Introduction
Many cellular components involved in gene regulation exist at low levels. For example, a genetic regulatory circuit may consist of one or two DNA copies of a gene and a few tens of DNA-binding transcription factor molecules, with random production of relatively short-lived mRNA transcripts that are each a template for the translation of a handful of protein molecules [25,31,34,35,77,85,93]. While the number and temporal/spatial distribution of these molecules are often important for a cell to carry out specific functions and, consequently, determine cell fate, such low copy numbers inevitably lead to random fluctuations, or noise, in their production and regulation (reviewed in [3,15,42,43,60,72]). In addition, these molecules are often influenced by their unique cellular environments, giving rise to further levels of cell-to-cell variation [17,55,62,63,87]. A deep understanding of how cell-fate decisions are made amidst these inevitable variations sometimes require the ability to measure how gene regulation is executed at the single-molecule and single-cell levels. In a recent demonstration of this requirement, a single-molecule mechanism of stochastic switching of E. coli cells to a lactose-metabolizing state was only revealed by in vivo single-molecule experiments [13].
The recent emergence of new experimental tools employing sensitive fluorescence detection in vivo has made it possible to visualize various aspects of gene regulation at the single-molecule level in the native, intracellular context. In this review, we will first describe general considerations for in vivo, single-molecule fluorescence detection of DNA, mRNA and protein molecules involved in gene regulation. We will then give an overview of the rapidly evolving suite of molecular tools available for observing gene regulation in vivo and discuss new insights they have brought into gene regulation. We focus mostly on experiments and methods in prokaryotic systems because many of the methods were first developed in bacterial cells, but we also touch upon on some single-molecule methods that are commonly used in higher organisms.
Considerations for achieving single-molecule detection in vivo
The principal challenge in visualizing single biomolecules in vivo lies in generating a signal above the background of cellular autofluorescence. Background fluorescence from unbound and non-specifically bound fluorescent probes poses an additional complication. Generally, single-molecule methods achieve a sufficient signal-to-background ratio by combining several strategies: optimizing excitation and detection, choosing fluorophores with appropriate photochemical properties and improving fluorescent signals.
Optimizing excitation and detection
Most single-molecule fluorescence experiments—especially those in living cells—require higher-power excitation sources and detectors with higher sensitivity and lower noise than those needed for ensemble fluorescence imaging. Requirements for excitation wavelength vary depending on the choice of the fluorophore, but generally fall in the visible range between 450 and 700 nm, with the most commonly used lines at 488, 514, 532, 561 and 632 nm. The illumination power density is usually around 0.5 kW/cm2 or higher for single molecule detection in vivo[69], which requires a total power output intensity of monochromatic light ~50 mW or higher (assuming that power loss in the optical pathway does not exceed ~50%). This is many times higher than the amount of power available in a ~40-nm band of visible light (i.e. a typical width for excitation filters) from mercury arc lamps and other excitation sources generally used for ensemble fluorescence imaging. Until recently, single-molecule experiments usually used large, expensive gas lasers and optically pumped dye lasers. Now, improvements in diode-pumped, solid-state lasers[53] (including new technologies such as optically pumped semiconductor lasers) have led to affordable lasers available at powers and wavelengths suitable for single-molecule imaging. Many microscope manufacturers now offer solid-state laser illumination options. Many user-friendly solutions, including Coherent's Obis and Cobolt's 04-01 series lasers, are now available commercially. Ideally, the excitation wavelength should match the peak of the fluorophore excitation spectrum; recent advances in supercontinuum lasers have made it possible to excite at any visible wavelength at powers high enough to detect single fluorescent protein molecules[74].
In addition to maximizing excitation, sensitive single-molecule imaging also requires the use of deep-cooled, electron-multiplying charge-coupled device (EM-CCD) cameras. These cameras have quantum efficiencies of ~90% in the visible light range (meaning that 9 in 10 photons incident on the CCD chip are detected) and are cooled to temperatures as low as –100° C to reduce thermal detector noise. EM-CCD cameras meeting these criteria that utilize the e2v CCD97 (with a 512 × 512 array of 16 μm × 16 μm pixels) or CCD201 (with a 1024 × 1024 array of 13 μm × 13 μm pixels) sensors are available from Roper, Andor, Hamamatsu and other vendors.
Using oil-immersion microscope objectives with high numerical apertures (>1.4) also improves detection efficiency. Large numerical apertures capture a larger fraction of emitted light and minimize cell background by reducing the depth of field. Background can also be reduced by using total internal reflection (TIR) microscopy, in which laser light is incident at an angle beyond the critical angle for total internal reflection and the sample is excited by an evanescent field that decays exponentially with increasing distance from the glass/cell interface[1]. However, TIR illumination generates an illumination field up to a few hundred nm from the glass/cell surface, while single-molecule studies of gene regulation generally require illumination of intracellular regions further away from the coverslip.
Simultaneous imaging of two fluorophores of different colors can be achieved by exciting both fluorophores and splitting the fluorescence image using a long-pass filter. This can be done using two EM-CCD cameras, or by projecting both images onto separate portions of a single EM-CCD sensor. A number of microscope add-ons that assist in image splitting, such as the Cairn Research OptoSplit and the Photometrics Dual Viewer, are commercially available. Multi-color experiments generally require multiple laser illumination sources and careful choices of fluorophores with non-overlapping emission spectra. However, recent experiments using single-wavelength[78] and supercontinuum[74] excitation have proven the possibility of multicolor imaging at the single-molecule level using only one light source.
