Abstract
BRAF mutations play a well-established role in melanomagenesis; however, without additional genetic alterations tumor development is restricted by oncogene-induced senescence (OIS). Here we show that mutations in the NF1 tumor suppressor gene cooperate with BRAF mutations in melanomagenesis by preventing OIS. In a genetically engineered mouse model, Nf1 mutations suppress Braf-induced senescence, promote melanocyte hyperproliferation, and enhance melanoma development. Nf1 mutations function by deregulating both PI3K and ERK pathways. As such, Nf1/Braf mutant tumors are resistant to BRAF inhibitors but are sensitive to combined MEK/mTOR inhibition. Importantly, NF1 is mutated or suppressed in human melanomas that harbor concurrent BRAF mutations, NF1 ablation decreases the sensitivity of melanoma cell lines to BRAF inhibitors, and NF1 is lost in tumors from patients following treatment with these agents. Collectively, these studies provide mechanistic insight into how NF1 cooperates with BRAF mutations in melanoma and demonstrate that NF1-inactivation may impact responses to targeted therapies.
Keywords: RAS, RAF, senescence, NF1, neurofibromin, melanoma, PI3K, mTOR
INTRODUCTION
Oncogene-induced senescence (OIS) is an irreversible growth arrest that is triggered by a variety of oncogenic signals (1). This form of senescence functions as a protective response to aberrant cell signaling and has been shown to restrict the progression of benign lesions such as melanocytic nevi, lung adenomas, neurofibromas, and prostatic intraepithelial neoplasia (2). Several mechanisms have been proposed to underlie OIS including excessive DNA damage (3–5), heterochromatin formation (6), negative feedback pathways (7, 8), and chemokine signaling (8–10). Notably, these mechanisms are not mutually exclusive and it is likely that they cooperate to establish a senescence response in different tissues.
OIS has been shown to be important for restricting melanoma development in response to activating BRAF mutations (11, 12). BRAF is mutated in 50–70% of human melanomas (reviewed in (13)). The most frequent BRAF mutation (BRAFV600E) results in a constitutively active kinase. Analysis of human lesions and mouse models has demonstrated that BRAFV600E mutations drive the development of benign nevi (14–16). However in the absence of additional mutations melanocytes within these nevi ultimately become senescent and do not progress to malignancy (11, 12, 15). Notably, a subset of genetic alterations found in human melanoma prevent BRAF-induced senescence, underscoring the importance of OIS as a mechanism of tumor suppression (15, 17). Nevertheless we still do not have a complete mechanistic or genetic understanding of how OIS is bypassed in melanoma or more generally in cancer.
We have previously shown that oncogenic RAF triggers a potent negative feedback signaling network that suppresses RAS and that this feedback loop plays an important role in OIS in vitro (7). Specifically, in response to constitutively activated RAF and MEK proteins, RAS becomes suppressed due to the upregulation of several direct negative RAS regulatory proteins and the concomitant inactivation of positive RAS regulators (7). Moreover RAF-induced RAS suppression substantially attenuates PI3K/AKT signaling, which contributes to OIS in this setting (7). These observations raise the intriguing possibility that mutational events that promote RAS activation might play an important role in preventing RAF-induced senescence. If so, then mutations in such genes might be expected to cooperate with BRAF mutations in human cancer.
The NF1 tumor suppressor gene encodes a RAS GTPase Activating Protein (RAS GAP) neurofibromin, which negatively regulates RAS by catalyzing the hydrolysis of RAS-GTP to RAS-GDP (18). Accordingly, RAS and downstream effector pathways are aberrantly activated in NF1-deficient tumors (18–20). NF1 is mutated in the familial cancer syndrome neurofibromatosis type 1 and has more recently been shown to be mutated or suppressed by proteasomal mechanisms in glioblastoma, lung cancer, and neuroblastoma (21–25); however, the full extent that NF1 loss may play in sporadic tumorigenesis is unknown. Because of its direct effects on RAS and known involvement in melanocyte biology, we investigated a potential role for NF1 in melanomagenesis. Our studies reveal distinct mechanisms by which NF1 mutations cooperate with different BRAF mutations in melanomas. Moreover, we have found that NF1-loss affects the therapeutic response to BRAF inhibitors.
RESULTS
Nf1 mutations rescue the inhibitory effects of constitutively activated RAF
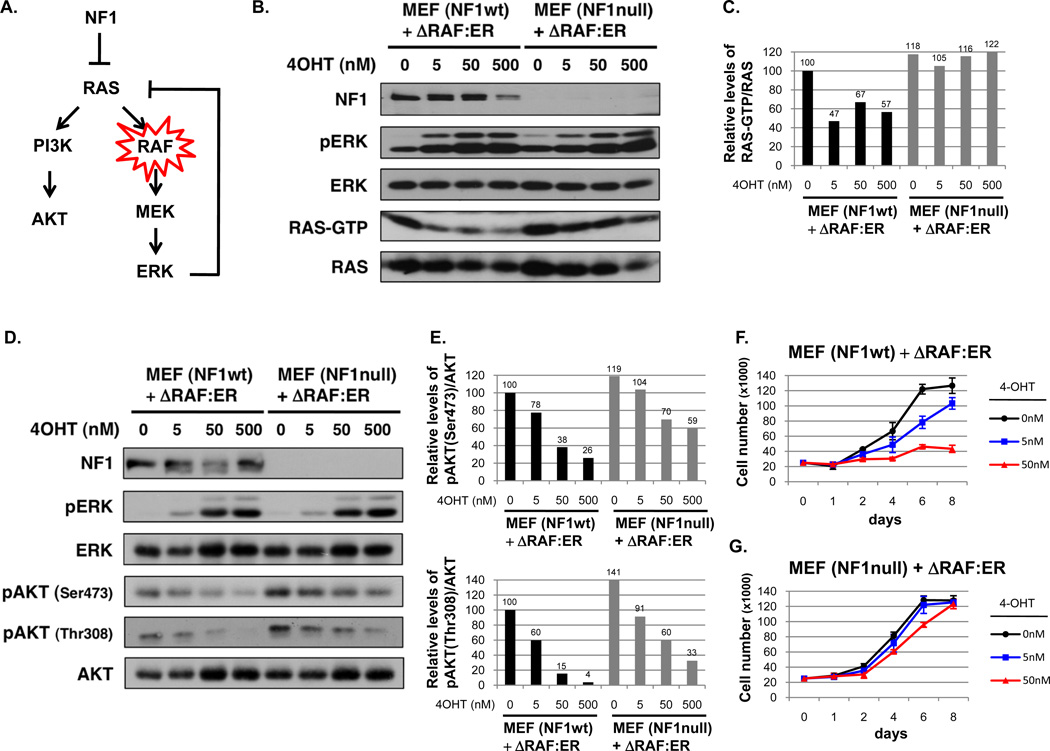
We previously showed that oncogenic RAF alleles potently suppress RAS and subsequent PI3K/AKT signaling and that this suppression is important for OIS in some settings (Fig. 1A) (7). Because NF1 encodes a direct negative regulator of RAS we reasoned that the effects of this feedback response might be counteracted by ablating NF1 expression. Wild-type and Nf1−/− mouse embryonic fibroblasts (MEFs) were stably infected with a hydroxy-tamoxifen (4-OHT) inducible, activated RAF construct (26). 4-OHT substantially suppressed RAS-GTP levels in wild-type cells, consistent with previous findings (Fig. 1B–C) (7). However, RAF activation had minimal suppressive effects on RAS activity in Nf1−/− cells (Fig. 1B–C). Similar effects were also observed in cells in which Nf1 expression was acutely ablated by shRNA sequences (Supplementary Fig. 1). Shortly thereafter AKT phosphorylation became substantially reduced in Nf1 wild-type cells at both Ser 473 and Thr 308 (Fig. 1D–E), however Nf1-deficiency significantly ameliorated this suppression (Fig. 1D–E). It should be noted that even in the absence of Nf1, RAF partially inhibited AKT phosphorylation, consistent with the known involvement of several redundant negative feedback signals (7). Notably, Nf1-loss caused a baseline activation of the PI3K/AKT pathway, as previously observed (Fig. 1D–E) (19, 20), and therefore minimized the net suppressive effects on AKT. Most importantly however, RAF activation exerted differential effects on proliferation in wild-type and Nf1-mutant cells. Consistent with previous observations oncogenic RAF caused a potent and irreversible growth arrest in Nf1 wild-type MEFs (Fig. 1F) (27, 28). However, RAF activation did not suppress the proliferation of Nf1-deficient cells (Fig. 1G), demonstrating that RAS suppression is a critical mediator of this inhibitory response.
