Abstract
IL-2 is a growth factor for activated T cells and is required for maintenance of naturally arising regulatory T cells (nTregs). Mice defective in IL-2/IL-2 receptor signaling pathways have impaired nTregs and suffer from lymphoproliferative disorders, suggesting that IL-2 is present and functional in healthy animals. However, the cellular source of IL-2 is currently unknown. To determine which cells produce IL-2 in healthy animals, we established mice carrying cre gene knock in at the il-2 locus (termed IL-2cre). When IL-2cre mice were crossed with EGFP reporter mice, EGFP was exclusively expressed by a fraction of CD4 T cells present in both lymphoid and non-lymphoid tissues. Live imaging of IL-2cre mice that carry the luciferase reporter showed concentrated localization of luciferase+ cells in Peyer's patches. These cells were not observed in new born mice but appeared within 3 days after birth. Reduction of antigen receptor repertoire by transgene expression reduced their number, indicating that recognition of environmental antigens is necessary for generation of these IL-2 producers in healthy animals. A substantial fraction of EGFP+ cells also produce IL-10 and IFN- , a characteristic profile of type 1 regulatory T cells (Tr1). The data suggest that a group of Tr1 cells have addition roles in immune homeostasis by producing IL-2 along with other cytokines and help maintaining Tregs.
Keywords: IL-2, IL-10, IFN-, Tr-1, Tregs
Introduction
IL-2 is a major growth factor for activated T cells and it is required for differentiation of activated naïve CD4 T cells into effector and memory CD4 T cells [1, 2]. Naïve CD4 T cells become activated after antigen recognition and begin to produce IL-2. Expression of IL-2 is tightly regulated in naïve CD4 T cells and requires an extensive period of antigen stimulation [3] . Most activated CD4 T cells die after 5-7 days of stimulation by activation induced cell death (AICD), and IL-2 also plays a critical role in AICD to induce cell death [4]. Moreover, IL-2 is essential for functional naturally arising regulatory T cells (nTregs) [5, 6]. il-2 knock-out mice as well as il-2r (known as CD25), il-2r (known as CD122), and stat5 (a signal transducer through IL-2-receptor) knock-out mice have profound defects in nTreg populations and undergo lymphoproliferation followed by lethal autoimmunity [reviewed by [5, 7]]. Since nTregs do not secrete IL-2, they depend on IL-2 from paracrine sources but the identity of cells that produce IL-2 in healthy animals and maintain functional nTregs is unknown.
Production of IL-2 in vivo has been studied using several different systems. One approach was the use of egfp knock-in at the il-2 locus [8]. In this system, the egfp gene was inserted in exon 1 of the il-2 gene. T cells from these mice expressed egfp in a manner indistinguishable from that of endogenous il-2 gene expression. Yet, these studies did not detect IL-2 expression when mice were not immunized with antigens, likely due to the lack of sensitivity of the system. Expression of IL-2 was also studied using transgenic mice that express EGFP under the control of the IL-2 promoter [9, 10]. These mice expressed IL-2 in response to TCR stimulation but the expression of egfp was somewhat different from the expression of the endogenous il-2 gene and again, these studies did not determine the presence of cells that express IL-2 in vivo under un-immunized conditions. Moreover, fate or localization of cells that produced IL-2 is currently unknown.
To identify which cells produce IL-2 in healthy animals, we established IL-2cre mice in which the cre gene was inserted into the il-2 locus. In IL-2cre mice, ex vivo cre gene expression was limited to activated T cells and the pattern of Cre expression mirrored endogenous il-2 gene expression. When IL-2cre mice were crossed with EGFP reporter mice, we found that EGFP is exclusively expressed in CD4 T cells in vivo. These EGFP+ CD4 cells, when re-stimulated ex vivo, produced IL-2 along with IFN and IL-10, but not IL-4, resembling the characteristics of IL-10-producing type 1 regulatory T cells (Tr1).
Materials and Methods
Generation of IL-2Cre mice
IL-2Cre knock-in mice were generated by inserting cre cDNA [11] into the il-2 gene just upstream of the first codon using genomic DNA fragment isolated from C57.BL/6 genomic DNA (liver) library. We placed an internal ribosomal entry sequence (IRES) into the 3’ of the cre gene to allow for the co-translation of the il-2 mRNA along with the cre mRNA. Genomic DNA extracted from tail was screened for genotyping by TaKaRa Ex Taq PCR system (TaKaRa, Otsu, Japan). Wild type il-2 allele was amplified with primers IL-2-5’: 5′-TGCCACACAGGTAGACTCTT-3′and IL-2cre -3’: 5′-GCTGTAGAGCTTGAAGTGGA-3′, in the following heating cycle: 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 minute; for 35 cycles. il-2cre allele was amplified with primers IL-2-5’ and IL-2-3’: 5’-ACGTTCTCCTTGCGGATGCG-3’, in the following heating cycle: 95°C for 30 sec, 54°C for 30 sec, and 72°C for 1 minute; for 35 cycles. All primers used in this study were synthesized by Integrated DNA Technology (Coralville, IA). PCR products were separated on 2% agarose gels and visualized by ethidium bromide staining. IL-2cre-PGK-neor mice were crossed with FLPeR mice on C57BL/6 background [purchased from Jackson Laboratory (Bar Harbor, ME)] to create the IL-2cre mice removed PGK-neor gene. IL-2cre mice were crossed with EGFP reporter Z/EG mice on C57BL/6 background (purchased from Jackson Laboratory to create the IL-2cre: Z/EG mice. For in vivo live imaging of T cells, IL-2cre mice were bred to luciferase reporter mice [FVB.129S6(B6)Gt(ROSA)26Sortm1(Luc)Kael/J, termed R26luc herein] from Jackson laboratory.
To fix the specificity of the TCR repertoire in CD4 T cells, IL-2cre: Z/EG mice were crossed with RAG-1 knock-out mice (Jackson Laboratory) and OT-II (Jackson Laboratory) TCR-transgenic (Tg) mice. CD4 T cells from OT-II mice express specific TCRs for OVA peptide 323-339.
All mice were maintained and bred under specific pathogen-free conditions under the approval of the Institutional Animal Care and Use Committee (IACUC) of the Medical College of Georgia and Loyola University Medical Center, Chicago.
Isolation of lymphoid and myeloid cells
To prepare single cell suspensions from thymus, spleen, mesenteric lymph nodes (mLN) and Peyer's patches (PP), the collected organs were incised and treated with 0.5% FCS-RPMI1640 medium containing 2 mg/ml collagenase D (Roche, Indianapolis, IN) and 30 g/ml DNase I (Roche) for 30 min at 37°C. The cells were suspended in 5% FCS-RPMI1640 medium containing 5 mM EDTA and were passed through 40 m cell strainer. Bone marrow cells were gently flushed into a tube using a syringe and a 25-gauge needle with 10 ml of 0.5% FCS-RPMI1640 medium. Red blood cells were removed by treatment with ACK buffer (Invitrogen, Carlsbad, CA).
For isolation of intraepithelial lymphocytes (IEL), small intestines were cut away from PP and the fat, and were opened longitudinally, washed to remove fecal content, and shaken in RPMI1640 medium containing 10% FCS and 1 mM DTT (Fluka, part of Sigma-Aldrich, St. Louis, MO) for 30 min at 37°C. IEL fractions were passed through a stainless steel tea strainer. For isolation of lamina propria lymphocytes (LPL), the remaining intestines were cut into small pieces and incubated with serum-free RPMI1640 medium containing 0.5 mg/ml Collagenase type 3 (Worthington, Lakewood, NJ) for 20 min at 37°C. The digested pieces including LPL or IEL fractions were passed through 40 m cell strainer and resuspended in 6 ml of 40% Percoll (GE Healthcare, Piscataway, NJ) and overlaid on 6 ml 70% Percoll in a 15 ml tube. Percoll gradient separation was performed by centrifugation at 800 g for 30 min at 25°C without braking. IEL or LPL were collected at the intermediate layer of the Percoll gradient, and were washed with RPMI1640 medium containing 10% FCS. For isolation of lung lymphocytes, lung lobes were dissected out, incised, and incubated in RPMI1640 medium containing 5% FCS and 5mM EDTA with gentle shake for 1 hour at room temperature, then were treated with Collagenase D (2 mg/ml, Roche) and DNaseI (30 g/ml, Roche) for 1 hour at 37°C. Digested pieces of lungs were passed through 40 m cell strainer and lymphoid cells were isolated by Percoll gradient performed as described above.