Choosing appropriate fluorophores
Choosing an appropriate fluorophore is a critical component of any single-molecule fluorescence experiment. Ideal fluorophores have strong emission in the yellow–red region of the visible spectrum, minimizing the influence of cellular autofluorescence, which is dominated by flavins and generally strongest at blue–green wavelengths[6]. The most commonly used fluorophores in in vivo gene regulation studies are fluorescent proteins, which have a wide range of photochemical properties. Several recent reviews have summarized the variety of available fluorescent proteins[21,27,70,91]. For fluorescent proteins to be used in gene regulation studies, the following properties need to be considered: brightness, photostability, maturation rate, and suitability as a fusion protein. In many cases bright (high quantum yield) and photostable (low photobleaching quantum yield) fluorescent proteins are preferred so that the tagged molecule can be tracked for a long period of time. Some methods require fluorescent proteins that are bright enough to be detected, yet can rapidly photobleach in order to detect newly synthesized protein molecules[34,93]. A fast chromophore maturation rate is also important for probing gene regulation: the rates of transcription and translation are on the time scale of minutes, so a fluorescent protein molecule that only becomes fluorescent hours after it is produced will be poorly suited to follow the dynamics of gene expression and regulation. The fastest maturing fluorescent protein thus far is a yellow fluorescent protein derivative named Venus[52], which was reported to mature with a half life of a few minutes both in vitro and in live E. coli cells[34,52,93]. Finally, fluorescent protein reporters must efficiently fold and mature under the growth conditions of a specific experiment and must not perturb the system under study by aggregating or changing the properties of molecules to which they are fused.
Many gene expression reporters use fluorescent proteins because of the ease of genetic manipulation and the lack of any need for exogenous cofactors. However, fluorescent proteins are often less photostable and less bright than organic fluorophores. They are also larger than synthetic organic dyes and can promote aggregation[96]. Recent advances in specific labeling of proteins with synthetic fluorophores have made it possible to visualize the distribution and dynamics of a variety of macromolecules using super-resolution microscopy (see discussions below). In designing an experiment employing organic dyes, the permeability of organic fluorophores through the cell membrane must be considered. In addition, background from non-specifically bound dye molecules needs to be minimized to allow single molecule detection. In the sections below we will discuss some specific cases where organic dyes were used to probe various aspects of gene regulation.
Improving fluorescent signals
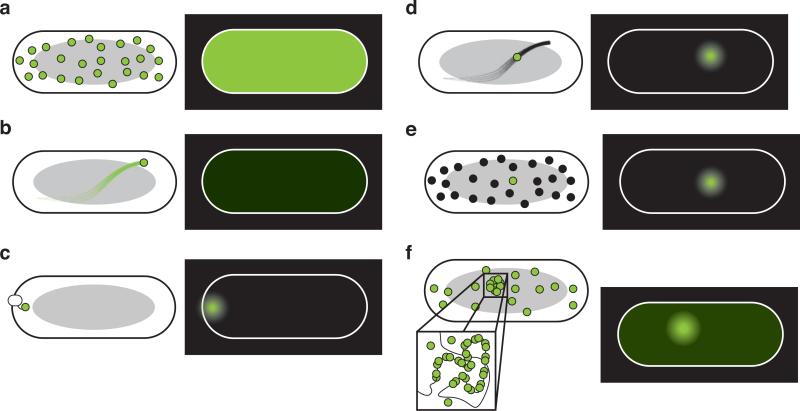
In general, fluorescent probes at high concentrations cannot be resolved with single-molecule resolution in living cells (Fig. 1A), and the signal from a diffusing fluorophore is usually undetectable under regular imaging conditions with an exposure time longer than a few milliseconds (one exception will be discussed below)[18,19] (Fig. 1B). This is because within the exposure time the fluorescence of a fast-diffusing molecule spreads across a large area and does not result in a clear, diffraction-limited fluorescent spot. However, a few strategies depicted in Figure 1 can be applied to improve the detection.
Figure 1.
Principles of detecting single biomolecules in living cells. Images in the left half of each part of the figure depict schemes for detecting fluorescent molecules; images to the right are mock, corresponding fluorescence images of rod-like bacterial cells (e.g. E. coli). Gray ovals indicate nucleoid. (a) High level of fluorescent probes. If many probes are present in the same cell and separated by less than the diffraction limit of light (~half the emission wavelength), single molecules cannot be distinguished and the whole cell appears bright. (b) Rapid diffusion of single-molecules. Here, a single fluorescent probe undergoes free diffusion in the cytoplasm. In the corresponding fluorescent image, the molecule's fluorescence is spread throughout the cell, resulting in a low level of fluorescence that is lower than the autofluorescent background. (c) Detection by localization. Single fluorescent probes can be detected if they are anchored to static or slowly diffusing structures (e.g. the cell membrane as shown here). (d) Stroboscopic illumination. Single, freely diffusing molecules can be detected if excitation light is briefly pulsed; here, a diffusing probe is illuminated by bright laser light for a fraction of an image frame and a diffraction limited fluorescent spot appears in the corresponding image. (e) Detection by stochastic activation. A single probe can be detected in a large population of probes if only one probe in a diffraction-limited area is fluorescent at a given time; this can be done using stochastic photoactivating or photoswitching probes. (f) Amplification using multiple probes. Single molecules can be detected if many probes bind one target molecule at many specific binding sites, creating a specific signal above the fluorescent background from nonspecifically bound and unbound probes. The use of large binding sites limits the spatial resolution of this technique.
First, if a fluorophore is anchored to a larger subcellular structure such as the cell membrane[34,93], an mRNA transcript[28,29] or chromosomal DNA[16,32], its signal is localized to a diffraction-limited spot that is easily detectable over background fluorescence (Fig. 1C). Second, single fluorescent probes expressed at very low levels can be detected without being anchored to a cellular structure by using stroboscopic illumination. In this illumination mode, the camera shutter is opened in the dark and an excitation laser is pulsed for only a fraction of the duration of the image frame (typically on the order of 1 ms). Because even freely diffusing, cytoplasmic molecules do not move very far in such a short period, molecular motion can be “frozen” in time, and hence a clear image of the molecule can be formed (Fig. 1D). Third, stochastic photoactivation or photoswitching methods such as Photoactiavated Localization Microscopy (PALM) and Stochastic Optical Reconstruction Microscopy (STORM) can be used to minimize the fluorescence background, so that even though the probes are present at high levels, only one molecule in a diffraction-limited area is fluorescent at one time. Therefore, single-molecule images can be obtained (Fig. 1E). Finally, fluorescent signals from single mRNA, DNA or protein molecules can be amplified by using many fluorophores [12,20,24,28,29,61,75]. In these methods, each single molecule of interest contains binding sites for multiple fluorophores, or directs the productions of multiple fluorophores; the aggregated signal from multiple probes is stronger than a single probe or the diffuse signal from unbound probes (Fig. 1F).