Figure 1. NF1 mutations rescue the inhibitory effects of activated RAF.
A) Model of negative feedback pathway and the role that NF1 could play in alleviating suppression. B) NF1 wild-type (wt) and null mouse embryonic fibroblasts (MEFs) expressing an inducible RAF construct (ΔRAF:ER) were treated with the indicated concentrations of hydroxy-tamoxifen (4-OHT) for 24 hours. Immunoblots of total cell lysates evaluating phospho-ERK, total ERK, RAS, and neurofibromin (NF1) are shown. RAS-GTP levels were assessed using a RAS pull-down assay and are quantified relative to total RAS levels in panel C. D) Immunoblots evaluating phospho-ERK and phospho-AKT in total cell lysates from cells treated with 4-OHT for 72 hours are shown. Relative phospho-AKT (Ser473) and phospho-AKT (Thr308) levels are quantified in panel E. F) Proliferation curve of NF1 wt and G) NF1 null MEFs expressing the inducible RAF construct after exposure to increasing concentrations of 4-OHT.
Compound mutations in Nf1 and BRaf promote melanocyte hyperproliferation in vivo
To investigate the potential cooperativity of NF1 and RAF mutations in a relevant tumorigenic setting we generated a mouse model to evaluate the effects of Nf1-loss in the presence of activating Braf mutations. As noted previously BRAF is mutated in 50–70% of human melanomas (13). Based on our in vitro findings and the fact that neurofibromin plays a well-established role in melanocytes (29), we evaluated the potential cooperativity of Braf and Nf1 mutations in the context of melanomagenesis.
Mice carrying a conditional inactivating mutation in Nf1 (Nf1flox/flox mice) (30) were crossed to mice with a conditional activating mutation in Braf (BrafCA/+ mice) (31). These animals were then crossed to a transgenic mouse strain harboring a tamoxifen (TM)-inducible Cre recombinase-estrogen receptor fusion transgene that is under the control of the melanocyte-specific tyrosinase promoter, designated Tyr::CreERT2 (32). Activation of CreER by TM in Tyr::CreER; BrafCA/+; Nf1flox/flox mice leads to melanocyte specific conversion of BrafCA to BrafV600E and the conversion of the Nf1flox alleles to Nf1 null alleles. Six genetic cohorts of animals were generated to evaluate the effects of Braf activation in the presence and absence of Nf1.
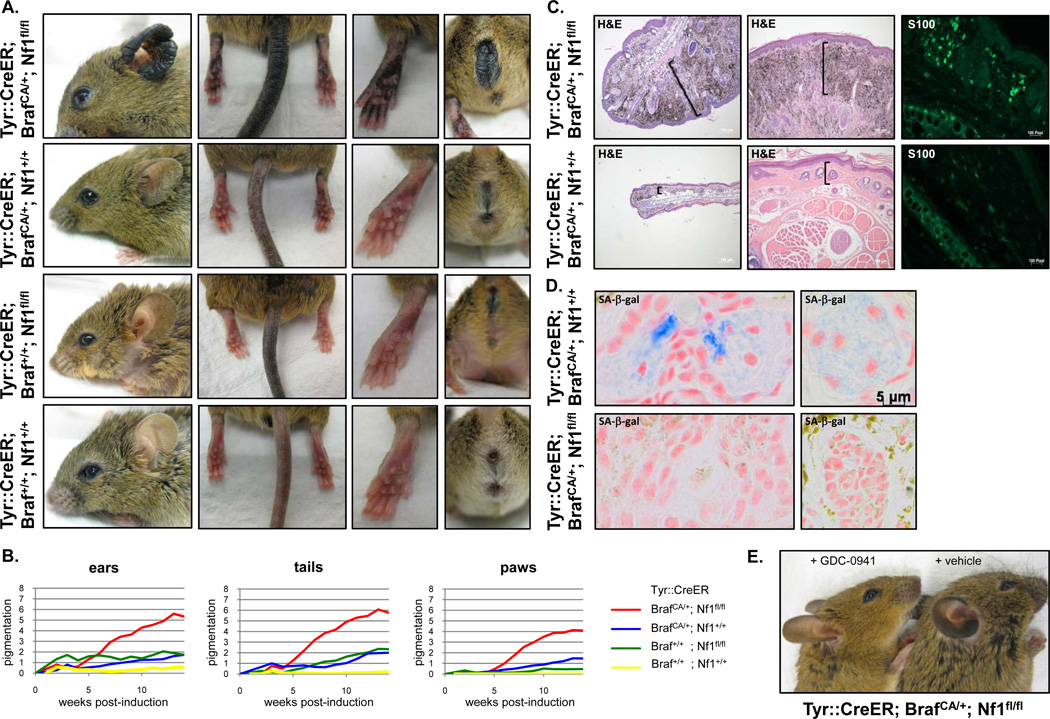
Mice were treated topically with TM 2–3 months after birth as previously described (15). After 4–5 weeks significant darkening of the tails, ears, eyelids, perianal regions, and paws was observed in the Tyr::CreER; BrafCA/+; Nf1flox/flox mice as compared to all other genotypes (Fig. 2A–B). While Tyr::CreER; BrafCA/+ animals exhibited subtle hyperpigmentation, the skin hyperpigmentation in the Tyr::CreER; BrafCA/+; Nf1flox/flox mice became dramatically more pronounced over time and was concurrent with visible thickening of the respective tissues (Fig. 2A–B). The histopathological features of these lesions were consistent with expansion of the dermis, the skin layer where murine melanocytes reside, and massive melanin deposition (Fig. 2C). An increased number of cells expressing the melanocyte marker S100 was observed, confirming excessive melanocyte hyperproliferation in Tyr::CreER; BrafCA/+; Nf1flox/flox mice as compared to the BrafCA/+ genotype (Fig. 2C). Importantly we found that deep dermal lesions derived from control Tyr::CreER; BrafCA/+ mice stained positive for senescence-associated (SA)-β-galactosidase (Fig. 2D, top panels), as has been shown in human nevi (12) and in lesions within the BrafV600E-driven mouse model described by Dhomen et al. 2009 (15). However, senescence was not observed in Tyr::CreER; BrafCA/+; Nf1flox/flox mice (Fig. 2D, bottom panels). These results are consistent with our cellular studies and indicate that mutations in Nf1 prevent Braf-induced senescence of melanocytes in mice, thereby rescuing the proliferative restriction and triggering excessive proliferation.
