molecular basis to explain the anti-proliferative action of Prox1 in HCC cells. Our data propose that Prox1 might be a molecular target for the development of new strategy for HCC treatment.
Materials and Methods
Cell culture and experimental reagents
Four HCC cell lines HepG2, Hep3B, Mahlavu and SK-HEP-1 were provided by Dr. Ming-Hong Tai (National Sun Yat-sen University; Kaohsiung, Taiwan). Cells were cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 containing 10% fetal calf serum and antibiotics. Flag-tagged Prox1 was a gift of You Hua Xie (Shanghai Institute for Biological Sciences; Shanghai, China). MYC-tagged Twist was provided by Dr. Lu-Hai Wang (National Health Research Institutes; Miaoli, Taiwan). p53 promoter-luciferase construct was obtained from Dr. Wen-Sen Lee (Taipei Medical University; Taipei, Taiwan). Lipofectamin2000 was purchased from Invitrogrn. Anti-Prox1 antibodies were purchased from UpState (07‑537), AngioBio (11‑002) and Abnova (PAB1919). Anti-actin antibody (MAB1501) was obtained from Millipore. Anti-Twist (H81), anti‑p53 (DO1) and anti-c-MYC (9E11) were purchased from Santa Cruz Biotechnology. shRNAs were purchased from the National RNAi Core Facility of Academic Sinica (Taipei).
Cell proliferation assay
pcDNA or Flag-tagged Prox1-transfected cells were subjected for cell proliferation assay by using BrdU incorporation assay purchased from Roche Applied Science. Standard procedure followed the manufacturer’s protocol.
RNA isolation, reverse transcription-polymerase chain reaction (RT-PCR) and western blot analysis
These assays were done as described previously.38 The primer sequences used for RT-PCR are shown in Figure S5.
Construction of mutant p53 promoter-reporter plasmids
The p53 promoter-luciferase construct containing the -291 to +71 region of human p53 gene was kindly provided by Dr. Lee WS (Taipei Medical University; Tapei, Taiwan) and was used to generate E-box site-mutated p53 promoter. Mutation was introduced into the E-box site located at -286/-281 bp region of p53 promoter by the QuikChange Site-directed mutagenesis kit according to the procedures of manufacturer (Stratagene). The sequences of the primers are forward: 5′-CCGAGCTCGGATCTGGCTGAGAGCAAACGC-3′ and reverse: 5′-GCGTTTGCTCTCAGCCAGATCCGAGCTCGG.-3′. The mutated nucleotides were indicated as boldface. Direct sequencing was performed to verify the success of each mutation.
Promoter activity assay
Promoter activity of p53 gene was analyzed as described previously.39 In brief, cells were plated onto 12‑well plates at the density of 100,000 cells/well and grown overnight. The wild-type or mutant promoter-luciferase construct was transfected into cells expressing luciferase (Luc) or Twist shRNA and pcDNA (control) or MYC-tagged Twist plasmid. After transfection, cells were incubated in medium containing 10% FCS for 48 h and promoter activity was determined by using the Firefly luciferase assay system (Promega Corp.) and normalized for the concentration of cellular proteins. Data of three independent experiments were expressed as mean ± SE. Paired results were evaluated by the Student’s t-test and p-value < 0.05 was considered significant.
Chromatin immunoprecipitation assay
pcDNA or Flag-tagged Prox1-transfected cells were fixed with 1% formaldehyde at 37°C for 10 min. Cells were harvested and chromatin immunoprecipitation assay was done as described previously.39 Anti-Twist, anti-Prox1 or anti-GATA-1 (negative control) antibodies were used for precipitating the protein/DNA complex. DNA fragments were recovered and subjected to PCR amplification by using the primers specific for the detection of -315 to -35 bp regions that contained the Twist (-287/-282) and Prox1 binding site (-191/-185, -89/-83 and -63/-57) in human p53 gene promoter. The sequences for the primers are forward: 5′- CATTTTAACTGATGAGAAGAAAGGA-3′, reverse: 5′-GGACAGTCGCCATGA CAAG-3′.
Senescence-associated β-galactosidase staining
pcDNA or Flag-tagged tagged Prox1-transfected cells were subjected for cellular senescence detection by using Senescence β-Galactosidase Staining Kit purchased from Cell Signaling Technology, Inc. Standard procedure followed the manufactory’s protocol. β-galactosidase-positive cells were counted 10 fields of each well and positive cells were presented on a light microscope.