In vivo single-molecule methods for probing different molecular species in gene regulation
In this section we review currently available methods for probing various molecular species important for gene regulation. In particular, we focus on methods that enable single-molecule detection of DNA (genes), transcription factors, mRNA and protein molecules. We also touch upon recent work probing the dynamics of single RNA polymerase and ribosome molecules in living cells. Using these methods, valuable information has been obtained regarding transcription and translation and how transcription factors act to regulate gene expression.
Visualizing chromosomal DNA dynamics
Specific labeling of chromosomal loci using tandem arrays of DNA binding sites with fluorescent protein fusions
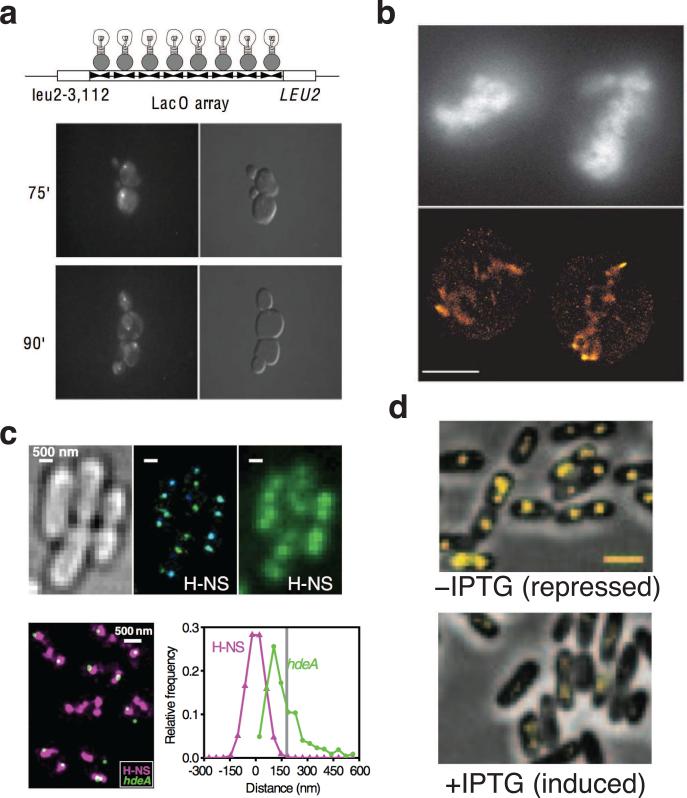
The earliest single-molecule studies of gene regulation in living cells observed the localization and dynamics of different chromosomal loci. This was accomplished by inserting hundreds of tandem transcription-factor binding sites such as lacO, tetO and parS into the chromosome and expressing fusion proteins containing both the corresponding transcription factor such as LacI, TetR and ParB and fluorescent protein moieties (Fig. 2A)[75]. By using many tandem binding sites, the signal from specifically bound fusion proteins is greater than the background fluorescence from unbound protein molecules (Fig. 1F). Hence, the labeled DNA segment can be easily detected as a distinct fluorescent spot. This method has been used to track the location, motion and large-scale movement of different chromosomal loci in C. crescentus [86] and E. coli cells [4,39,89]. It was also combined with the biochemical assay chromosome conformation capture (3C) to model the three-dimensional structure of the C. crescentus chromosome [81].
Figure 2.
Single-molecule imaging of chromosomal DNA and transcription factors. (a) The localization of homologous chromosomal loci was visualized in synchronized budding yeast cells (adapted from [9]). Top, an array of lacO binding sites was inserted into the LEU2 locus of the yeast chromosome; each half-site in the array can bind a GFP-LacI fusion protein (shown as a light bulb). Bottom, cells at 75 min show single, bright fluorescent spots; at 95 min, relatively dim fluorescent spots appear in both mother and daughter cells, indicating separation of sister chromatids (left, GFP fluorescence; right, DIC). (b) The distribution of chromosomal DNA in mitotic chicken cells was imaged with super-resolution using non-specifically bound, intercalating fluorescent probes (adapted from [3]). Scale bar 5 μm. (c) Top, the histone-like protein H-NS was imaged with super-resolution in E. coli cells (adapted from [10]) (left, brightfield; center, super-resolution reconstruction with spot color indicating z position from blue to green; right, widefield fluorescence image). Bottom, H-NS clusters co-localize with the hdeA gene which is regulated by H-NS (left, overlay of H-NS super-resolution image and an image of TetR-eYFP molecules bound to a 6×tetO array near the hdeA locus; right, distribution of hdeA spot positions relative to nearest H-NS clusters. (d) Strongly bound fusions of fluorescent protein and the Lac repressor were observed in the absence of the inducer IPTG, but only weak nucleoid-localized fluorescence was observed upon IPTG addition because of rapid diffusion of unbound and non-specifically bound fluorescent probes (adapted from [6]).
While the use of hundreds of tandem binding sites gives rise to a strong fluorescent signal against cellular background, the accuracy of this method in determining the position of a DNA segment is limited by diffraction, because the long tandem binding sites span several kilobases, exceeding the size of a diffraction-limited spot. A recent study overcame this problem by using only six tandem repeats and co-expressing a low-level of the corresponding TetR-eYFP fusion. Consequently, a spatial resolution of ~35 nm was achieved in detecting the position of the resulting fluorescence spots [88]. The position of the labeled gene was then analyzed against the cellular localization of the corresponding transcription factor to reveal the regulation of the gene by the transcription factor. Here, the low expression level of the fusion protein was key to achieve low cellular background so that distinct, diffraction-limited fluorescent spots could be detected.
Non-specific labeling of DNA using organic dyes
A second class of methods to probe the structure and dynamics of chromosomal DNA is to visualize the spatial distribution of DNA-binding dyes using single-molecule, super-resolution microscopy methods such as stochastic optical reconstruction microscopy (STORM[64]) and derivatives such as dSTORM[44]. Using these super-resolution methods, DNA can be non-specifically labeled with a DNA-binding organic dye. The fluorescence of the dye can reversibly transition between a fluorescent and metastable dark states; the dye may also reversibly bind to DNA, enhancing its fluorescence (Fig. 2B)[22]. Thus, not all dye molecules will fluoresce at the same time. By optimizing experimental conditions, the number of fluorescent dye molecules in a diffraction-limited area can be limited to only one at a time. The diffraction-limited fluorescence intensity profile of this single dye molecule can then be fit to a Gaussian function to extract its centroid position with a resolution on the order of 10 nm. The intercalating cyanine dye YOYO-1 was first used to image purified DNA in vitro with a resolution of ~40 nm[23]. Later another DNA major-groove-binding dye, PicoGreen, was used to map the organization of the E. coli chromosome both in fixed[67] and live[5] cells.