Figure 2. Nf1 and Braf mutations cooperate in vivo and lead to enhanced melanocyte proliferation.
A) Phenotype of mice with the indicated genotypes 7 months after Tyr::CreER induction. B) Quantification of pigmentation phenotype of different genotypes over time following Tyr::CreER induction. C) Histological images of ears (left panels) and tails (middle panels) from mice with the specified genotypes 9 months after tamoxifen treatment. S100 expression was evaluated by immunofluorescent staining (right panels). D) Sections of ear stained for eosin and senescence-associated (SA)-β-galactosidase (light blue) from tamoxifen treated Tyr::CreER; BrafCA/+; Nf1+/+ (top panel) and Tyr::CreER; BrafCA/+; Nf1flox/flox mice (bottom panel) mice. Note the blue staining and cell morphology differences. E) Tyr::CreER; BrafCA/+; Nf1flox/flox mice were treated daily with the PI3K inhibitor GDC-0941 (150mg/kg) or vehicle for 8 weeks after Tyr::CreER induction. The hyperpigmentation phenotype was prevented by GDC-0941 treatment.
PI3K is a well-known effector of RAS (33). We and others have previously shown that NF1 loss triggers the activation of the PI3K/AKT/mTOR pathway through RAS (19, 20, 34). Moreover, Nf1 mutations minimize the suppressive effects of Braf mutations on this pathway (Fig. 1D–E). Notably, we found that the PI3K inhibitor GDC-0941 prevented the melanocytic hyperplasia in Tyr::CreER; BrafCA/+; Nf1flox/flox mice (Fig. 2E), demonstrating that Nf1 loss was mediating its effects in melanocytes, in part, by permitting/enhancing the activation of this pathway.
Nf1 and BRaf mutations cooperate to promote melanomas in mice
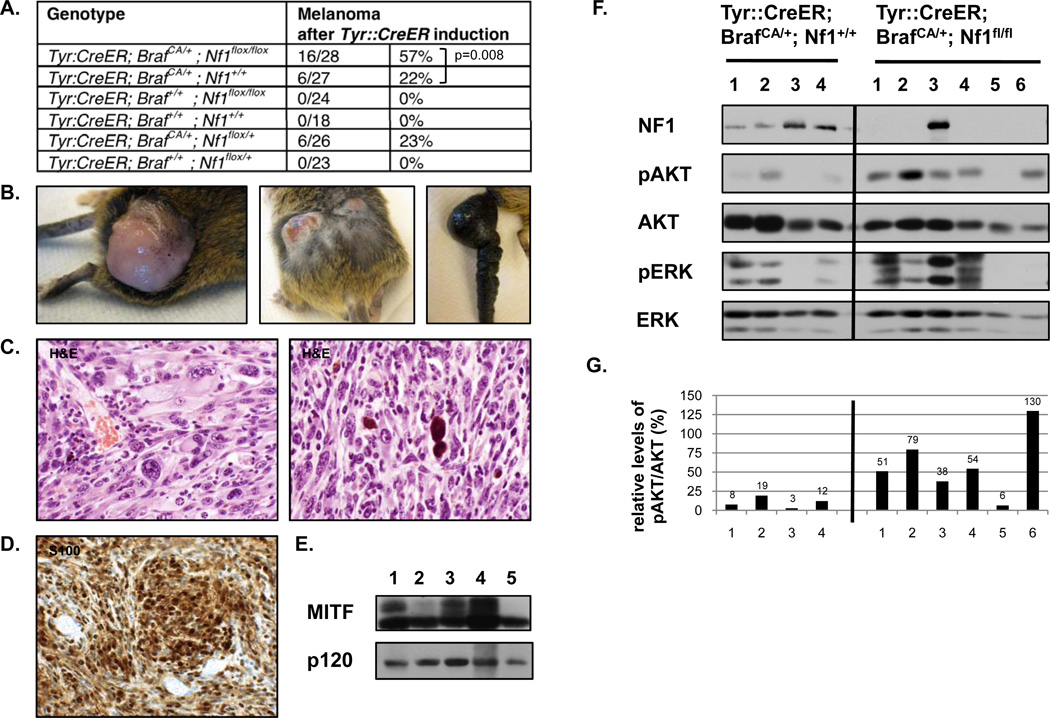
If OIS does in fact restrict tumor development then Nf1/Braf mutant mice would be expected to be more prone to developing melanomas. It has been previously reported that a subset of BrafV600E mice develop melanomas, presumably due to the stochastic acquisition of additional genetic alterations (15). Consistent with this observation 22% (6/27) of the BrafV600E mice in our cohort developed melanomas; however, 57% (16/28) of the Tyr::CreER; BrafCA/+; Nf1flox/flox mice developed melanomas (p=0.008, Chi Square test) (Fig. 3A–E). One Braf/Nf1 mutant mouse developed two melanomas, which was never observed in Braf mutant animals, however melanomas from both genotypes grew at similar rates. These observations support the hypothesis that Nf1 loss prevents Braf-induced senescence in vivo and therefore plays a role early in tumor development rather than in progression. However effects on metastasis could not be evaluated in this model, as animals from both genotypes needed to be euthanized due to primary tumor size. While pigmented melanocytes were occasionally observed on the exterior these melanomas were typically hypopigmented (Fig. 3B). All Braf/Nf1 tumors displayed histological and cytological features typical of malignant melanomas. Tumors were highly cellular with most showing a fascicular growth pattern (Fig. 3C). The degree of pleomorphism was variable. The majority of the tumor cells were amelanotic, however many tumors showed occasional clusters of pigmented cells and pigment containing macrophages (melanophages) (Fig. 3C, right). All tumors involved the dermis as well as subcutaneous soft tissue and ulceration of the tumor surface was seen in the majority of cases. Tumors expressed both S100 (Fig. 3D) and MITF (Fig. 3E), two markers typically used to diagnose human melanomas. Melanomas from both genotypes were further evaluated by immunoblot (Fig. 3F). One tumor that developed in Tyr::CreER; BrafCA/+; Nf1flox/flox mice did not efficiently excise Nf1. However in general higher levels of phospho-AKT were typically observed in Braf/Nf1 mutant versus Braf mutant tumors (Fig. 3F–G), consistent with the observation that Nf1 mutations enhance PI3K/AKT activation and contribute to tumorigenesis, in part, via this pathway (19, 20, 34) (Fig. 2E).
Figure 3. Nf1 and Braf mutations cooperate to promote melanomagenesis in mice.
A) Genotypes and tumor phenotypes of experimental animals and control groups. B) Pictures showing tumors from tamoxifen (TM) treated Tyr::CreER; BrafCA/+; Nf1flox/flox mice. Note the pigmented area on the surface of the tumor (left panel), the presence of more than one lesion (middle panel) and the remarkable thickening of the tail (right panel). C–D) Representative histological images (400× magnification) of tumors from TM-induced Tyr::CreER; BrafCA/+; Nf1flox/flox mice. C) Tumor sections are stained with hematoxylin and eosin (H&E). Tumors show marked cytologic atypia and pleomorphism, including bizarre cells, with irregular chromatin distribution and irregular nuclear outlines (left panel). Occasionally, melanophages containing extensive brown pigment were observed (right panel). D) The vast majority of neoplastic cells stain positive for S100. E) MITF protein expression in primary tumor tissue from TM-treated Tyr::CreER; BrafCA/+; Nf1flox/flox mice was confirmed by immunoblotting. F) Protein levels of NF1, phospho-AKT and phospho-ERK in distinct tumors from TM-induced Tyr::CreER; BrafCA/+; Nf1+/+ (left) and Tyr::CreER; BrafCA/+; Nf1flox/flox (right) mice, as determined by immunoblotting. Relative phospho-AKT levels are quantified in panel G.