Anchorage-independent growth assay
1 × 104 cells were suspended in complete medium containing 0.3% agar. The cells were then placed into 6-well plates containing a solid agar bottom composed of complete medium and 0.7% agar. The cultural plates were grown in incubator and the number of colony formation was counted by crystal violet staining at 3 weeks after seeding.
Tumor xenografts in BALB/c nude mice and immunohistochemal analysis
Animal studies were approved by the Animal Care and Ethics Committee of the National Sun Yat-Sen University (Kaohsiung, Taiwan). Five-week-old male athymic nude mice (BALB/c-nu/nu) were obtained from the National Laboratory Animal Center. On day 1, two groups (n = 5 for each group) of mice were subcutaneously injected with pcDNA or Flag-tagged Prox1-transfected SK-HEP-1 cells (1 × 106 cells in 0.1 mL PBS) on the right dorsal flank. Tumor diameter was measured with a caliber ruler every 3 d. Tumor volume (mm3) was estimated by using the formula: volume = (shortest diameter)2 × (longest diameter) × 0.5. After continuous observation for 2 weeks, mice were sacrificed and the tumors were dissected. Weight of tumors were measured and embedded in paraffin using routine procedures and subjected for immunohistochemical (IHC) analysis. The paraffin-embedded tumor sections were obtained from nude mice. IHC protocol was performed as previous described.40
Statistical analysis
Student’s t-test was used to evaluate the difference between various experimental groups. Differences were considered to be significant at p < 0.05.
Supplementary Material
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgment
We thank Dr. M.H. Tai, Dr. Y.H. Xie, Dr. L.H. Wang and Dr. W.S. Lee for providing experimental materials. This study was supported by the following grants: (1) NSC 99–2628-B-400–004-MY3 from National Science Council, Republic of China; (2) DOH 101-TD-C-111–002 and DOH 101-TD-C-111–004 from Department of Health, Taiwan, Republic of China.
Glossary
Abbreviations:
- Prox1
prospero-related homeobox 1
- HCC
hepatocellular carcinoma
- CDKIs
cyclin-dependent kinase inhibitors
- EMT
epithelial-mesenchymal transition
- ChIP
chromatin immunoprecipitation
- IHC
immunohistochemistry
Supplemental Material
Supplemental material may be found here:
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/23293
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Acosta JC, Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol. 2012;22:211–9. doi: 10.1016/j.tcb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113:3613–22. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 5.Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 6.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 7.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci U S A. 2007;104:13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol 2010; 12:54-9; sup pp 1-14. [DOI] [PubMed] [Google Scholar]
- 11.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-M. [DOI] [PubMed] [Google Scholar]
- 12.Zinovieva RD, Duncan MK, Johnson TR, Torres R, Polymeropoulos MH, Tomarev SI. Structure and chromosomal localization of the human homeobox gene Prox 1. Genomics. 1996;35:517–22. doi: 10.1006/geno.1996.0392. [DOI] [PubMed] [Google Scholar]
- 13.Petrova TV, Nykänen A, Norrmén C, Ivanov KI, Andersson LC, Haglund C, et al. Transcription factor PROX1 induces colon cancer progression by promoting the transition from benign to highly dysplastic phenotype. Cancer Cell. 2008;13:407–19. doi: 10.1016/j.ccr.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Elsir T, Eriksson A, Orrego A, Lindström MS, Nistér M. Expression of PROX1 Is a common feature of high-grade malignant astrocytic gliomas. J Neuropathol Exp Neurol. 2010;69:129–38. doi: 10.1097/NEN.0b013e3181ca4767. [DOI] [PubMed] [Google Scholar]
- 15.Dadras SS, Skrzypek A, Nguyen L, Shin JW, Schulz MM, Arbiser J, et al. Prox-1 promotes invasion of kaposiform hemangioendotheliomas. J Invest Dermatol. 2008;128:2798–806. doi: 10.1038/jid.2008.176. [DOI] [PubMed] [Google Scholar]
- 16.Leu MH, Huang CC, Pan MR, Chen HH, Hung WC. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0832. [DOI] [PubMed] [Google Scholar]
- 17.Versmold B, Felsberg J, Mikeska T, Ehrentraut D, Köhler J, Hampl JA, et al. Epigenetic silencing of the candidate tumor suppressor gene PROX1 in sporadic breast cancer. Int J Cancer. 2007;121:547–54. doi: 10.1002/ijc.22705. [DOI] [PubMed] [Google Scholar]