In vivo Live Imaging
Luciferase reporter mice (R26luc) with or without IL-2cre knock in allele were first shaved on the ventral side, and then cleaned with 70% ethanol. Mice were injected with 100μl of 25mg/ml of Luciferin (D-Luciferin Firefly potassium salt, Caliper Life Sciences, Hopkinton, MA USA) intraperitoneally. Mice were anesthetised with 2% isoflurane and placed in the Xenogen IVIS 100 (Caliper LifeSciences, Hopkinton, MA USA) 10 minutes after injection. Live images were taken at 5 minute intervals. To determine the location of sites emitting light, mice were euthanized and the abdominal cavity was exposed.
Cell sorting and cell culture
Naïve CD4+CD62L+ T cells were purified from spleen using a mouse CD4+CD62L+ T cell isolation kit II (Miltenyi Biotec, Auburn, CA: purity >95%). To collect EGFP+ CD4 T cells (CD3+CD4+EGFP+), memory CD4 T cells (CD3+CD4+CD62Llow EGFP-) and naïve CD4 T cells (CD3+CD4+CD62Lhigh EGFP-), each fraction was collected from spleen using FACS Aria (Becton Dickinson, San Jose, CA: purity >97%).
The sorted naïve CD4 T cells were stimulated with plate-bound anti-CD3 antibodies (5 g/ml) and soluble anti-CD28 antibodies (2 g/ml) for 2 days, and were further cultured without stimulation and with exogenous IL-2 (10 ng/ml). The induced EGFP+ CD4 T cells were analyzed by Accuri C6 flow cytometer (Accuri cytometers, Ann Arbor, MI) or FACS Canto (Becton Dickinson). The induced EGFP+ and EGFP- CD4 T cells were purified by FACS Aria (Becton Dickinson).
Cell lysis and Western blotting
The cells were lysed in SDS sample buffer (containing 2%SDS, 5% beta-mercaptoethanol, 5% glycerol, and 62 mM Tris, pH 6.5), boiled, and frozen at -80°C until analysis. Lysates from an equal number of cells were loaded in each well, followed by electrophoresis and blotting onto PVDF membrane. Membranes were probed with antibodies specific for Cre (Novagen, Madison, WI) and -actin (Sigma, St Louis, MO), followed by the appropriate HRP-conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA) and enhanced chemiluminescence (GE Healthcare).
Induction of Th1/Th2 differentiation
Isolated naïve CD4+CD62L+ T cells were activated with plate-bound anti-CD3 (5 g/ml; 2C11; BioLegend, San Diego, CA) and soluble anti-CD28 (2 g/ml; 37.51; BioLegend) in the presence of polarizing cytokines or/and blocking antibodies. For Th1 polarization, anti-IL-4 (10 g/ml; 11B11; eBioscience, San Diego, CA) and IL-12 (10 g/ml; Peprotech, Rocky Hill, NJ) were used. For Th2 polarization, anti-IFN (10 g/ml; XMG1.2; eBiosicence) and IL-4 (10 ng/ml; Peprotech) were used. After 7 days, cells were re-stimulated phorbol 12-myristate 13-acetate (PMA, Sigma) (50 ng/ml) and ionomycin (1 M, Sigma) for 4 hr and the expression of IL-4-, IFN -, and IL-10-producing cells were analyzed by intracellular cytokine staining.
Antibodies and flow cytometry
Anti-CD4-PE (GK1.5), anti-CD44-PE (IM7), anti-CD25-PE (PC61), anti-ICOS-PE (15F9), anti-CD127-PE (A7R34), anti-CD4-PE-Cy7 (GK1.5), anti-CD4-APC (GK1.5), anti-CD3-APC (145-2C11), and anti-V 2-PE (B20.1) antibodies were purchased from eBioscience. Anti-CD62L-PE (MEL-14), anti-CCR7-PE (4B12), and anti-CD4-PE-Cy5 (GK1.5) were obtained from BioLegend. Anti-CD69-PE (H1.2F3) and Streptavidin-PE, Streptavidin-APC were purchased from BD Pharmingen (San Diego, CA). Stained cells were acquired on a FACS Canto flow cytometer (Becton Dickinson) or an Accuri C6 flow cytometer (Accuri Cytometers) and analyzed on FlowJo software (Tree Star, Ashland, OR). Gates were set based on the data obtained from unstained cells or cells stained with isotype control antibodies.
Intracellular cytokine staining
Freshly isolated CD4+T cells were stimulated with PMA (50 ng/ml) and ionomycin (1 M) at 37°C for 4 h before addition of monensin (2 M, Sigma) for inhibition of protein export and then were cultured for an additional 2 h. Cells were fixed and permeabilized and then were stained with anti-CD4-PE-Cy7 (GK1.5), anti-IFN -PE (XMG1.2), anti-IL-4-Alexa Flour 647 (11B11) (all eBioscience), anti-IL-10-PE (JES5-16E3), anti-IL-10-Alexa Flour 647 (JES5-16E3), and anti-IL-2-PerCP-Cy5.5 (JES6-5H4), (all BioLegend). Stained cells were analyzed using a FACS Canto flow cytometer (BD Biosciences) and data were analyzed by Flowjo software (Tree Star).
Cytokines
CD4 T cells were purified from spleen by a cell sorter, and were stimulated with plate-bound anti-CD3 antibodies (at the indicated concentration) and soluble anti-CD28 antibodies (2 g/ml) for 2 days. Cytokines (IL-2, IL-4, IFN and IL-10) were measured using the Bio-Plex Pro Mouse Cytokine Th1/Th2 Assay from Bio-Rad Laboratories (Hercules, CA).
Antibiotics treatment of mice
To determine the effect of microbial flora, IL-2Cre/GFP mice were treated with antibiotics in the drinking water according to the procedure reported previously [12]. 8 weeks old mice were fed 4 weeks with water containing the cocktail of antibiotics [ampicillin 1g/L, (Sigma), vancomycin 500mg/L, (Abbott Labs), neomycin sulfate 1g/L (Pharmacia/Upjohn), and metronidazole 1g/L (Sidmack Labs)]. After the treatment, mice were sacrificed and Peyer's patches form each animal was harvested and analyzed for the presence of GFP+ cells using flow cytometry.