Nf1 mutations desensitize melanomas to BRAF inhibitors
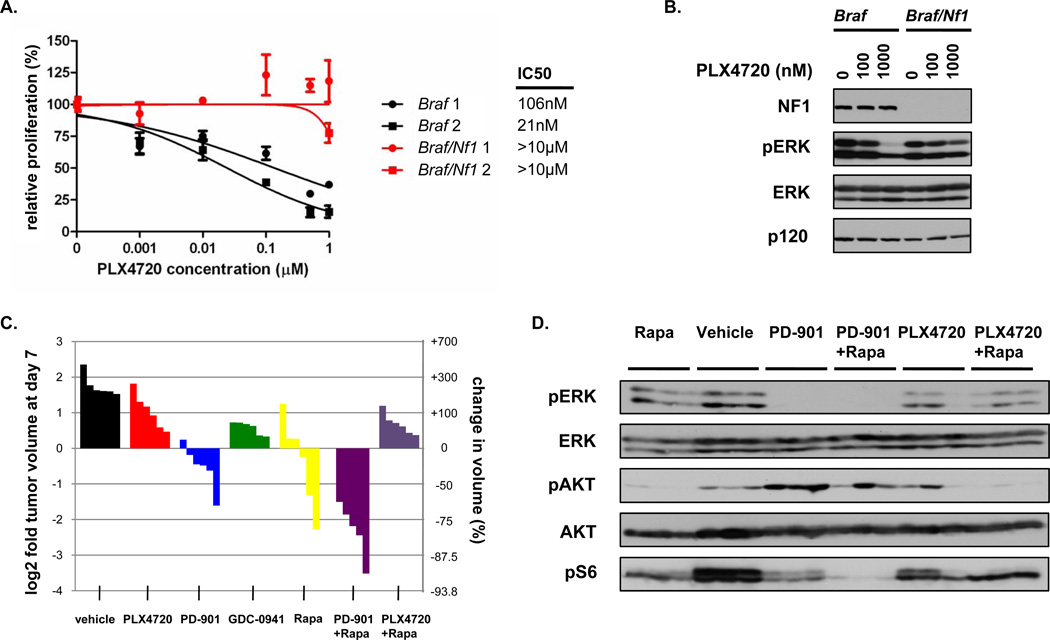
The mutant BRAF selective inhibitor PLX4032 (vemurafenib, Plexxikon/Roche) promotes the regression of human melanomas that harbor activating BRAF mutations and has been approved for treating human melanomas (35). To investigate the sensitivity of melanomas harboring compound mutations in Braf and Nf1 to BRAF inhibitors and other targeted agents, we first isolated cells from Braf and Braf/Nf1 mutant tumors. As expected, cells from two independently derived Braf mutant melanomas were sensitive to the PLX4032 analogue, PLX4720 (Fig. 4A). However, cells from Braf/Nf1 mutant tumors were insensitive to this agent (Fig. 4A, IC50>10µM). Biochemical studies confirmed that increasing concentrations of PLX4720 effectively suppressed phospho-ERK levels in Braf mutant cells (Fig. 4B). Notably, however, phospho-ERK was not as effectively suppressed in Braf/Nf1 mutant cells (Fig. 4B).
Figure 4. Nf1 mutations densensitize mouse melanomas to BRAF inhibitors.
A) Dose sensitivity curves for PLX4720 are shown for two independently derived cell lines from Braf mutant (black) and Braf/Nf1 mutant (red) melanomas. B) Pharmacodynamic analysis of phospho-ERK in Braf and Braf/Nf1 cells in vitro after a 24-hour incubation with 0, 100, and 1000nM PLX4720. C) Waterfall plot depicting tumor growth of Braf/Nf1 mutant allografts after 7 days of treatment with vehicle (black), PLX4720 (red), PD0325901 (blue), GDC-0941 (green), rapamycin (yellow), PD0325901/rapamycin (purple), and PLX4720/rapamycin (violet). Each bar represents a different tumor. Both Braf/Nf1 mutant cell lines shown in Fig. 4A were injected into three mice each, resulting in a total of 6 tumors per treatment arm. D) In vivo pharmacodynamic analysis of phospho-ERK, phospho-AKT, and phospho-S6 in Braf/Nf1 mutant allografts treated with Rapamycin (Rapa), Vehicle, PD0325901 (PD-901), PD0325901/Rapamycin, PLX4720, and PLX4720/Rapamycin. Lysates were prepared from tumor tissues that were harvested 2 hours post administration of the respective treatment from 3 tumors shown in Fig. 4C.
Next we evaluated sensitivity to these agents in vivo. While allografts from several independent Braf/Nf1 mutant melanomas were readily established, cells from BRaf mutant mouse melanomas never successfully grew as allografts. As such we focused on using Braf/Nf1 allografts to determine whether we could identify a more effective therapy for these genetically distinct tumors. Two weeks after injection, when tumors were growing in log phase, mice were randomly divided into different treatment groups. Similar to our in vitro analysis, Braf/Nf1 mutant melanomas were relatively insensitive to PLX4720 (Fig. 4C, red) and phospho-ERK was not efficiently inhibited in tumors in vivo (Fig. 4D). This finding is consistent with the observation that induction of tumor regression by PLX4032 requires almost complete suppression of ERK signaling (36). In contrast, these tumors were more sensitive to the MEK inhibitor PD0325901 (Fig. 4C, blue), which effectively suppressed phospho-ERK in vivo (Fig. 4D). Together with our in vitro studies this insensitivity suggests that neurofibromin loss may enhance ERK activity via a BRAF-independent mechanism, which will be discussed further below.
Because our in vivo experiments demonstrated that melanocyte hyperproliferation in Tyr::CreER; BrafCA/+; Nf1flox/flox mice could be rescued by the PI3K inhibitor GDC-0941, we evaluated the effects of GDC-0941 on tumor growth, and found that it had a slight growth suppressive effect on these melanomas (Fig. 4C, green). However given that mTOR has been shown to be an important effector in NF1 mutant tumors we also assessed the mTOR inhibitor rapamycin (20, 34, 37). Rapamycin had a more pronounced effect as compared to GDC-0941, but more importantly it synergized with PD0325901 to promote tumor regression (Fig. 4C, purple). In contrast, rapamycin did not promote tumor regression when combined with PLX4720 (Fig. 4C, violet). Taken together these results suggest that Nf1 mutations can desensitize Braf mutant melanomas to PLX4720. Based on the biochemical function of neurofibromin and the preclinical data presented here, therapies aimed at targeting both MEK and mTOR may represent an alternative therapeutic approach for Braf/Nf1 mutant tumors.
