Abstract
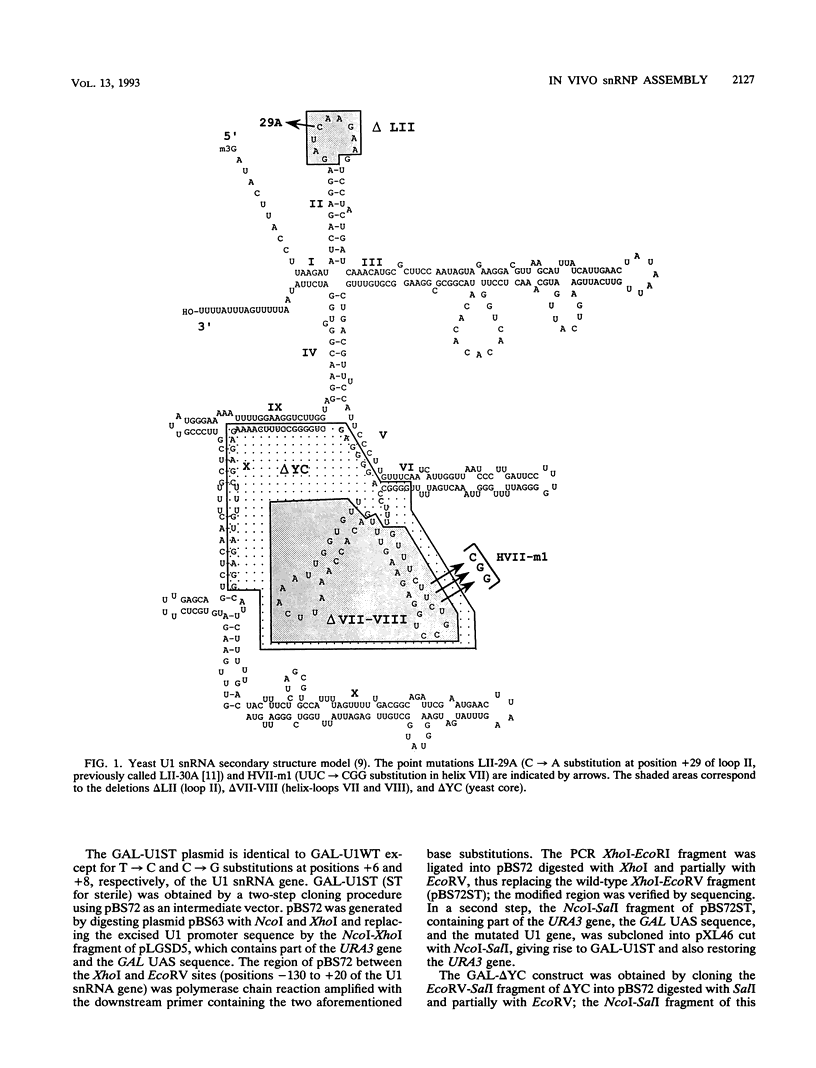
Two highly conserved regions of the 586-nucleotide yeast (Saccharomyces cerevisiae) U1 small nuclear RNA (snRNA) can be mutated or deleted with little or no effect on growth rate: the universally conserved loop II (corresponding to the metazoan A loop) and the yeast core region (X. Liao, L. Kretzner, B. Séraphin, and M. Rosbash, Genes Dev. 4:1766-1774, 1990). To examine the contribution of these regions to U1 small nuclear ribonucleoprotein particle (snRNP) activity, a competitor U1 gene, encoding a nonfunctional U1 snRNA molecule, was introduced into a number of strains carrying a U1 snRNA gene with loop II or yeast core mutations. The presence of the nonfunctional U1 gene lowered the growth rate of these mutant strains but not wild-type strains, consistent with the notion that mutant U1 RNAs are less active than wild-type U1 snRNAs. A detailed analysis of the U1 snRNA levels and half-lives in a number of merodiploid strains suggests that these mutant U1 snRNAs interact with U1 snRNP proteins less well than do their wild-type counterparts. Competition for protein factors during snRNP assembly could account for a number of previous observations in both yeast and mammalian cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fabrizio P., McPheeters D. S., Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989 Dec;3(12B):2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S. M., Mattaj I. W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990 Aug 10;62(3):569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Scherly D., Mattaj I. W. Multiple domains of U1 snRNA, including U1 specific protein binding sites, are required for splicing. EMBO J. 1990 Apr;9(4):1237–1244. doi: 10.1002/j.1460-2075.1990.tb08231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Kazmaier M., Mattaj I. W. In vitro assembly of U1 snRNPs. EMBO J. 1987 Nov;6(11):3479–3485. doi: 10.1002/j.1460-2075.1987.tb02672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Heinrichs V., Bach M., Winkelmann G., Lührmann R. U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5' splice site. Science. 1990 Jan 5;247(4938):69–72. doi: 10.1126/science.2136774. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Krol A., Rosbash M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc Natl Acad Sci U S A. 1990 Jan;87(2):851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Rymond B. C., Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987 Aug 14;50(4):593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Liao X. L., Kretzner L., Seraphin B., Rosbash M. Universally conserved and yeast-specific U1 snRNA sequences are important but not essential for U1 snRNP function. Genes Dev. 1990 Oct;4(10):1766–1774. doi: 10.1101/gad.4.10.1766. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986 Sep 12;46(6):905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Fabrizio P., Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989 Dec;3(12B):2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- Nelissen R. L., Heinrichs V., Habets W. J., Simons F., Lührmann R., van Venrooij W. J. Zinc finger-like structure in U1-specific protein C is essential for specific binding to U1 snRNP. Nucleic Acids Res. 1991 Feb 11;19(3):449–454. doi: 10.1093/nar/19.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Schena M., Yamamoto K. R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988 Aug 19;241(4868):965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- Shulman R. W., Warner J. R. Ribosomal RNA transcription in a mutant of Saccharomyces cerevisiae defective in ribosomal protein synthesis. Mol Gen Genet. 1978 May 3;161(2):221–223. doi: 10.1007/BF00274191. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Jones M. H., Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987 Sep 18;237(4821):1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Kivens W. J., Guthrie C. More than half of yeast U1 snRNA is dispensable for growth. Nucleic Acids Res. 1991 Dec 11;19(23):6367–6372. doi: 10.1093/nar/19.23.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Mutational analysis of the interactions between U1 small nuclear RNA and pre-mRNA of yeast. Gene. 1989 Oct 15;82(1):145–151. doi: 10.1016/0378-1119(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Udem S. A. Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972 Mar 28;65(2):243–257. doi: 10.1016/0022-2836(72)90280-x. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Ohshima Y. Three types of rat U1 small nuclear RNA genes with different flanking sequences are induced to express in vivo. Eur J Biochem. 1988 May 16;174(1):125–132. doi: 10.1111/j.1432-1033.1988.tb14071.x. [DOI] [PubMed] [Google Scholar]
- Yuo C. Y., Weiner A. M. Genetic analysis of the role of human U1 snRNA in mRNA splicing: I. Effect of mutations in the highly conserved stem-loop I of U1. Genes Dev. 1989 May;3(5):697–707. doi: 10.1101/gad.3.5.697. [DOI] [PubMed] [Google Scholar]