Visualizing DNA-binding protein dynamics
The spatial distribution and dynamics of a group of nucleoid-associated proteins, transcription factors and histone protein have been visualized using single-molecule methods. In two studies, the nucleoid-associated proteins, HU, H-NS, Fis, IHF, StpA in E. coli (Fig. 2C)[88] and HU in C. crescentus[40] were fused to the photoactivatable fluorescent protein mEos2[48] or photoswitchable fluorescent protein eYFP[8]. The intracellular distributions of these fusion proteins were then investigated using the super-resolution imaging method photoactivation localization microscopy (PALM[7]). Similar to the principle of STORM, the fluorescence of the photoactivatable or photoswitchable fluorescent protein molecules can be activated or switched on one at a time in a diffraction-limited area by using a low activation/switching laser intensity. The positions of individual molecules can then be determined with high accuracy and superimposed to generate a super-resolution image. It was found that most varieties of nucleoid-associated proteins have a largely random distribution in fixed E. coli and C. crescentus cells, but that the H-NS protein, a global transcription factor, forms clusters with an average size of ~160 nm that co-localize with genes known to be sequestered by H-NS (Fig. 2C)[88]. In another study, the histone protein H2B of mammalian cells was fused with E. coli dihydrofolate reductase (eDHFR), which binds non-covalently with a trimethoporin chemical tag (TMP) that is conjugated to a photoswitchable organic fluorophore ATTO655[90]. The membrane-permeable TMP-ATTO655 dye was introduced into cells by being included in the growth media. In this study, individual histone-core proteins spaced 100 nm were visualized and their dynamics were tracked in live cells using the dSTORM super-resolution imaging method.
Two studies investigated the kinetics of the lac repressor LacI binding to its specific DNA binding site lacO by fusing LacI to the yellow fluorescent protein Venus. Because non-specifically bound LacI-Venus molecules undergo rapid diffusion, only specifically bound LacI-Venus molecules will form distinct, diffraction-limited fluorescent spots. The key to this assay is that the expression level of LacI-Venus must be stringently controlled—the cellular fluorescence level from the non-specifically bound LacI-venus molecules needs to be sufficiently low to distinguish the specifically bound ones, which only have one or a few specific binding sites. Using this method, the authors monitored time-dependence of the fraction of cells showing distinct fluorescent spots, which is indicative of LacI-Venus molecules unbinding DNA after induction (Fig. 2D)[16,32] and successfully dissected the DNA searching mechanisms of LacI by comparing to in vitro investigations of the “hopping” and “sliding” mechanisms [9,68] and to theoretical studies on how macromolecular crowding in vivo influences this mobility[41].
Visualizing mRNA and dynamics of transcription
Probing mRNA expression and transportation in living cells
In some ways, visualizing mRNA at the single-molecule level is a simpler task than visualizing DNA. Its single-stranded structure allows one to use highly specific oligonucleotide probes, and many RNA folding motifs are recognized specifically by proteins with strong binding affinities. However, relatively short mRNA lifetimes (especially in prokaryotic cells) introduce technical difficulties in experiments; live-cell experiments must quickly and specifically label mRNA molecules with mature fluorescent molecules. Several methods have arisen to combat these challenges.
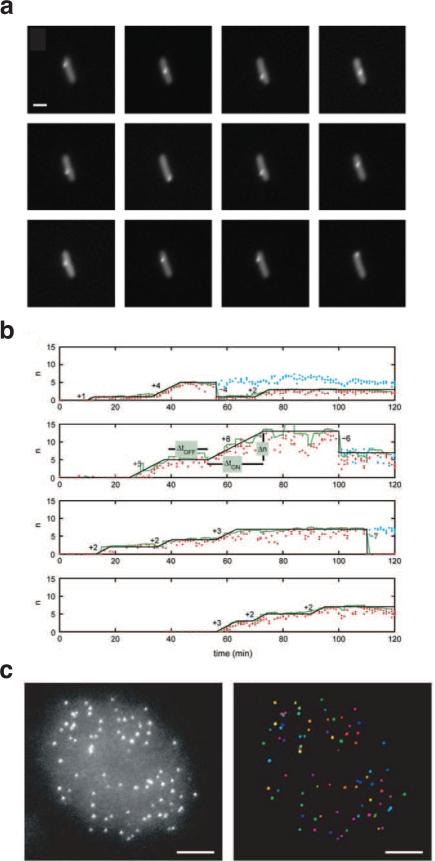
The earliest experiments visualizing mRNA in living cells utilized a similar principle to achieve signal over background as that used to localize chromosomal loci discussed above. By introducing a tandem array of MS2 binding motif sequences (specific RNA sequences bound by the phage MS2 coat protein) into an mRNA of interest and co-expressing an MS2-EGFP fusion protein, the localization and dynamics of mammalian mRNA were found to vary depending on an mRNA “zip code” sequence[24]. This method was also used to track mRNA within the nucleus[38] and its transportation through the nuclear pore complex[51]. In bacterial cells this method was employed to track mRNA localization dynamics(Fig. 3A)[28] and follow the production of single mRNA molecules in real time, which revealed that multiple mRNA molecules were produced in short transcriptional bursts(Fig. 3B)[29]. A recent experiment utilized the bacteriophage PP7 coat protein in a similar way to label nascent transcripts and observed the activity of single, elongating RNA polymerase molecules[38].
Figure 3.