NF1 is suppressed and/or mutated in human melanoma
Given these compelling mouse phenotypes we next investigated whether NF1 is lost or mutated in human melanomas. Like p53, neurofibromin can be inactivated by both genetic and proteasomal mechanisms (23). We first evaluated neurofibromin expression in a panel of melanoma cell lines. Four out of eleven cell lines exhibited little or no neurofibromin expression (Fig. 5A). Sequence analysis confirmed that three of these cell lines harbored loss-of-function mutations in the NF1 gene (Supplementary Table 1). It should be noted that two of the NF1 alterations (p.K1290K in A375 cells and p.K2307K in WM3670 cells) abolish splice donor sites (c.3870G>A and c.6921G>A, respectively), result in defective splicing and disrupt the NF1 transcript. Sequence analysis of the cDNA of NF1 enabled the detection of these aberrant splicing events, however, both NF1 mutations may have been categorized as ‘non-deleterious’ silent alterations using exome sequencing approaches. We then mined publically available databases and identified numerous additional NF1 mutations in human cell lines (Supplementary Table 1). Analysis of primary melanomas also confirmed the presence of somatic NF1 mutations (Supplementary Table 2). In many, but not in all cases, BRAF mutations were also present. These findings suggest that while NF1 mutations can cooperate with activating BRAF mutations, they may also play a broader role in melanomagenesis. Interestingly, however, while we expected to find coincidental mutations with BRAFV600E, we also observed NF1 mutations in cells that harbored inactivating BRAF mutations (Supplementary Table 1), a point which will be discussed below.
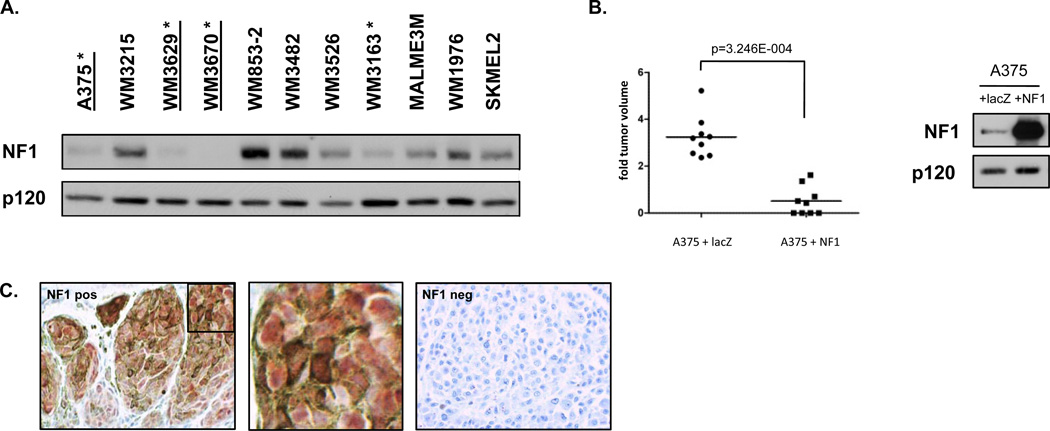
Figure 5. NF1 protein expression in human melanomas.
A) NF1 protein levels in a panel of 11 human melanomas cell lines as determined by immunoblotting. Four lines (with asterisks) exhibit low or no NF1 expression. Three lines (underlined) harbor NF1 mutations. B) NF1 reconstitution in A375 cells potently suppresses the growth of xenografts in mice. Tumor size is shown on left, protein expression in right panel. C) Tissue microarray analysis of NF1 in human malignant and metastatic melanomas. Positive NF1 staining in nevus is shown in left panel. A magnification of the inset box is shown in the middle panel. Negative NF1 staining in human melanoma is shown in the right panel.
To complement the mouse modeling studies and confirm a functional role for NF1 inactivation in human melanomas we reconstituted neurofibromin in A375 cells, which harbor compound NF1 and BRAFV600E mutations. Full-length neurofibromin expression potently suppressed the growth of xenografts in mice (p=3.246E-004, Mann Whitney U test), consistent with the notion that NF1-inactivation plays a causal role in tumor development (Fig. 5B). As noted above, the NF1 tumor suppressor is frequently inactivated by proteasomal mechanisms in human cancer (23). As such mutational analysis may underestimate the frequency of NF1/neurofibromin loss in tumors. Therefore we performed immunohistochemical analysis on human melanoma tumor arrays. No visible neurofibromin expression was observed in 15% (6/39) of melanomas and 18% (6/34) of the metastatic melanomas (Fig. 5C and Supplementary Table 3). It should be noted that only a complete absence of staining was scored as negative in this analysis. Therefore excessive but incomplete neurofibromin destruction was not considered, but could still play a role in tumor development.
NF1 loss and BRAF inhibitors in human tumors
Preclinical studies in mouse tumors suggested that Nf1 loss can desensitize Braf mutant melanomas to BRAF inhibitors. To evaluate this possibility in human tumors we genetically ablated neurofibromin expression with lentivrial shRNA sequences in human melanoma cell lines and found that NF1 suppression decreased the sensitivity of WM3526 cells to PLX4720 by 11-fold (Fig. 6A and Supplementary Fig. 2). As noted previously A375 cells harbor a loss of function NF1 mutation and express very low levels of neurofibromin. Because these cells remain somewhat sensitive to BRAF inhibitors we further reduced neurofibromin expression with shRNA sequences and found that NF1 suppression also decreased the sensitivity of these cells to BRAF inhibitors (Fig. 6B and Supplementary Fig. 2). It should be noted that while the acute ablation of NF1 in established melanoma cell lines desensitized these cells to PLX4720, tumors that naturally developed in the absence of Nf1 were much more resistant to this agent. We hypothesize that this difference may reflect inherent differences in pre-existing signaling networks and/or cooperating mutations in these established tumor cells, which did not evolve in the absence of NF1. Nevertheless, consistent with observations in mouse tumors, PLX4720 was much less effective at suppressing phospho-ERK in cells in which NF1 was ablated, indicating that NF1 loss was promoting BRAF-independent ERK signaling in these tumor cells as well (Fig. 6C).
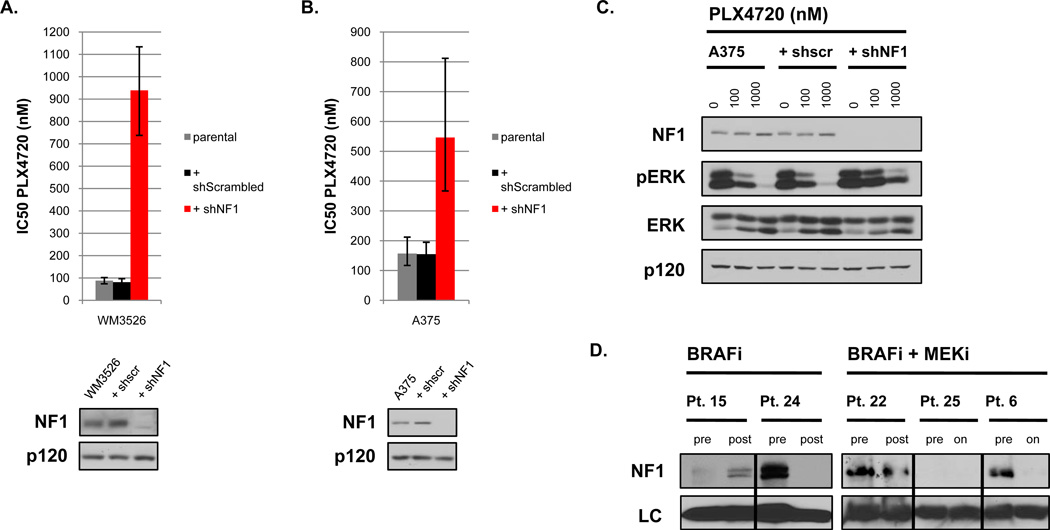
Figure 6. NF1 levels mediate sensitivity to BRAF inhibition.