Results
Generation of IL-2cre: Z/EG mice
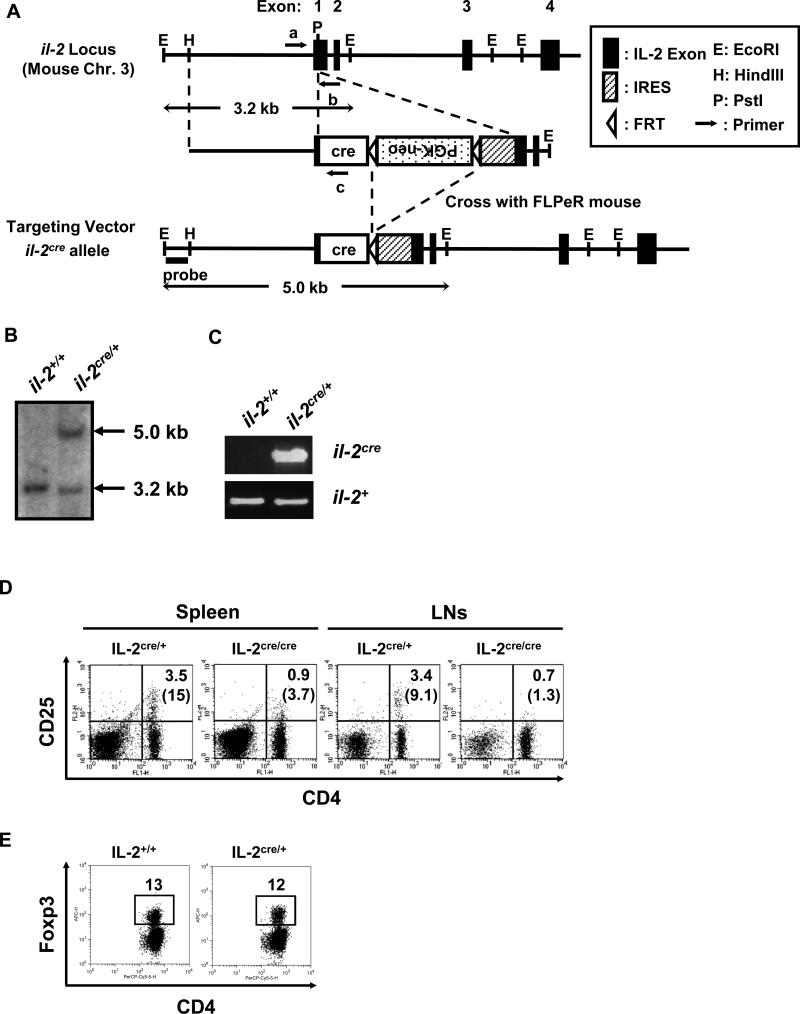
To determine the cells that underwent IL-2 expression in vivo under unimmunized physiological conditions, we established IL-2cre mice that carry an il-2 allele with the cre gene knocked in (Fig. 1). The cre gene was inserted 5’-upstream of the first codon of il-2. There is an internal ribosome entry sequence (IRES) at the 3’ of the cre gene that allows co-translation of Cre with IL-2. This insertion was intended to enable il-2 to be expressed even when mice are homozygous for the cre knock-in. Contrary to our expectations, il-2cre/cre homozygous mice develop a lymphoproliferative disorder (over four times higher cell numbers in spleen than the control in average) similar to that observed in il-2 knock out (KO) mice, accompanied by a loss of CD4+CD25+ cells (Fig. 1D) [7]. The data indicated that insertion of cre gene reduced expression of IL-2 and/or disturbed the function of cells that play roles in homeostasis of nTregs in vivo. Therefore, we used heterozygous (il-2cre/+) mice harboring a single-copy of cre in this study to determine the cells that express IL-2 without complications arise from reduction of nTregs. The level of Foxp3+ cells in il-2cre/+ mice was comparable to that observed in wild-type mice, indicating that the cre knock-in does not adversely affect or attenuate the development and/or maintenance of nTregs (Fig. 1E).
Figure 1. Generation of IL-2cre knock-in mice.
(A) Design of cre knock-in at il-2 locus. Mouse il-2 locus is schematically presented (closed boxes show exons from the wildtype gene) with relevant restriction enzyme sites; the targeting vector and the il-2cre allele (after removal of the FRT- flanked neo gene) are also shown. Arrows represent primers used for genotyping in (C). (B) Southern blotting analysis of tail genomic DNA for il-2cre knock-in. DNA hybridization was performed with the outside probe indicated in (A). The 3.2 kb band corresponds to the DNA fragment derived from wildtype il-2 allele, and the 5.0 kb band corresponds to the il-2cre allele after homologous recombination with the targeting vector. (C) PCR genotyping of tail genomic DNA for IL-2cre mice. Exon 1 with inserted cre (il-2cre) was detected by the primers a and c (shown in Fig. S1A). Exon 1 from wildtype allele (il-2+) was detected by the primers a and b. (D) Spleen and lymph nodes from IL-2cre/+ and litter mate IL-2cre/cre mice were analyzed for the expression of CD4 and CD25. Numbers indicate percentages of CD4+CD25+ cells among total cells. Parenthesized numbers indicate the percentages of CD25+ cells among CD4+ T cells. (E) Expression of Foxp3 by splenic CD4 T cells from wildtype and IL-2cre/+ mice.
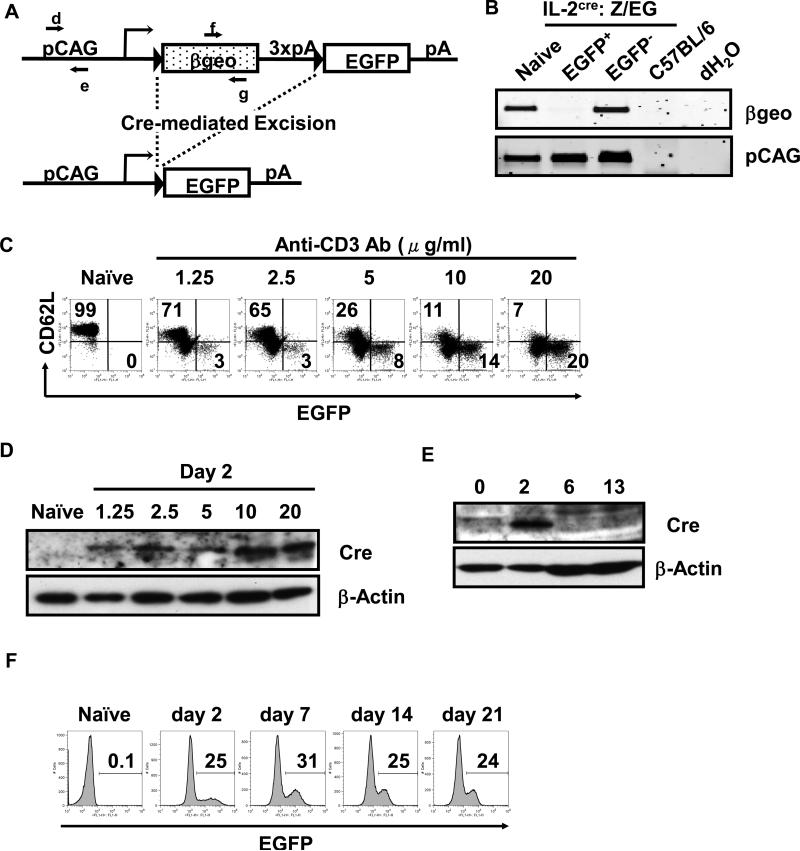
To determine if the IL-2cre mouse is an effective system to monitor the fate of cells that express IL-2, we bred IL-2cre mice with mice that carry an egfp reporter gene with a floxed transcriptional stop cassette (Z/EG mice, purchased from Jackson Laboratories). CMV-β-actin fusion promoter drives expression of egfp in Z/EG mice in a tissue non-specific manner. There is a transcriptional stop cassette (3xpA) located at the 5’-side of the egfp gene (Fig. 2A). When Cre is absent, this reporter gene expresses the β-geo gene (-galactosidase and neomycin resistance fusion gene) but the transcript is terminated at the site of the stop cassette and as such, no EGFP will be expressed. However, when Cre is present, the region that is flanked by the loxP sequence, including the transcription stop cassette, will be removed and induce the expression of EGFP. Since the promoter is constitutively active in a tissue non-specific manner, once cells express EGFP, they will remain EGFP+ and all cells that were derived from the cells that initially produced Cre (and thus IL-2) will remain EGFP+. To determine if EGFP expression from IL-2cre: Z/EG mice require deletion of the stop cassette, naïve CD62Lhigh CD4 T cells were stimulated ex vivo with anti-CD3 and anti-CD28 antibodies to induce IL-2 expression and were separated into EGFP+ and EGFP- fractions by a cell sorter (Fig.2B). PCR analysis showed removal of β-geo gene in EGFP+ T cells but not in induced EGFP+ cells, showing that the removal of the stop cassette correlates with EGFP expression. Expression of Cre and GFP expression by T cells correlated with the strength of stimulation. When naïve CD62Lhigh CD4 T cells from IL-2cre: Z/EG mice were stimulated with a graded dose of plate-bound anti-CD3 antibodies and soluble anti-CD28 antibodies, the frequency of EGFP+ cells as well as the level of Cre protein increased in an anti-CD3 dose-dependent manner (Fig. 2C and D). Cre protein became clearly detectable after 2 days of stimulation but was undetectable at day 6 and 13 after stimulation (Fig. 2E). Unlike Cre protein, EGFP expression was maintained among cells cultured with exogenous IL-2 for 21 days as expected (Fig. 2F). The data indicate that EGFP expression is maintained among the cells that once expressed IL-2 and suggest that the lineage of cells derived from the initial IL-2+ cells can be tracked by the system (Fig. 2F).