- 18.Nagai H, Li Y, Hatano S, Toshihito O, Yuge M, Ito E, et al. Mutations and aberrant DNA methylation of the PROX1 gene in hematologic malignancies. Genes Chromosomes Cancer. 2003;38:13–21. doi: 10.1002/gcc.10248. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi M, Yoshimoto T, Shimoda M, Kono T, Koizumi M, Yazumi S, et al. Loss of function of the candidate tumor suppressor prox1 by RNA mutation in human cancer cells. Neoplasia. 2006;8:1003–10. doi: 10.1593/neo.06595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudas J, Mansuroglu T, Moriconi F, Haller F, Wilting J, Lorf T, et al. Altered regulation of Prox1-gene-expression in liver tumors. BMC Cancer. 2008;8:92. doi: 10.1186/1471-2407-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudas J, Papoutsi M, Hecht M, Elmaouhoub A, Saile B, Christ B, et al. The homeobox transcription factor Prox1 is highly conserved in embryonic hepatoblasts and in adult and transformed hepatocytes, but is absent from bile duct epithelium. Anat Embryol (Berl) 2004;208:359–66. doi: 10.1007/s00429-004-0403-4. [DOI] [PubMed] [Google Scholar]
- 22.Sosa-Pineda B, Wigle JT, Oliver G. Hepatocyte migration during liver development requires Prox1. Nat Genet. 2000;25:254–5. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 23.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–22. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 24.Shimoda M, Takahashi M, Yoshimoto T, Kono T, Ikai I, Kubo H. A homeobox protein, prox1, is involved in the differentiation, proliferation, and prognosis in hepatocellular carcinoma. Clin Cancer Res. 2006;12:6005–11. doi: 10.1158/1078-0432.CCR-06-0712. [DOI] [PubMed] [Google Scholar]
- 25.Laerm A, Helmbold P, Goldberg M, Dammann R, Holzhausen HJ, Ballhausen WG. Prospero-related homeobox 1 (PROX1) is frequently inactivated by genomic deletions and epigenetic silencing in carcinomas of the bilary system. J Hepatol. 2007;46:89–97. doi: 10.1016/j.jhep.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 26.Castanon I, Baylies MK. A Twist in fate: evolutionary comparison of Twist structure and function. Gene. 2002;287:11–22. doi: 10.1016/S0378-1119(01)00893-9. [DOI] [PubMed] [Google Scholar]
- 27.Wigle JT, Oliver G. Prox1 function is required for the development of the murine lymphatic system. Cell. 1999;98:769–78. doi: 10.1016/S0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 28.Risebro CA, Searles RG, Melville AA, Ehler E, Jina N, Shah S, et al. Prox1 maintains muscle structure and growth in the developing heart. Development. 2009;136:495–505. doi: 10.1242/dev.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang TM, Hung WC. Transcriptional repression of TWIST1 gene by Prospero-related homeobox 1 inhibits invasiveness of hepatocellular carcinoma cells. FEBS Lett. 2012;586:3746–52. doi: 10.1016/j.febslet.2012.08.034. [DOI] [PubMed] [Google Scholar]
- 30.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–28. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 31.Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–17. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 34.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 35.Gullaud M, Delanoue R, Silber J. A Drosophila model to study the functions of TWIST orthologs in apoptosis and proliferation. Cell Death Differ. 2003;10:641–51. doi: 10.1038/sj.cdd.4401222. [DOI] [PubMed] [Google Scholar]
- 36.Shiota M, Izumi H, Onitsuka T, Miyamoto N, Kashiwagi E, Kidani A, et al. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 2008;27:5543–53. doi: 10.1038/onc.2008.176. [DOI] [PubMed] [Google Scholar]
- 37.Duncan MK, Cui W, Oh DJ, Tomarev SI. Prox1 is differentially localized during lens development. Mech Dev. 2002;112:195–8. doi: 10.1016/S0925-4773(01)00645-1. [DOI] [PubMed] [Google Scholar]
- 38.Liu LT, Chang HC, Chiang LC, Hung WC. Histone deacetylase inhibitor up-regulates RECK to inhibit MMP-2 activation and cancer cell invasion. Cancer Res. 2003;63:3069–72. [PubMed] [Google Scholar]
- 39.Hsu MC, Chang HC, Hung WC. HER-2/neu represses the metastasis suppressor RECK via ERK and Sp transcription factors to promote cell invasion. J Biol Chem. 2006;281:4718–25. doi: 10.1074/jbc.M510937200. [DOI] [PubMed] [Google Scholar]
- 40.Pan MR, Chang HC, Wu YC, Huang CC, Hung WC. Tubocapsanolide A inhibits transforming growth factor-beta-activating kinase 1 to suppress NF-kappaB-induced CCR7. J Biol Chem. 2009;284:2746–54. doi: 10.1074/jbc.M806223200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.