Hybridization approaches to imaging single mRNA transcripts. (a) A single mRNA transcripts was tracked within a living E. coli cell for 6 min (scale bar 1 μm) by incorporating an array of 96 MS2 binding sequences into transcripts and co-expressing GFP-MS2 fusion proteins (adapted from [4]). (b) In an experiment similar to that in b, the appearance of fluorescent spots corresponding to mRNA transcripts bearing MS2 arrays was observed in growing E. coli colonies (adapted from [5]). Data from four single-cell time traces is shown here; red dots indicate the number of transcripts within one cell, determined from the time-averaged fluorescence intensities of spots within a cell (green lines); black lines indicate how this data was quantized by eye to follow transcription bursting in cell lineages, with burst duration (ΔtON), size (Δn) and time between bursts (ΔtOFF) indicated on the figure; blue dots show time traces in sister cells after cell division. (c) Transcripts were visualized in Chinese hamster ovary (CHO) cells as single, diffraction-limited spots using SM-FISH (left, flattened image of 3D image stack; scale bar 5 μm) that could be automatically detected as single molecules (right, individual mRNA transcripts randomly colored)(adapted from [8]).
Similar to the specific chromosomal DNA labeling method discussed above, the key to successful detection of single MS2-labeled mRNA molecules is to ensure that the fluorescent cellular background arising from unbound MS2 fusion fluorescent proteins is low. This is often achieved by fine-tuning the expression level of the fusion protein, which is not an easy task—especially in mammalian cells. To overcome the background problem, a new method was recently developed in which the mRNA of interested was engineered to have 96 repeat binding sites for a molecular beacon (reviewed in [10]). Molecular beacons are single-stranded, hairpin-shaped DNA or DNA analog molecules that have a fluorescent dye and a quencher conjugated at either end. Freely molecular beacons have minimal fluorescence because the quencher and the fluorescent dye are in close contact in the hairpin structure. Upon hybridization with a complementary mRNA strand, molecular-beacon molecules become fluorescent due to the opening of the hairpin. Using this method, the transport of single mRNA-protein (mRNAP) complexes from transcription sites to nuclear pores was tracked[83]. Note that in this assay molecular beacons must be microinjected into cells; so far, this is not possible in prokaryotic cells.
Two other methods, which are not yet at the single-molecule level, take different approaches to minimize fluorescence background. In one method, bi-molecular fluorescence complementation[26] is used to label mRNA transcripts. Here, RNA molecules containing an RNA aptamer recognized by the eukaryotic initiation factor eIF4a were imaged by co-expressing two fragments of eIF4a fused to two fragments of GFP[82]. The GFP chromophore only becomes fluorescent when both halves of eIF4a bind the RNA aptamer and the two GFP fragments form a complete fluorescent complex. GFP fragments not brought together by binding to the RNA aptamer are not fluorescent, minimizing cellular background. This study did not achieve sufficient brightness for single-molecule resolution, but another recent study demonstrated that a split GFP molecule can be detected at the single-molecule level in living cells[57]. One factor to consider for this method is that the slow maturation rate of the split GFP might limit how much dynamic information can be learned about mRNA synthesis.
In the second method, an RNA aptamer that binds specifically to a cell-permeable, GFP fluorophore-like dye was developed. The dye is not fluorescent in the absence of RNA-aptamer binding. Upon binding, the RNA aptamer mimics structural contacts of GFP to its encaged fluorophore to activate the dye's fluorescence. The resulting RNA-dye complex, termed Spinach, has a molar brightness of ~50% of eGFP[54]. One advantage of this method is that the RNA apatamer is much smaller in size (~100 bases) than the tandem ms2 sequence or 96 molecular beacon binding sites used in other studies, which could introduce less perturbation to native RNA structure and dynamics.
All the above methods require the engineering of native RNA sequences in order to introduce the binding site for a specific labeling fluorophore. The addition of these tags may change the structure and lifetime of the mRNA of study. A new method overcame this problem by targeting a multiply labeled tetravalent RNA imaging probe (MTRIP) to native mRNA sequences [66]. In this method, multiple (three on average) fluorophores (Cy3B or Atto647N) were conjugated to one oligonucleotide which has a 5′ biotin tag. Four of the oligonucleotides were then bound to streptavidin to form one MTRIP probe and delivered into cells via pores formed by a bacterial toxin; permeabilization was reversed by exchanging the media for one lacking toxin. Once inside the cell, these probes specifically recognized and hybridized with target mRNA molecules. Because each MTRIP probe contained on average 12 fluorophores, the fluorescence single was greatly enhanced and correspondingly single mRNA molecules could be detected.
Among the methods discussed above, the ones that using molecular beacons or oligonucleotides labeled with organic dyes are not amenable to prokaryotic systems, since it is difficult to deliver these probes into bacterial cells using microinjection. However, recent studies have shown that it may be possible to partially permeablize a bacterial cell by detergent treatment to get exogenous fluorescent probes into bacterial cells while minimally perturbing cell physiology[71].
Counting single mRNA molecules using fluorescence in situ hybridization (SM-FISH)
The SM-FISH method, which can only be performed on fixed cells, offers higher spatial resolution, labeling specificity and detection sensitivity than the live-cell mRNA hybridization assays[59]. In this method, cells are fixed, permeabilized and one or more short oligonucleotides each conjugated to a fluorescent dye are added to hybridize with complimentary target mRNAs (Fig. 3C). After hybridization, cells are extensively washed to minimize unbound probes. Detection of single mRNA molecules was first achieved using 5 probes complimentary to the β-actin transcript[20]. The number of oligonucleotide probes has ranged from one [77] to as many as 48 targeting non-overlapping sequences on the same transcript[59]. The design and synthesis of the latter is now commercially available from Biosearch Technologies. Recent work has shown that SM-FISH is applicable in numerous experimental systems including bacteria[73,94], yeast[76,95] mammalian tissue culture[61], tissue extracted from whole mice[30,37], C. elegans[49,65] and Drosophila[45] with very robust and reproducible results.
The use of multiple probes helps discriminate against non-specifically bound fluorescent probes[61], but single-probe SM-FISH measurements offer an easy way to quantify the copy numbers of mRNA molecules, because each single probe corresponds to one single mRNA molecule[77]. When multiple probes are used, the number of mRNA molecule in large eukaryotic cells is quantified by counting the number of separate fluorescent spots—in these large cells individual mRNA molecules are often dispersed and do not overlap. In small bacterial cells, when multiple probes are used, the number of mRNA transcripts is quantified by dividing the total fluorescence intensity of individual spot to the expected fluorescence intensity of a single transcript[73,94]. Note that the fluorescence intensity of a single transcript does not necessarily correspond to the expected fluorescence intensity of all probes added together, because frequently a transcript is only labeled at a fraction of all possible probe-binding sites. In our lab, working with E. coli cells, we found that ~30 non-overlapping probes in fairly stringent washing conditions reproducibly results an average of ~6 probes bound to each target mRNA. This enables reasonably quantized intensities for fluorescent spots corresponding to single or multiple mRNA molecules.