Figure 2. Cre-mediated homologous recombination in CD4 T cells from IL-2cre: Z/EG mice and expression of EGFP by activated T cells.
(A) A schematic representation of Z/EG (lacZ/EGFP) transgene. The Z/EG transgene consists of pCAG promoter (a hybrid promoter between cytomegalo virus (CMV) enhancer and chicken -actin promoter), a loxP-flanked geo (lacZ/neomycin-resistance) fusion gene, three SV40 polyadenylation sequences (3 x pA), and egfp gene. In the Z/EG mouse because the egfp gene is inserted downstream of the 3’ of the geo and 3x pA, EGFP will not be expressed until Cre excises the geo gene. When the il-2 promoter is activated, the Cre protein is produced and causes the deletion of the DNA fragment flanked by the loxP sequence via homologous recombination between two loxP sites, leading to expression of EGFP. The primers d and e are designed to detect pCAG promoter. The primers f and g are designed to detect geo. (B) PCR analysis of genomic DNA from EGFP+ T cells and control samples. Naïve CD62Lhigh CD4 T cells from IL-2-Cre: Z/EG mice were purified by sorting on a flow cytometer, stimulated with plate-bound anti-CD3 antibodies and soluble anti-CD28 antibodies for 2 days, and were cultured without further stimulation with exogenous IL-2 (10 ng/ml) for 11 days. These cells were then sorted into EGFP+ and EGFP- populations by flow cytometry, and were subjected to isolation of genomic DNA. The tail DNA of wild type C57BL/6 mice and dH2O only samples were used as negative control. Genomic DNA templates were subjected to PCR with primers f and g (shown in A) for detection of geo. The pCAG promoter fragment was amplified with primers d and e (shown in Fig. 2A) to show the amount of DNA used for the analysis. (C) Expression of EGFP by naïve or activated CD4 T cells. Naïve CD62Lhigh CD4 T cells were purified from IL-2cre: Z/EG mice and stimulated with graded doses of plate-bound anti-CD3 antibodies (at the concentration indicated above each panel) and soluble anti-CD28 antibodies (2 g/ml) for 2 days. The numbers indicate the percentage of cells within upper left (CD62L+ EGFP-) and lower right quadrant (CD62L- EGFP+) of the panel. (D) Western Blotting analysis of Cre expression by naïve or activated CD4 T cells. Naïve CD62Lhigh CD4 T cells from IL-2cre mice were stimulated with plate-bound anti-CD3 antibodies (concentrations indicated above each lane) and soluble anti-CD28 antibodies (2 g/ml) for 2 days. -actin blot was used as a loading control. (E) Time course analysis of Cre expression by activated CD4 T cells. Naïve CD62Lhigh CD4 T cells from IL-2cre: Z/EG mice were stimulated by plate-coated anti-CD3 antibodies (20 g/ml) and soluble anti-CD28 antibodies (2 g/ml) for 2 days and then cultured without stimulation in the presence of exogenous IL-2 for days shown above each lane (2~13 days). Total cell lysates were analyzed for the expression of the Cre protein. -actin blot was used to determine the level of protein loaded in each lane. (F) Time course analysis of EGFP expression by activated CD4 T cells. Naïve CD62Lhigh CD4 T cells isolated from IL-2cre: Z/EG mouse spleens were stimulated as in (C) for 2 days (day 2 sample). After this stimulation period, cells were removed from stimulation and maintained in the presence of exogenous IL-2. Cells were harvested 7, 14, or 21 days (as indicated above each panel) after start of initial stimulation and analyzed for EGFP expression by flow cytometry. The numbers indicate the percentage of EGFP+ among CD4+ cells.
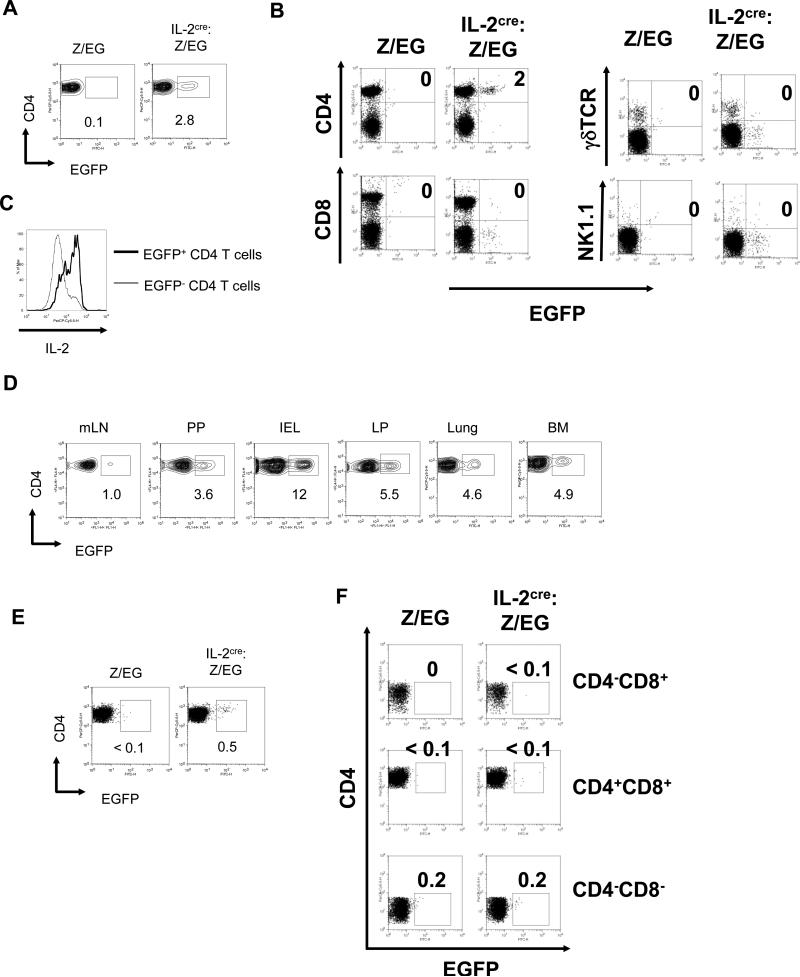
Decrease of Foxp3+ Tregs in IL-2cre homozygous mice (Fig.1D) indicated that cells that express IL-2 in healthy mice play a critical role in homeostasis of nTregs. To determine the presence of naturally existing IL-2 producers, we next characterized EGFP expression in secondary lymphoid organs from healthy mice maintained under specific pathogen free (SPF) conditions (Fig. 3). Mice that carry the Z/EG transgene alone showed no EGFP+ cells in the spleen, suggesting that there is no leaky expression from this reporter. On the other hand, mice that carry the IL-2cre knock-in and the Z/EG reporter demonstrated the presence of EGFP+ cells in the splenic CD3+CD4+ population. Approximately 3% of CD3+CD4+ cells in the spleen from IL-2cre: Z/EG mice were EGFP+ (Fig. 3A). We only observed EGFP+ populations among conventional CD3+CD4+ populations, but not among CD8+, γδ+ or NK1.1+ CD3+ cells (Fig.3B). To determine if EGFP+ CD4 T cells still produce IL-2, we tested their ability to produce IL-2 after a brief stimulation (4 hours) with PMA plus ionomycin. Intracellular staining with anti-IL-2 antibody showed a higher level of IL-2 expression by EGFP+ cells compared to EGFP- CD4 T cells (Fig.3C), suggesting that a majority of EGFP+ cells maintain the ability to produce IL-2 upon re-stimulation.