A) Bar graph representing IC50 values for PLX4720 in the BRAFV600E mutant melanoma cell line WM3526 in the absence or presence of a short hairpin RNA (shRNA) specific for NF1. The doubling time of these cells did not change in the presence of shRNA sequences. B) A similar graph representing IC50 values for PLX4720 in the BRAFV600E mutant melanoma cell line A375. C) Pharmacodynamic analysis of phospho-ERK in A375 cells with or without residual NF1 expression after a 24-hour incubation with 0, 100, and 1000nM PLX4720. D) NF1 protein levels in tumor biopsies from patients (Pt.) before and after treatment with a BRAF inhibitor (BRAFi) (vemurafenib) or combined BRAF/MEK inhibitors (BRAF/MEKi) (dabrafenib + trametinib). Where indicated samples were collected from relapsed biopsies (post) or from residual tumor tissue while patients were still on treatment (on). Total levels of GAPDH (BRAFi samples) and HSC70 (BRAF/MEKi samples) served as loading controls (LC).
Finally, we were able to obtain frozen tissue from 5 sets of pre- and post-treatment tumor biopsies from patients treated with a BRAF inhibitor (vemurafenib) or combined BRAF and MEK inhibitor (dabrafenib + trametinib). Notably, 2 out of 5 tumors expressed very little or no neurofibromin prior to treatment, consistent with observation that NF1/neurofibromin is lost or suppressed in human melanomas at a relatively frequent rate (Fig. 6D). However in 2 of the remaining 3 tumors where neurofibromin was robustly expressed prior to treatment, neurofibromin was no longer expressed in tumors following treatment (Fig. 6D). Taken together with preclinical studies in the mouse model and genetic studies in human melanoma cell lines, these observations further support the hypothesis that NF1/neurofibromin suppression may play an important role in mediating resistance to BRAF inhibitors. Notably, in glioblastoma neurofibromin appears to be more frequently lost by proteasomal mechanisms (23). Therefore future studies aimed at assessing NF1/neurofibromin loss in response to therapies will likely require analysis of both protein expression and genetic alterations.
NF1 mutations cooperate with BRAF mutations by activating both K- and HRAS
Our central hypothesis is that loss of NF1 alleviates the suppression of RAS imposed by activated BRAF. To identify the RAS isoforms that are critically regulated by neurofibromin in melanomas we performed both gain- and loss-of-function studies. In melanoma cells that retain neurofibromin expression RNAi-mediated suppression of neurofibromin resulted in the activation of H- and KRAS, but not NRAS (Fig. 7A). Conversely, reconstituting melanoma cell lines with an active neurofibromin fragment suppressed H- and KRAS activity but not NRAS activity (Fig. 7B). Finally shRNA-mediated suppression of either H- or KRAS suppressed the ability of NF1-deficient melanoma cells harboring an active BRAF mutation to proliferate and form colonies in soft agar, whereas NRAS-specific shRNA sequences had no effect (Fig. 7C–E). These results suggest that neurofibromin loss/suppression activates H- and KRAS in melanomas and that both of these isoforms are critical for the tumorigenicity of these cancer cells.
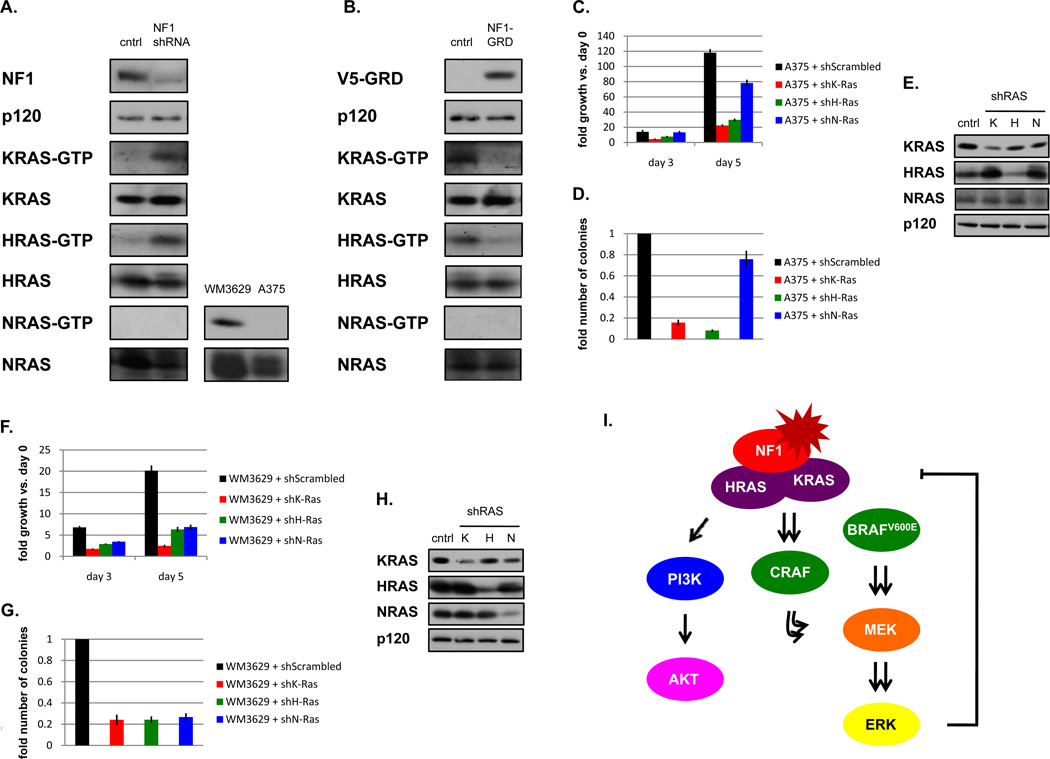
Figure 7. NF1 critically regulate KRAS and HRAS in melanomas.
A) (left) In WM1976 melanoma cells that retain NF1 expression, RNAi mediated suppression of NF1 results in the activation of H- and KRAS, but not NRAS. (bottom right panels) NRAS-GTP assay confirming that NRAS is active in melanoma cells with an activating NRAS mutation (WM3629), but not in melanoma cells wild-type for NRAS (A375). B) Ectopic expression of the catalytically active NF1 fragment (GAP-related domain, GRD) suppresses H- and KRAS, but not NRAS activity in melanoma cells (MALME3M). RAS-GTP levels were assessed using a RAS pulldown assay. Immunoblots on total cell lysates are shown. C) shRNA mediated suppression of H- or KRAS, but not NRAS, suppresses the ability of A375 cells to proliferate and D) form colonies in soft agar. E) Confirmation of RAS isoform-specific knockdown by shRNAs in A375 cells. F) shRNA mediated suppression of H-, K- and NRAS suppresses the ability of WM3629 cells to proliferate and G) form colonies in soft agar. H) Confirmation of RAS isoform-specific knockdown by shRNAs in WM3629 cells. I) Suggested model for the distinct roles of NF1 in melanomagenesis.