The SM-FISH method is often used to measure the copy number distributions of RNA molecules in fixed cells and to gain insight into underlying transcription mechanisms. Constitutive expression at a constant rate results in a Poisson distribution of mRNA copy numbers, while greater-than-Poisson variance could arise from various sources of biomolecular noise or pulsatile transcriptional mechanisms[79]. Pulsatile transcription, or transcriptional bursting, has been inferred from eukaryotic RNA distributions using SM-FISH for ribosomal RNA transcription in yeast[76] and mRNA transcription in mammalian cells[61], where it was found to likely depend upon local chromatin remodeling. One study in yeast used SM-FISH to find that transcription from some promoters proceeded at a constant rate while others were bursty[95]. Transcriptional bursting has also been found in E. coli using SM-FISH[73,94], but the molecular mechanism of bursting is unknown[50]. By changing the portion of a transcript targeted by hybridization or using two-color imaging, the SM-FISH method has also been applied to identify roles for noncoding RNA in yeast transcription regulation[11], study mRNA splicing[84] and investigate regulation of mRNA decay[80]. In these experiments, single transcripts in vivo were first identified as isolated fluorescent spots. Next, by choosing the correct combination of probe sequences and label colors, one can determine whether one or more sequences appear in a given transcript molecule. For example, a two-color experiment used differently colored SM-FISH probes to label the 5′ end of a transcript and the rest of the transcript, making it possible to distinguish between nascent and mature transcripts, which was required to estimate the rates of both transcription and transcript decay[80].
Recent developments are pushing the capabilities of SM-FISH. One study used a multicolor, combinatorial approach of SM-FISH to simultaneously measure the distribution of 32 different mRNA transcripts in a single cell[47]. In this approach, a set of probes with a unique combination of colors was designed against each target mRNA transcript. The identity of a transcript is then ascertained by measuring the relative intensity of different colors in a fluorescent spot. To further enhance this method's multiplex capabilities, photoswitchable dyes such as those used in super-resolution imaging method STORM were used to enable the identification of single mRNA molecules’ positions with ~20 nm resolution. In another study, SM-FISH was combined with other fluorescence methods to simultaneously visualize E. coli and C. crescentus mRNA transcripts (using SM-FISH) and their genetic locus (using an array of lacO sites and a fusion of a fluorescent protein and the lac repressor). The authors found that the mRNA was strongly co-localized with its encoding gene[46]. This implied that, for this gene, mRNA diffusion from the site of transcription occurred at a much lower rate than mRNA degradation.
Visualizing Protein expression and dynamics of translation
Probing translation dynamics
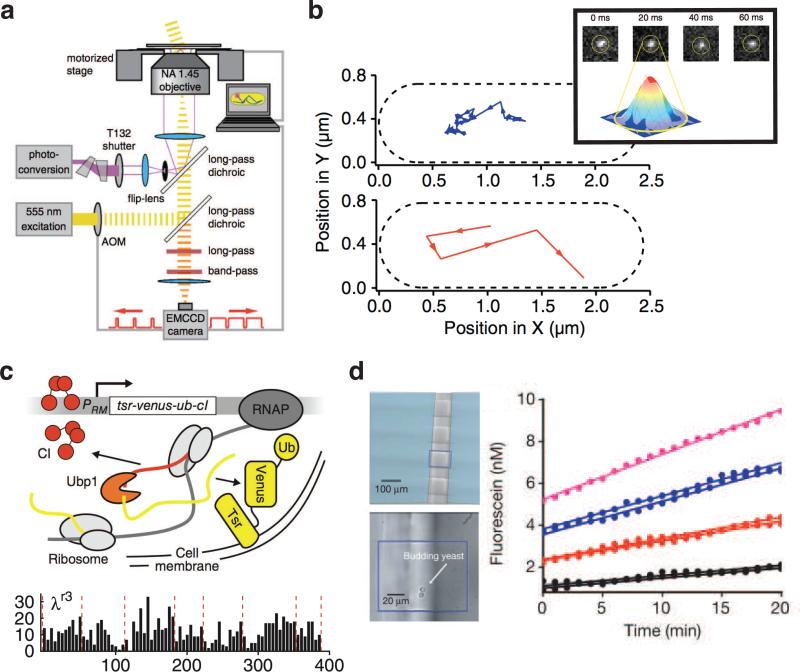
Single-molecule methods have been applied to study the cell's regulatory response to amino-acid starvation. The photoactivatable fluorescent protein Dendra2 was fused to RelA, an enzyme that catalyzes the production of a molecule that triggers physiological remodeling of bacteria in the stringent response[58]. The free diffusion of RelA molecules in cytoplasm was imaged using stroboscopic illumination (Fig. 1D and 4A). In stroboscopic illumination, the sample is illuminated for only a fraction of the total exposure time (on the order of 1 ms)[92]. Stroboscopic illumination “freezes” the molecule's action in time and a clear fluorescence image of the molecule can be obtained. Using this method the authors found that under normal conditions, RelA molecules were sequestered by the large ribosome complex and diffused at the same rate as the a ribosomal subunit, but upon starvation RelA was released into the cytoplasm and diffused at the same rate as free mEos2 (Fig. 4B)[18]. Here, using the photoactivatable Dendra2 and mEos2 as well as stroboscopic illumination were key: photoactivation allowed only one molecule at a time to be tracked with minimal fluorescent background while stroboscopic illumination enabled tracking freely diffusing protein molecules in the cytoplasm.
Figure 4.