Figure 3. Presence of EGFP+ CD4 T cells in IL-2cre: Z/EG mice.
(A) Expression of EGFP by CD3+CD4+ cells in spleens from Z/EG control mice and IL-2cre: Z/EG mice. The numbers indicate the percent of cells within each gated region. (B) Expression of EGFP by different subset of lymphocytes from spleen of IL-2cre: Z/EG were analyzed by surface antigen staining for CD4, CD8, TCR, and NK1.1 as indicated. (C) Expression on IL-2 by EGFP- and EGFP+ CD4 T cells. Splenic CD4 T cells from IL-2cre: Z/EG were stimulated ex vivo by PMA and ionomycin for 4 hours. Expression of IL-2 was determined by intracellular staining and flow cytometry. (D) Expression of EGFP by CD3+CD4+ cells isolated from mesenteric lymph nodes (mLN), Peyer's patches (PP), intraepithelial lymphocyte of small intestine (IEL), and lamina propria of small intestine (LP), lung and bone marrow (BM) of IL-2cre: Z/EG mice. The numbers indicate the percent of cells within each gated region. (E) Expression of EGFP by CD4+ single positive thymocytes isolated from Z/EG control mice and IL-2cre: Z/EG mice. CD4+CD8- cells were gated by analysis software from total thymocytes and analyzed for EGFP expression. (F) Expression of EGFP by non-CD4+ CD8- thymocytes as analyzed in (E).
In addition to spleen, we found EGFP+ CD4 T cells in many other tissues including bone marrow (BM), mesenteric lymph nodes (mLN), and Peyer's patches (PP) (Fig. 3D). Most notably, we found a high frequency of EGFP+ cells in non-lymphoid mucosal tissues such as lung and lamina propria (LP). All EGFP+ cells were found in CD4+ T cell fractions. The intraepithelial lymphocyte (IEL) population also contained CD4+EGFP+ T cells at a higher frequency than secondary lymphoid organs. A small fraction of thymocytes was also EGFP+ though it is not clear if they are newly generated T cells or circulating mature T cells (Fig. 3E and F). These cells were present only in the CD4 single positive population and present as a minor fraction (0.5% of CD4+CD8- cells) (Fig. 3E) but not in the double-negative (CD4-CD8-), the double-positive (CD4+CD8+), or the CD8 single positive population (Fig. 3F).
In vivo detection of IL-2Cre+ cells by live animal imaging
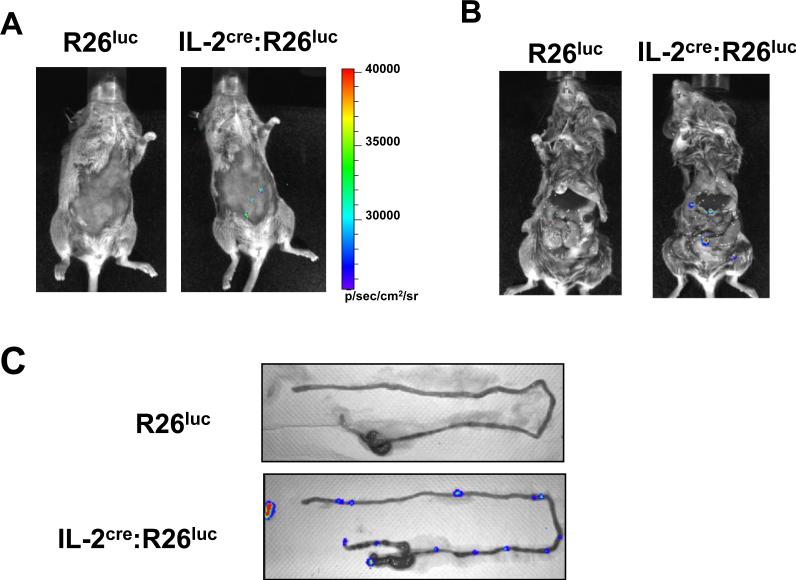
The presence of T cells that produce IL-2 under healthy conditions was also confirmed by another reporter mouse system that produces luciferase in response to Cre expression (R26luc mouse). The basic design for these reporter animals is the same as that described for the EGFP reporter mice. In brief, a transcription stop cassette surrounded by the loxP cassette sequences represses expression of luciferase gene in these animals. Cells that express Cre lose the stop cassette and start expressing luciferase continuously from the ROSA26 gene promoter. Luciferase produces bioluminescence that can be detected in live animals when the substrate (luciferin) is present [13]. We used this system to detect the presence of cells producing/produced IL-2 in healthy animals that did not undergo immunization or pathological conditions (Fig. 4A). When R26luc mice were injected with luciferin, luciferase-producing cells were undetectable. In contrast, IL-2cre:R26luc mice showed the presence of several light-emitting regions in the abdominal area detectable from the outside of mice. To determine the exact locations of luciferase producing cells, the abdominal cavity was exposed (Fig. 4B). The bright spots were observed along the small intestine. Close examination of the small intestine clearly showed these cells are present in PPs from IL-2cre:R26luc mice, but not from R26luc mice (Fig. 4C). The data from EGFP reporter mice showed that the IEL or LP contains a higher frequency of Cre+ cells than PPs. The difference is likely due to the fact that luciferase detection reflects the density of cells expressing luciferase in situ while EGFP+ cells are analyzed after the ex vivo purification process using a Percoll gradient and reflects the frequency among purified cells. Alternatively, luciferin may not be distributed effectively into the LP or the mucosal surface of intestine. Hence, the luciferase system might underestimate the level of Cre+ cells in certain tissues.
Figure 4. Detection of IL-2cre expressing cells by whole animal imaging analysis.
(A,B) Bioluminescence imaging of R26luc (left) and IL-2cre: R26luc mice (right) was performed at 10 minutes after intraperitoneal injection (IP) of D-luciferin. Mice were first observed alive after shaving of the fur (A), then euthanized and the abdominal cavity was exposed (B). Detected luminescence is shown as pseudo color with the color scale shown on the side. (C) Ex vivo bioluminescent images of mLN and intestine from R26luc (upper panel) or IL-2cre: R26luc mice (lower panel) 15~20 minutes after the injection of D-luciferin. Detected luminescent is shown as in A.