As shown in Supplementary Table 1, NF1 mutations were also detected in cells that harbored inactivating BRAF mutations. While the kinase activity of these BRAF proteins is compromised, they have been proposed to function as scaffolds that translocate CRAF to the membrane and promote CRAF activation downstream of KRAS (38). Notably, the oncogenic effects of these mutants are significantly enhanced in the presence of KRASG12D (38). Similarly, an NF1 mutation might be expected to function as an alternative mechanism of activating KRAS, and also potentiate the effects of these BRAF mutants. To evaluate the contribution of RAS isoforms in human melanomas we examined WM3629 cells. Interestingly, in addition to harboring the kinase-dead BRAFD594G mutation and an NF1 deletion, this cell line also possesses an activating NRASG12D mutation (39, 40). The WM3670 line similarly harbors an inactivating BRAF mutation, as well as NF1 and NRASG12D mutations (39, 40). Consistent with the presence of these mutations and our previous observations, NRAS, KRAS, and HRAS are all activated in WM3629 cells. Moreover, shRNA-mediated ablation of all three RAS isoforms, H-, K-, and NRAS, suppressed the ability of WM3629 cells to proliferate and form colonies in soft agar (Fig. 7F–H). These results further demonstrate that neurofibromin critically regulates H- and KRAS in melanomas and suggest that in the presence of NRAS mutations all three RAS isoforms cooperate with inactivating BRAF mutations to promote tumorigenesis.
DISCUSSION
This study establishes several distinct roles for NF1 in melanomagenesis. First, our data suggest that in the presence of activating Braf mutations, Nf1-loss prevents Braf-induced senescence of melanocytes. In mice, this results in excessive melanocyte hyperproliferation and ultimately enhances melanoma development. Importantly, NF1 mutations co-occur with activating BRAF mutations in human melanomas and neurofibromin reconstitution potently suppresses the growth of human melanoma cells as xenografts, further supporting a causal role for NF1 loss in melanomagenesis. However, we have found that NF1 mutations can also cooperate with inactivating BRAF mutations in melanomas. While these BRAF mutations are less common, in this setting we found that NF1 mutations co-occur with NRAS mutations and that all three RAS isoforms are required for the tumorigenic properties of these cells. Thus, NF1 mutations can contribute to melanoma development in at least two genetic settings via distinct mechanisms.
We also found that in the context of activating BRAF alleles NF1 mutations contribute to tumorigenesis, in part, by promoting the activation of the PI3K/AKT/mTOR pathway. Notably, the PTEN tumor suppressor is commonly mutated or lost in human melanomas (41). Mouse modeling studies and human tumor analysis suggest that BRAF and PTEN mutations cooperate in melanomagenesis (14). More recently, PTEN loss has been shown to prevent BRAF-induced senescence in mice and in human melanocytes, further supporting the notion that co-activation of these pathways is important for preventing OIS (17). Our data demonstrate that NF1 loss is another important mechanism by which the PI3K pathway can become inactivated in melanomas. It should be noted however, that NF1 and PTEN mutations do not appear to be mutually exclusive in melanomas. Moreover, we have found that NF1 loss also results in the activation of multiple RAS isoforms and potentiates ERK activation independent from activating BRAF mutations. Taken together these observations suggest that NF1 loss contributes to melanomagenesis by enhancing the activation of both PI3K and ERK signaling, which may be important in the context of selecting effective therapies (Model presented in Fig. 7I).
Several mechanisms have been reported to mediate the resistance of BRAF mutant melanomas to BRAF inhibitors (42–46). Additional mechanisms are likely to be discovered and currently one prevalent mechanism of resistance has not emerged. We have shown that Nf1 mutations confer resistance to PLX4720 in Braf-mutant mouse melanomas, however these tumors are sensitive to combined MEK/mTOR inhibitors. Importantly, RNAi-mediated NF1 suppression also decreases the sensitivity of human melanoma cell lines to BRAF inhibitors and most notably NF1/neurofibromin is lost in a subset of relapsing and residual tumors from patients exposed to BRAF inhibitors. The observation that NF1 is also mutated or lost in some naive primary tumors is consistent with the hypothesis that NF1-inactivation may contribute to both de novo and acquired resistance. However, while NF1 mutations can be detected in human melanomas, the true frequency of NF1-loss may be difficult to assess, because like PTEN and P53, the NF1 protein is frequently inactivated by proteasomal destruction (23). Immunohistochemical analysis of tumor microarrays indicates that neurofibromin expression is completely absent in 15–18% of melanomas, however a more quantitative evaluation of protein levels may be required to accurately evaluate its expression/suppression before and after drug treatment. To date, much of the resistance to BRAF inhibitors appears to be driven by events that activate ERK through mechanisms that circumvent or decrease dependency on BRAF. However, aberrant activation of the PI3K/AKT pathway has also been implicated in resistance to BRAF inhibitors (46–48). In this respect NF1 (loss) is uniquely poised, as it activates the ERK pathway through its effects on KRAS and HRAS and at the same time enhances PI3K/AKT/mTOR signaling.
METHODS
Cell Culture Techniques, Infections and Proliferation Curves
Wild-type and NF1 null mouse embryonic fibroblasts (MEFs) were generated as described (49). Cells were stably infected with a pBabe retroviral vector containing the 4-hydroxy tamoxifen (4-OHT) inducible estrogen receptor RAF-1 (ΔRAF:ER) construct (28). This construct is an estrogen receptor-RAF-1 fusion molecule, and contains the ligand-binding domain of the estrogen receptor fused to the activated RAF kinase domain of RAF-1. Cells were split to an equal density 16hr prior to harvesting in DMEM medium containing 1% fetal calf serum (FCS) and different concentrations of 4-OHT (0, 5, 50 or 500nM). For proliferation studies, cells were counted at seeded at a density of 25 × 104 in a 12-well format in DMEM medium containing 10% FCS and different concentrations of 4-OHT (0, 5, 50nM). Cells were counted the day after plating and every other day after until uninduced cells reached confluency. Media containing fresh 4-OHT was changed every 48hrs. The melanoma cell lines were obtained from the ATCC and the Garraway lab (46). No additional authentication was done by the authors. All melanoma cell lines were grown in RPMI medium supplemented with 10% FCS, penicillin and streptomycin. For dose-response curves, cells were seeded in triplicate in a 6-well format. Cells were counted the day after plating when PLX4720 (Calbiochem) was added as well as after 3 days when DMSO treated cells reached confluency. To allow comparison of IC50’s between cell lines growth inhibition was calculated at 5 population doublings for every cell line. Neurofibromin activity was reconstituted using the previously described NF1-GAP related domain (GRD) construct (20) or an expression vector containing full length NF1 cDNA. Neurofibromin expression was abolished using short hairpin RNA’s specific to NF1 (20).
Preparation of Protein Lysates and Western Blotting
RAS-GTP levels were detected using a RAS activation assay, following the manufacturer’s instructions (EMD Millipore). Tumor lysates were homogenized and extracted with boiling 1% SDS buffer. Resulting protein lysates were quantified and run according to validated immunoblot procedures with the following antibodies: phospho-AKT (Ser473, #4060, Cell Signaling), phospho-AKT (Thr308, #9275, Cell Signaling), AKT (#9272, Cell Signaling), phospho-ERK (Thr202/Thr204, #4370, Cell Signaling), ERK (#9102, Cell Signaling), phospho-S6 (Ser235/236, #2211, Cell Signaling), S6 (#2317, Cell Signaling), NF1 (#A300-140A, Bethyl Laboratories), p120 (#G12920, Trans Labs), RAS (#05-516, Upstate), KRAS (#sc-30, Santa Cruz), HRAS (sc-520, Santa Cruz), NRAS (#sc-519, Santa Cruz) and MITF (#M3621, Dako). Immunoblots were quantified with Image J software.