Expression and dynamics of single protein molecules involved in gene regulation. (a) Illustration of stroboscopic illumination of single, photoactivated fluorescent probes. Probes are activated to become fluorescent by a UV beam. The EM-CCD camera directs short pulses of illumination light that last a fraction of the image frame time by triggering an acousto-optic modulator (AOM) that modulates the excitation beam (adapted from [2]). (b) The positions of single RelA protein molecules were tracked using stroboscopic illumination (adapted from [2]) and fitting fluorescent spots to a two-dimensional Gaussian function (top right). Molecules observed slowly in normal growth conditions (middle) and rapidly under conditions of amino-acid starvation (bottom). (c) The production of single transcription factor molecules was following in growing cell lineages using the CoTrAC method (adapted from [7]). Top, in the CoTrAC method, a protein of interest such as the λ repressor CI (red) is expressed in translational fusion with a membrane-localized fluorescent probe (here, Tsr-Venus, yellow) with the Ubiquitin sequence inserted in between. The Ubiquitin hydrolase Ubp1 (orange) is co-expressed and co-translationally cleaves the nascent polypeptide chain so that CI molecules can be detected by counting single Venus probes without affecting transcription regulation by CI. Bottom, CoTrAC was used to create time traces of the number transcription molecules produced in every 5-min time interval in cell lineages; the x axis indicates elapsed time in min and the y axis indicates the number of protein molecules produced; dashed red lines indicate cell division times. (d) The quantized expression of β-galactosidase enzymes from one or a few cells was observed in in single microfabricated chambers (top left). Bottom left, budding yeast cell in one chamber. Right, the total fluorescence signal within a chamber increased linearly over time, with the slope corresponding to the number of active enzyme molecules in each chamber (adapted from [1]).
Very recently, spatial distributions of RNA polymerase and ribosome complexes were mapped in living E. coli cells using a superresolution imaging method [2]. The ribosomal subunit S2 was fused to eYFP and localized at the single-molecule level. This was done by first photobleaching the initial fluorescent population using strong laser illumination and then localizing/tracking individual YFP molecules that return to the fluorescent state either spontaneously or by UV activation [8,14]. Similarly, the β’ RNA polymerase subunit was fused to yGFP[33] and its spatial distribution was mapped. In this study, S2-YFP and β’-yGFP molecules were excited by 15-ms laser pulses during 30-ms imaging frames. The 15-ms laser pulse was sufficiently long so that only ribosomes and RNA polymerase complexes tethered to their template molecules were visualized. This is because protein complexes not bound to larger nucleic-acid templates generally diffuse too far within 15 ms to create a localized, diffraction-limited fluorescent spot. By comparing the spatial distributions of both ribosome and RNA polymerase with that of the nucleoid with high resolution, the authors found RNA polymerase molecules are distributed near cytoplasmic membrane throughout most of the cell but not at the cellular end caps, suggesting that “transertion”, the coupling of transcription, translation and membrane-protein insertion, can account for radial, but not axial, expansion of the nucleoid.
Probing protein production
Recently, the expression of single protein molecules in live bacterial cells was followed in real time. In these studies, a bright, fast-maturing yellow fluorescent protein Venus[52] was fused to the membrane-bound E. coli serine receptor Tsr (Fig. 4C)[34,77,93]. The membrane-bound Tsr sequence targets newly produced Venus molecules to the membrane; since the diffusion of these membrane-targeted molecules is greatly slowed, they can be easily detected as individual, diffraction-limited spots (Fig. 1C). Hence, the activity of the promoter driving Tsr-Venus fusion expression was followed in real time by counting the number of newly produced Tsr-Venus molecules on the membrane[93]. Once detected, fluorescent molecules were photobleached so that any fluorescent spots appearing in the subsequent image come from newly synthesized fluorescent protein molecules. Using this method, burst-like protein production was directly observed[34,93] and fluctuations in protein production were analyzed [77]. Note that the measurement of protein production, rather than concentration, is particularly important as it can be used to distinguish between possible mechanisms of gene expression that produce indistinguishable concentration distributions[36,56].
Probing autoregulation of transcription-factor without disrupting function
Studies employing direct fusions of fluorescent proteins to molecules of interest can be problematic, especially for transcription factors, which are often small, with both termini containing important functional surfaces for DNA-binding or multimerization. The single-molecule gene expression reporter Tsr-Venus must be targeted to membrane to achieve single-molecule detection sensitivity, which is not compatible with probing the production of transcription factors or proteins that requires specific, non-membrane localization. To overcome this problem, a new method called Co-Translational Activation by Cleavage (CoTrAC) was developed[34]. The Tsr-Venus reporter was expressed in fusion with a transcription factor (the bacteriophage λ repressor CI) with the Ubiquitin (Ub) sequence inserted in between to create a single polypeptide chain Tsr-Venus-Ub-CI (Fig. 4C)[34]. The Tsr-Venus-Ub chain was cleaved from CI by co-expressing the Ubiquitin hydrolase Ubp1, which specifically recognize the Ubiquitin sequence and cleaves at the last glycine residue of Ubiquitin, leaving a transcription factor with a native peptide sequence. In this way, the number of CI molecules expressed was measured by counting the number of membrane-bound Tsr-Venus-Ub molecules without CI autoregulatory functions being perturbed (Fig. 4C)[34].
Indirect, single-molecule observation of protein production
Lastly, it is possible to detect protein expression at the single-molecule level by detecting enzymatic activity of single expressed protein molecules. In one such study, β-galactosidase was used as the gene expression reporter, and its expression was detected at the single-molecule level by monitoring the hydrolysis of the fluorogenic substrate fluorescein-di-β-D-galactopyranoside in microfluidic chambers where single E. coli, yeast or embryonic mouse cells was confined (Fig. 4D)[12]. The rate of increase of the fluorescence level of the microfludic chamber was proportional to the number of active β-galactosidase enzyme molecules expressed from the cell within the chamber; with fluorescence-increase rates being quantized and evenly spaced for chambers with 1 to 3 enzyme molecules. Because the reaction product in this experiment diffuses out of the cell, the method is limited to studying a single cell per isolated well. The development of a reporter that can be retained inside the cell could make it possible to observe gene expression in growing cell colonies.