Ontogeny of EGFP+ CD4 T cells
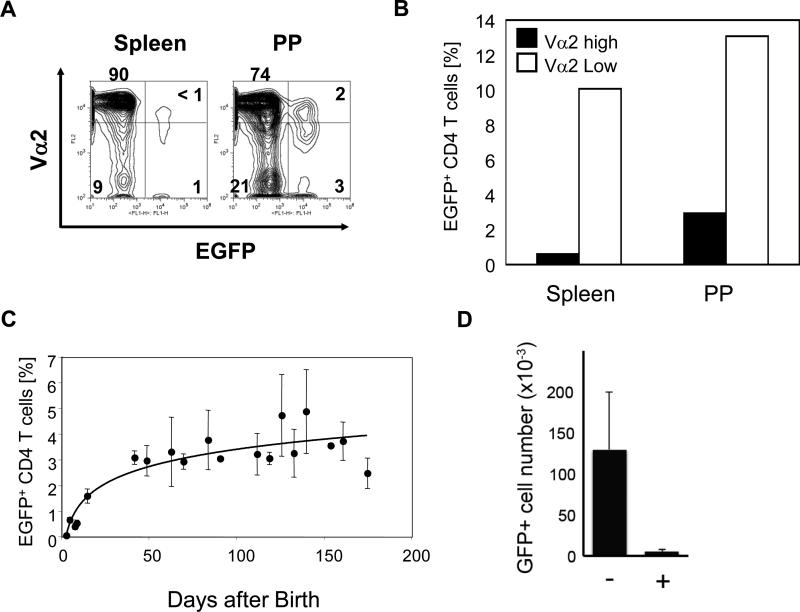
The data presented above show that a major source of IL-2 in vivo in healthy animals is CD4+ T cells. Production of IL-2 may be independent from antigen recognition. Alternatively, IL-2 expression may be induced by antigen-recognition by T cells. If generation of EGFP+ CD4 T cells requires recognition of antigens, then we expect a reduced frequency of EGFP+ CD4 T cells in mice with a restricted TCR repertoire due to decrease of antigen-reactive T cells. On the other hand, if IL-2 production is antigen-independent, limitations in TCR repertoire would not affect the expression of IL-2 and EGFP. Expression of transgenic TCRs limits the diversity of TCRs expressed by mature T cells due to allelic exclusion of tcr gene rearrangement [14]. Thus, IL-2Cre: Z/EG mice were crossed with TCR transgenic mice (OT-II, specific to chicken ovalbumin (OVA), expressing Vα2/Vβ5 [15]) with Rag1-deficiency. Due to unidentified causes we were not able to obtain RAG1 homozygous OT-II:IL-2cre:Z/EG mice. However, in Rag1 heterozygous OT-II: IL-2cre: Z/EG mice, CD4+ T cells that dominantly express OT-II-derived TCR (Vα2high) had a significantly lower percentage of EGFP+ CD4 T cells compared to T cells expressing non-transgenic TCR (Vα2low/negative) (Fig. 5A and B). Moreover, the peak expression level of V 2 cells among EGFP+ cells was lower than that of EGFP- cells both in spleen and PPs, suggesting they express endogenous TCR chain. Together, these data suggest that antigen recognition is required for expression of IL-2 by CD4 T cells.
Figure 5. Effect of TCR repertoire and age on the presence of EGFP+ CD4 T cells.
(A) Kinetics of the appearance of EGFP+ CD4 T cells during mouse development. Percentage of EGFP+CD4+ T cells among splenic CD4+ T cells from individual IL-2cre: Z/EG mice were analyzed at the indicated age and shown by closed circles (at least three mice were analyzed for each age group). Trend curves are fitted with using curve fit analysis from Excel (Microsoft). (B) CD4 T cells from spleen and Peyer's patches of Rag1+/-:OT-II:IL-2cre:Z/EG mice were analyzed for expression of V 2 and EGFP. The gate was set to divide cells into V 2high /V 2low/- groups and EGFP+/EGFP- groups. Percentage of EGFP+ cells were determined for V 2high and V 2low/- groups. (C) Percentage of EGFP+ cells among V 2high or V 2low CD4+ T cells from OT-II: Rag1-/+: IL-2cre: Z/EG mice. Data are representative of three independent experiments. (D) Number of EGFP+ cells from PPs of mice treated with water containing antibiotics (+) or water alone (-). Mice were treated for 4 weeks with antibiotics and analyzed (n=3).
Antigens recognized by these EGFP+ CD4 T cells may be derived from the environment such as commensal bacterium or food or from self. Alternatively, antigen could be derived from self-tissues. If they recognize self-antigens, EGFP+ CD4 T cells may be generated in utero. On the other hand, if they are derived from the environment, we will observe EGFP+ cells only after birth of animals. To discriminate these two possibilities, we examined when during mouse development EGFP+ CD4 T cells are generated (Fig. 5C). EGFP+ CD4 T cells were not detectable in the spleen of newborn IL-2cre: Z/EG mice (1-3 days after birth). The earliest time when EGFP+ CD4 cells became detectable was 5 days after birth. The frequency of EGFP+ CD4 cells increased with age during the lactation period and remained constant at about 3% up to 170 days after birth. Moreover, treatment of mice with antibiotics to reduce commensal bacterium resulted in a significant reduction in EGFP+ cells from PPs (Fig. 5D). Together, these data show that generation of EGFP+ CD4 cells require exposure of T cells to environmental factors after birth and suggest that IL-2 is produced in response to environmental antigens.
Function and phenotype characterization of EGFP+ CD4 cells in IL-2cre: Z/EG mice
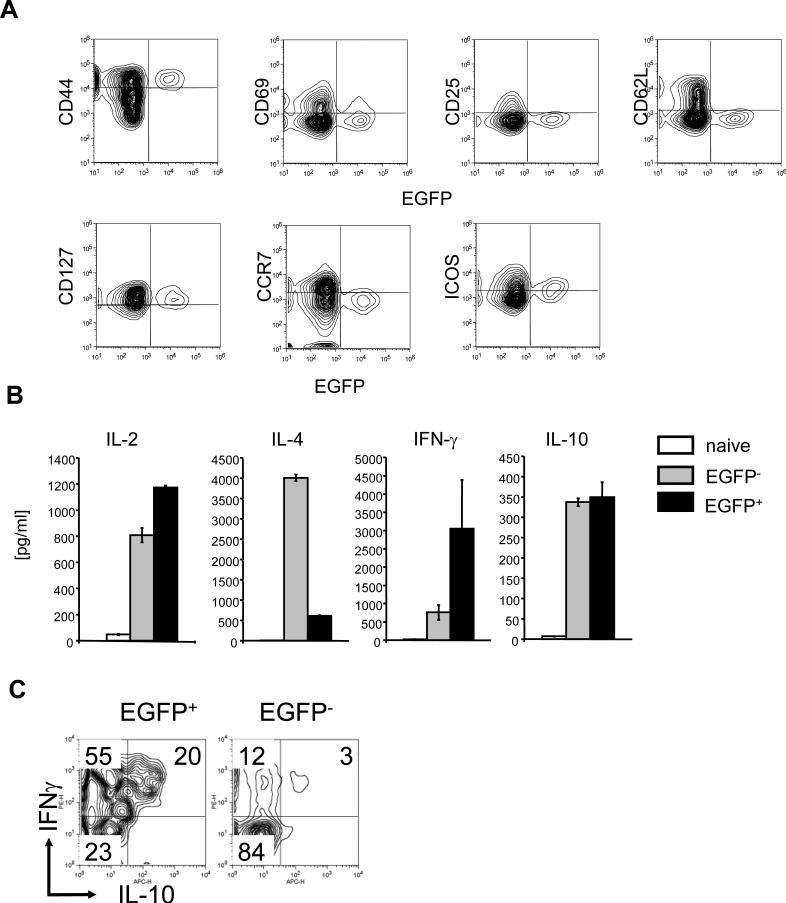
Antigen-stimulation of T cells results in surface phenotype changes. Expression of CD44/CD127 expression and loss of CD62L is a hallmark for T cell that underwent recent antigen-stimulation and cells with this phenotype are considered as “effector memory” T cells. As expected from the antigen- requirement for EGFP expression shown above, EGFP+ CD4 T cells are CD44+CD62L- and CD127+. EGFP+ CD4 T cells also expressed inducible T cell co-stimulator (ICOS), an activation-induced co-stimulatory molecule. However, they did not express early activation markers such as CD69 and CD25 or CCR7, a marker for central memory T cells. These surface antigen expression by EGFP+ CD4 T cells is similar to that of so-called “effector-memory” CD4 T cells. It has been shown previously that mice maintained under SPF conditions have the CD4 T cell population with memory-phenotype cells[16]. Thus, a fraction of these residential effector-memory T cells is a source of IL-2 under healthy conditions.