Experimental Animals
Animal procedures were approved by the Center for Animal and Comparative Medicine in Harvard Medical School in accordance with the NIH Guild for the Care and Use of Laboratory Animals and the Animal Welfare Act. A breeding scheme was set up where mice carrying a conditional Nf1 allele (Nf1flox/flox mice) (30) were crossed to mice with a conditional activating mutation in Braf (BrafCA/+ mice) (31) and a transgenic mouse strain harboring a tamoxifen (TM)-inducible Cre recombinase-estrogen receptor fusion transgene that is under the control of the melanocyte-specific tyrosinase promoter, designated Tyr::CreERT2 (32). Genotyping was performed by PCR as described for every model (30–32). To activate Tyr::CreER mice were treated topically with freshly prepared tamoxifen (T5648, Sigma) 2–3 months after birth. Tamoxifen (20mg/ml in 100% ethanol) was applied to a small section of skin on the shaven backs and the treatment schedule consisted of 4 treatments over 7 days. For all genotypes pigmentation levels were quantified weekly on a scale from 0 (no pigmentation) to 8 (high pigmentation). To study the contribution of the PI3K/AKT pathway in the observed hyperpigmentation phenotype in the Tyr::CreER; BrafCA/+; Nf1flox/flox mice, animals were treated daily with the PI3K inhibitor GDC-0941 (150mg/kg, oral gavage) for 8 weeks after Tyr::CreER induction.
Xenograft Studies and Treatments
For cancer cell xenograft experiments nude mice were inoculated subcutaneously with 3 × 106 human or mouse melanoma cells. Tumor volumes were calculated by measuring length and width of the lesions and with the formula [(length) × (width)2 × 0.52]. Murine Nf1/Braf mutant melanoma cells formed rapidly growing tumors. Two weeks after injection, when tumors were growing in log phase, mice were randomly divided into different treatment groups that were administered either the PI3K inhibitor GDC-0941 (150mg/kg, oral gavage (OG)), the mTOR inhibitor rapamycin (5mg/kg, intraperitoneal injection (IP)), the MEK inhibitor PD0325901 (10mg/kg, OG), the BRAF inhibitor PLX4720 (2.5mg/kg, IP) or the combination of both PD0325901 or PLX4720 and rapamycin. Mice were treated daily for 7 days.
Histology, Immunohistochemistry and SA-β-gal staining
Tissues were fixed in formalin, embedded in paraffin and stained with hematoxylin and eosin (H&E) where indicated. Standard immunohistochemistry and immunofluorescence protocols were followed for S100 (Z0311, Dako, 1:400) staining. An alkaline phosphatase staining method was used for NF1 (ab30325, Abcam, 1:300) immunohistochemistry. Antigen unmasking was performed by pressure cooking in citrate buffer (pH6). A human melanoma tissue micorarray (ME804a, US Biomax Inc) was used for evaluation of NF1 expression in malignant and metastatic melanomas. Senescence associated (SA)-β-gal staining was performed on fresh mouse tissue. Small pieces were fixed for 30 min in 3% paraformaldehyde, incubated in SA-β-gal staining solution as described (50), and then embedded and sectioned.
NF1 Mutational analysis and Data mining
The entire NF1 coding region of 11 human melanoma cell lines (Fig. 5A) was amplified in 5 overlapping RT-PCR fragments and used as the template for direct sequencing, essentially as described (51). Copy number analysis by multiplex ligation-dependent probe amplification (MLPA) was performed as described (52). The nomenclature of the mutations is based on NF1 mRNA sequence NM_001042492.2, with 1 being the first nucleotide of the ATG start codon. Next, publicly available resources providing information on somatic mutations implicated in human melanoma cell lines (53, 54) and primary tumors (55–57) were mined.
Patient Samples
Patients with metastatic melanoma containing BRAFV600E mutation (confirmed by genotyping) were enrolled on clinical trials for treatment with a BRAF inhibitor (vemurafenib) or combined BRAF + MEK inhibitor (dabrafenib + trametinib) and were consented for tissue acquisition per IRB-approved protocol. Tumor biopsies were performed pre-treatment (day 0), at 10–14 days on treatment, and/or at time of progression if applicable. Formalin-fixed tissue was analyzed to confirm that viable tumor was present via hematoxylin and eosin (H&E) staining. Additional tissue was snap frozen and stored in liquid nitrogen.
Supplementary Material
Acknowledgements
We thank M. McMahon and K. Haigis for providing the BrafCA/+ mice; and L.Chin for providing the Tyr::CreER mice and L. Garraway for cell lines.
Financial support: OM is a postdoctoral fellow of the Research Foundation Flanders (FWO). KC and the majority of this work was supported by the NCI (R01 CA111754) and the Ludwig Center at DF/HCC.
Footnotes
There are no conflicts of interest to disclose
SIGNIFICANCE
This study elucidates the mechanism by which NF1 mutations cooperate with different BRAF mutations in melanomagenesis and demonstrates that NF1-loss may desensitize tumors to BRAF inhibitors.
REFERENCES
- 1.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 2.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 4.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 5.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 7.Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 11.Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 13.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 17.Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernards A, Settleman J. GAPs in growth factor signalling. Growth Factors. 2005;23:143–149. doi: 10.1080/08977190500130480. [DOI] [PubMed] [Google Scholar]
- 19.Dasgupta B, Yi Y, Chen DY, Weber JD, Gutmann DH. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- 20.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch E, Cherwinski H, Peterson D, McMahon M. Mutations of critical amino acids affect the biological and biochemical properties of oncogenic A-Raf and Raf-1. Oncogene. 1997;15:1021–1033. doi: 10.1038/sj.onc.1201270. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Schepper S, Boucneau J, Lambert J, Messiaen L, Naeyaert JM. Pigment cell-related manifestations in neurofibromatosis type 1: an overview. Pigment Cell Res. 2005;18:13–24. doi: 10.1111/j.1600-0749.2004.00206.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–267. doi: 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- 33.Ramjaun AR, Downward J. Ras and phosphoinositide 3-kinase: partners in development and tumorigenesis. Cell Cycle. 2007;6:2902–2905. doi: 10.4161/cc.6.23.4996. [DOI] [PubMed] [Google Scholar]
- 34.Johannessen CM, Johnson BW, Williams SM, Chan AW, Reczek EE, Lynch RC, et al. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 35.Ji Z, Flaherty KT, Tsao H. Targeting the RAS pathway in melanoma. Trends Mol Med. 2012;18:27–35. doi: 10.1016/j.molmed.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Raedt T, Walton Z, Yecies JL, Li D, Chen Y, Malone CF, et al. Exploiting cancer cell vulnerabilities to develop a combination therapy for ras-driven tumors. Cancer Cell. 2011;20:400–413. doi: 10.1016/j.ccr.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28:85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin AF, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 42.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71:2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–454. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messiaen LM, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 52.Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, et al. Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer. 2006;45:265–276. doi: 10.1002/gcc.20289. [DOI] [PubMed] [Google Scholar]
- 53.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.