Effector-memory T cells consisted from different type of effector T cells that are characterized by the cytokine production profile (e.g. Th1, Th2). Thus, we examined if these EGFP+ CD4 T cells maintain the ability to produce IL-2 and if they make other cytokines than IL-2. When stimulated ex vivo with anti-CD3 and anti-CD28 antibodies, EGFP+ CD4 T cells produced a higher level of IL-2 than naïve CD4 T cells or EGFP- memory (CD44+CD62L-) CD4 T cells (Fig. 5B). EGFP+ cells also produced a significantly higher level of IFN- than EGFP- memory T cells. Conversely, IL-4 production by EGFP+ CD4 T cells was substantially lower than that of EGFP- memory CD4 T cells. The data suggested that EGFP+ cells are Th1-like memory T cells.
A subset of memory T cells express high levels of IFN-γ along with immunoregulatory cytokine IL-10 and play a critical role in modulation of immune responses[17-19]. They were termed Tr1 cells[20]. Since EGFP+ cells may be an important subset of cells for immune regulation, we determined whether or not EGFP+ CD4 T cells are classical Th1 type effector cells or Tr1 cells by testing their IL-10 expression. Activated EGFP+ CD4+ cells produced a higher level of IL-10 compared to naïve CD4 T cells but the level was comparable to that of EGFP- cells (Fig. 6B). IL-10 may be produced by a subset of EGFP+ CD4 T cells distinctive from IFN- producing T cells. Alternatively, IL-10 may be produced by T cells that also express IFN- . To discriminate between these possibilities, we performed single cell analysis (Fig. 6C). About 20% of EGFP+ CD4 cells produced IL-10 and almost all EGFP+ IL-10+ cells also were IFN + (Fig. 6C). About 50% of EGFP+ cells produced IFN- but not IL-10. In contrast, only 3% of EGFP- memory cells produced IL-10 and IFN- . These data show that EGFP+ CD4 T cells are heterogeneous in cytokine production and a substantial portion of these cells co-produce IFN , IL-10, and IL-2 in healthy mice under SPF conditions.
Figure 6. Surface antigen phenotypes and cytokine expression by EGFP+ CD4 T cells from IL-2cre: Z/EG mice.
(A) Expression of EGFP and cell surface antigens (CD44, CD69, CD25, CD62L, CD127, CCR7, and ICOS) by CD4+ T cells from spleen from IL-2cre: Z/EG mice were analyzed by flow cytometry. Data are representative of three independent experiments. (B) Cytokine production by naive, EGFP- memory, or EGFP+ CD4 T cells. CD62Lhigh EGFP- CD4 T cells (Naïve), CD62Llow EGFP- CD4 T cells (EGFP-) and EGFP+ CD4 T cells (EGFP+) were purified from spleen of IL-2cre: Z/EG mice by a flow cytometric cell sorter (FACS Aria) and were stimulated with plate-bound anti-CD3 antibodies soluble anti-CD28 antibodies for 2 days. The culture supernatants were harvested and were analyzed for IL-2, IL-4, IFN and IL-10 by Th1/Th2 Bio-Plex cytokine assay system (Bio-Rad laboratories). Data are representative of six independent experiments. (C) Single cell analysis of IFN , IL-4 and IL-10 expression by EGFP+ CD4 T cells. EGFP+CD4+ cells from IL-2cre: Z/EG mouse spleens were stimulated in vitro with PMA and ionomycin. Cells were stained to determine intracellular cytokine levels and analyzed by flow cytometry. Data are representative of four independent experiments.
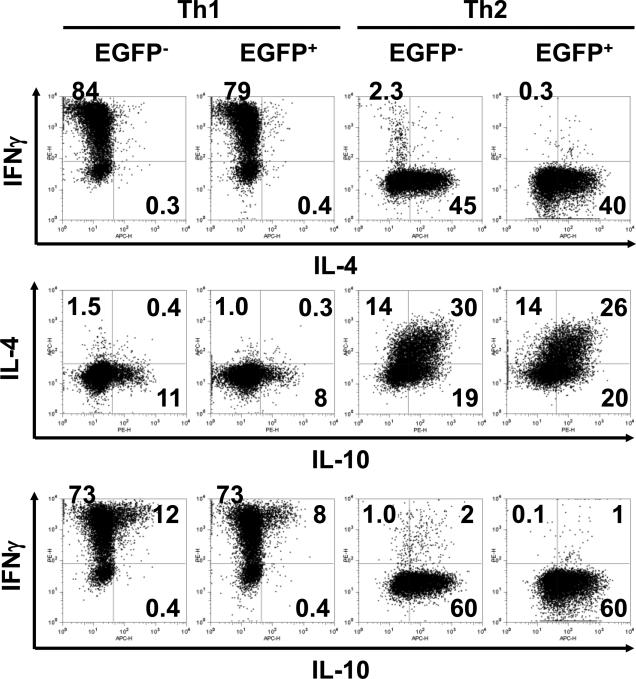
One caveat of these data is that our knock-in system caused T cells to differentiate preferentially into IFN -dominated Th0/Th1 cells. To test this possibility, we examined expression of EGFP by naïve CD4 T cells that were induced ex vivo to be Th1/Th2. After polarizing culture, EGFP+ CD4 T cells were found in both IFN + IL-4- cells (in Th1-polarizing culture) as well as in IFN - IL-4+ cells (in Th2-polarizing culture) (Fig. 7). The pattern of differentiation of EGFP+ CD4 T cells was comparable to that of EGFP- CD4 T cells. These data show that cre knock-in did not cause biased differentiation of naïve CD4 T cells and that EGFP+ CD4 T cells produce cytokine profile reflects how these cells differentiated and/or were maintained in vivo.
Figure 7. In vitro Th1/Th2 differentiation of CD4 T cells from IL-2cre: Z/EG mouse splenocytes.
Purified naïve CD62Lhigh CD4 T cells were stimulated with plate-bound anti-CD3 plus soluble anti-CD28 antibody under Th1 or Th2-polarizing condition (see materials and methods). After 7 days, cells were re-stimulated PMA and ionomycin for 4 hours and expression of IL-4, IFN- and IL-10 by EGFP+ and EGFP- T cells was analyzed by intracellular cytokine staining. Data are representative of three independent experiments. The numbers indicate the percent of cells within quadrant panels.
Discussion
Our newly developed IL-2cre knock-in mouse enabled us to detect that a subset of CD4 T cells produce IL-2 or had produced IL-2 previously in healthy animals. CD4 T cells that produced IL-2 are not detectable at birth but appear within 5 days after birth, suggesting that environmental factors play a critical role in their generation. The number of these cells continued to increase during the mouse development without pathological outcomes and stabilized after 50 days. These cells have effector-memory T cell-like surface antigen phenotype of CD44+CD62L-. These cells predominantly co-produced IFN- and a subsequent fraction of them also produced IL-10, suggesting that some of them are Tr1-like T cells.
Based on our data showing that IEL contains the highest frequency of EGFP+ cells, antigens present in the intestinal tract, such as food components and/or commensal bacteria, are likely candidates that induce CD4 T cells to produce IL-2. A number of studies have demonstrated that commensal bacterium play pivotal roles in maintenance of immunological homeostasis and development of gut associated lymphoid tissues (reviewed in [21, 22]). Loss or changes of gut microbiota is often associated with reduced resistance to infections and altered immune responses against foreign and/or self-antigens[23, 24]. The effect from mucosal environment was mainly studied on Th17 and Tregs, but the influence on T cells that produce IL-2 has not been well characterized. Changes in the bacterial flora could affect the maintenance of these IL-2 producing CD4 T cells and cause changes in the homeostasis of the mucosal immune system either by directly altering their functions and/or by indirectly affecting maintenance of other cell types such as Tregs.
A significant portion of these IL-2 producers can express IFN and importantly, about 20% of the IFN producing cells also express IL-10. These IL-10/IFN co-producers have been previously found in a subset of Tr1 cells that are induced by IL-27 [19] and their immunoregulatory role has been described [17, 18, 25]. Our data show that a subset of these IL-10 and IFN- co-producers also produces IL-2. The data indicate the possibility that upon antigen stimulation, these cells could produce IL-10 for direct suppression of other lymphocytes and myeloid cells while producing IL-2 to support growth of Foxp3+ Tregs. This hypothesis explains the phenomenon reported previously where IL-10 producing effector cells were required for maintenance of Tregs to protect mice from colitis[26]. Though IL-10 can directly work on Tregs to enhance their functions, IL-2 may also help Treg expansion and maintenance.
It should be noted that unlike Tregs, these IL-2 producing cells are not CD25+, suggesting that they do not depend on IL-2 for their own maintenance. Instead, they are highly positive for CD127 (IL-7R ). Thus, the maintenance of the cells would require IL-7. Recently it has been shown that loss of IL-7 causes reduction in Foxp3+ Tregs in periphery[27]. Since Tregs are negative/low for expression of IL-7 receptor [28], reduction of Tregs in IL-7 knock out mice may be due to the loss of other cells that are required for maintenance of Tregs. One possible candidate is the IL-2 producing CD4 T cells identified herein by EGFP expression. Loss of IL-7 could reduce the number of IL-2 producing T cells and therefore reduce the level of IL-2 in vivo, resulting in loss of Tregs.
How IL-2+ T cells are initially generated is currently unknown. These cells may be generated in the thymus as we identified a small fraction of CD4+ single positive thymocytes that are EGFP+. It has been shown that orally introduced antigens can be presented to thymocytes (reviewed by [29]). Alternatively, thymic EGFP+ cells may be circulating T cells generated in the mucosal environment. Since EGFP+ cells become detectable well after birth (~5 days) and they all express CD44 but not CD62L, a majority of them appear to have undergone stimulation by the antigens originated from the exogenous environment. CD103+ DCs present in intestinal mucosa induced Foxp3+ Tregs from naïve T cells in a vitamin A dependent manner [30]. A subset of antigen presenting cells may be specialized in generating CD4 T cells that produce IL-2 from naïve cells. Presence of other cytokines such as IL-27, which induces Tr1 cells, may also contribute to generation of IL-2 producers [31]. Understanding how IL-2-producing cells are generated and maintained under healthy conditions could provide critical information to support homeostasis of the regulatory wing of the immune system.
Highlights.
IL-2Cre/EGFP reporter mice demonstrate the presence of naturally existing CD4 T cells that produce IL-2 in variety of tissues of healthy mice.
EGFP+ (a surrogate marker for IL-2) cells are CD4+,CD44+, CD62L-, and ICOS+.
The frequency of cells produce IL-2 detected in this system is highest in lamina propria.
EGFP+ cells are not detectable in newborn mice and become detectable 5 days after birth.
A significant fraction of EGFP+ cells also produce IL-10 and IFN-γ and resembles Tr1 cells.
Acknowledgments
The authors thank Drs. Chris Wiethoff and Britte Beaudette-Zlatanova for critical reading of the manuscript, Dr. Arata Takeuchi, Akiko Takumi and Dr. Takashi Saito for the isolation of lamina propria lymphocytes, Mariko Takami and Nada Daher for the mice genotyping, and Dr. Patricia Simms (Loyola FACS core facility) for cell sorting. This work was supported by NIH RO1 AI055022(M.I), PRESTO grant (Japan Science and Technology Agency) (M.I.), The Uehara memorial foundation research fellowship (M.Y.), and Japan society for the promotion of science (JSPS) postdoctoral fellowships for research abroad (M.Y.).
Abbreviations used in this paper
- EGFP
enhanced green fluorescent protein
- nTregs
naturally arising regulatory T cells
- SPF
specific pathogen free
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Belz GT, Masson F. Interleukin-2 tickles T cell memory. Immunity. 32:7–9. doi: 10.1016/j.immuni.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nature reviews Immunology. 2009;9:480–90. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwashima M. Kinetic perspectives of T cell antigen receptor siganling. Immunol Review. 2003:192. doi: 10.1034/j.1600-065x.2003.00024.x. [DOI] [PubMed] [Google Scholar]
- 4.Dooms H, Abbas AK. Revisiting the role of IL-2 in autoimmunity. European journal of immunology. 40:1538–40. doi: 10.1002/eji.201040617. [DOI] [PubMed] [Google Scholar]
- 5.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. The Journal of experimental medicine. 2005;201:723–35. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 33:153–65. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Naramura M, Hu RJ, Gu H. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 1998;9:209–16. doi: 10.1016/s1074-7613(00)80603-2. [DOI] [PubMed] [Google Scholar]
- 9.Saparov A, Wagner FH, Zheng R, Oliver JR, Maeda H, Hockett RD, et al. Interleukin-2 expression by a subpopulation of primary T cells is linked to enhanced memory/effector function. Immunity. 1999;11:271–80. doi: 10.1016/s1074-7613(00)80102-8. [DOI] [PubMed] [Google Scholar]
- 10.Yui MA, Hernandez-Hoyos G, Rothenberg EV. A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J Immunol. 2001;166:1730–9. doi: 10.4049/jimmunol.166.3.1730. [DOI] [PubMed] [Google Scholar]
- 11.Koresawa Y, Miyagawa S, Ikawa M, Matsunami K, Yamada M, Shirakura R, et al. Synthesis of a new Cre recombinase gene based on optimal codon usage for mammalian systems. J Biochem. 2000;127:367–72. doi: 10.1093/oxfordjournals.jbchem.a022617. [DOI] [PubMed] [Google Scholar]
- 12.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG., Jr Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Molecular imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 14.Spicuglia S, Pekowska A, Zacarias-Cabeza J, Ferrier P. Epigenetic control of Tcrb gene rearrangement. Seminars in immunology. 2010;22:330–6. doi: 10.1016/j.smim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 16.Lefrancois L, Marzo AL. The descent of memory T-cell subsets. Nature reviews Immunology. 2006;6:618–23. doi: 10.1038/nri1866. [DOI] [PubMed] [Google Scholar]
- 17.Haringer B, Lozza L, Steckel B, Geginat J. Identification and characterization of IL-10/IFN-gamma-producing effector-like T cells with regulatory function in human blood. J Exp Med. 2009;206:1009–17. doi: 10.1084/jem.20082238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jankovic D, Kugler DG, Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. 3:239–46. doi: 10.1038/mi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, et al. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nature immunology. 2007;8:1380–9. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 20.Gregori S, Goudy KS, Roncarolo MG. The cellular and molecular mechanisms of immuno-suppression by human type 1 regulatory T cells. Frontiers in immunology. 2012;3:30. doi: 10.3389/fimmu.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishio J, Honda K. Immunoregulation by the gut microbiota. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–74. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–85. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 24.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabrysova L, Nicolson KS, Streeter HB, Verhagen J, Sabatos-Peyton CA, Morgan DJ, et al. Negative feedback control of the autoimmune response through antigen-induced differentiation of IL-10-secreting Th1 cells. The Journal of experimental medicine. 2009;206:1755–67. doi: 10.1084/jem.20082118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–84. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim GY, Ligons DL, Hong C, Luckey MA, Keller HR, Tai X, et al. An In Vivo IL-7 Requirement for Peripheral Foxp3+ Regulatory T Cell Homeostasis. J Immunol. 2012;188:5859–66. doi: 10.4049/jimmunol.1102328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203:1701–11. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitacre CC, Song F, Wardrop RM, 3rd, Campbell K, McClain M, Benson J, et al. Regulation of autoreactive T cell function by oral tolerance to self-antigens. Annals of the New York Academy of Sciences. 2004;1029:172–9. doi: 10.1196/annals.1309.033. [DOI] [PubMed] [Google Scholar]
- 30.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. The Journal of experimental medicine. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stumhofer JS, Hunter CA. Advances in understanding the anti-inflammatory properties of IL-27. Immunology letters. 2008;117:123–30. doi: 10.1016/j.imlet.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]