Future directions
Recent developments in single-molecule methods have provided a wealth of information regarding various aspects of gene regulation in vivo. Nevertheless, the field will benefit from further development in single-molecule fluorescent reporters. For one, in higher eukaryotic cells there is no single-molecule reporter of protein expression similar to the Tsr-Venus reporter in E. coli cells. Secondly, there is no fast-maturing, red fluorescent protein with comparable maturation kinetics to the yellow fluorescent protein Venus. Having such a fluorescent protein will enable simultaneous, single-molecule probing of two genes, offering great potential to reveal the dynamics of mutually regulatory genes. Finally, in vivo, single-molecule assays focus on the detection of individual transcription factor, mRNA and protein molecules, while studies directly probing dynamics of single RNA polymerase and ribosome complexes in living cells are much fewer. There are many unanswered questions that can potentially be addressed by modulating the activity of these enzymes before and during synthesis and observing single-molecule dynamics in vivo. Particularly, there is a wealth of single-molecule, in vitro experiments for both RNA polymerase and ribosome; it will be interesting to see the extent to which in vitro results are reproducible within a living cell. Ultimately, results from in vivo, single-molecule experiments will play important roles in determining how gene regulation robustly controls cellular function and cell fate in spite of the low copy numbers of genes, mRNA transcripts and transcription factors.
References
- 1.Ambrose EJ. A Surface Contact Microscope for the study of Cell Movements. 1956;178:1194–1194. doi: 10.1038/1781194a0. Published Online: 24 November 1956; | Doi:10.1038/1781194a0. [DOI] [PubMed] [Google Scholar]
- 2.Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Molecular Microbiology. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balázsi G, van Oudenaarden A, Collins JJ. Cellular Decision Making and Biological Noise: From Microbes to Mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benke A, Manley S. Live-Cell dSTORM of Cellular DNA Based on Direct DNA Labeling. ChemBioChem. 2012;13:298–301. doi: 10.1002/cbic.201100679. [DOI] [PubMed] [Google Scholar]
- 6.Benson RC, Meyer RA, Zaruba ME, McKhann GM. Cellular autofluorescence--is it due to flavins? J Histochem Cytochem. 1979;27:44–8. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 7.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nature Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, et al. Nonspecifically bound proteins spin while diffusing along DNA. Nature Structural & Molecular Biology. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Bogaard PTC, Tyagi S. Using Molecular Beacons to Study Dispersal of mRNPs from the Gene Locus. In: Hancock R, Walker JM, editors. The Nucleus. Humana Press; 2008. pp. 91–103. [DOI] [PubMed] [Google Scholar]
- 11.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, et al. Single-Cell Analysis Reveals that Noncoding RNAs Contribute to Clonal Heterogeneity by Modulating Transcription Factor Recruitment. Molecular Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 13.Choi PJ, Cai L, Frieda K, Xie XS. A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science. 2008;322:442–6. doi: 10.1126/science.1161427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–8. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 15.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elf J, Li G-W, Xie XS. Probing Transcription Factor Dynamics at the Single-Molecule Level in a Living Cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic Gene Expression in a Single Cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 18.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. PNAS. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English BP, Sanamrad A, Tankov S, Hauryliuk V, Elf J. Tracking of individual freely diffusing fluorescent protein molecules in the bacterial cytoplasm. 2010. arXiv:1003.2110.
- 20.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–90. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–43. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 22.Flors C. DNA and chromatin imaging with super-resolution fluorescence microscopy based on single-molecule localization. Biopolymers. 2011;95:290–297. doi: 10.1002/bip.21574. [DOI] [PubMed] [Google Scholar]
- 23.Flors C, Ravarani CNJ, Dryden DTF. Super-Resolution Imaging of DNA Labelled with Intercalating Dyes. ChemPhysChem. 2009;10:2201–2204. doi: 10.1002/cphc.200900384. [DOI] [PubMed] [Google Scholar]
- 24.Fusco D, Bertrand E, Singer RH. Imaging of single mRNAs in the cytoplasm of living cells. Prog Mol Subcell Biol. 2004;35:135–50. doi: 10.1007/978-3-540-74266-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh I, Hamilton AD, Regan L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J. Am. Chem. Soc. 2000;122:5658–5659. [Google Scholar]
- 27.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 28.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. PNAS. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-Time Kinetics of Gene Activity in Individual Bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 30.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 Cooperatively Determine the Mammary Stem Cell State. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guptasarma P. Does replication-induced transcription regulate synthesis of the myriad low copy number proteins of Escherichia coli? Bioessays. 1995;17:987–97. doi: 10.1002/bies.950171112. [DOI] [PubMed] [Google Scholar]
- 32.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac Repressor Displays Facilitated Diffusion in Living Cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 33.Hansen FG, Atlung T. YGFP: a spectral variant of GFP. BioTechniques. 2011;50:411–412. doi: 10.2144/000113691. [DOI] [PubMed] [Google Scholar]
- 34.Hensel Z, Feng H, Han B, Hatem C, Wang J, Xiao J. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nature Structural & Molecular Biology. 2012;19:797–802. doi: 10.1038/nsmb.2336. [DOI] [PubMed] [Google Scholar]
- 35.Holland MJ. Transcript abundance in yeast varies over six orders of magnitude. J Biol Chem. 2002;277:14363–6. doi: 10.1074/jbc.C200101200. [DOI] [PubMed] [Google Scholar]
- 36.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat. Genet. 2011;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-Time Observation of Transcription Initiation and Elongation on an Endogenous Yeast Gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–43. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee SF, Thompson MA, Schwartz MA, Shapiro L, Moerner WE. Super-Resolution Imaging of the Nucleoid-Associated Protein HU in Caulobacter crescentus. Biophysical Journal. 2011;100:L31–L33. doi: 10.1016/j.bpj.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G-W, Berg OG, Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nature Physics. 2009;5:294–297. [Google Scholar]
- 42.Li G-W, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 44.van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nature Protocols. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 45.Little SC, Tkačik G, Kneeland TB, Wieschaus EF, Gregor T. The Formation of the Bicoid Morphogen Gradient Requires Protein Movement from Anteriorly Localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llopis PM, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, et al. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature Methods. 2012 doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–3. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Middelkoop TC, Williams L, Yang P-T, Luchtenberg J, Betist MC, Ji N, et al. The thrombospondin repeat containing protein MIG-21 controls a left–right asymmetric Wnt signaling response in migrating C. elegans neuroblasts. Developmental Biology. 2012;361:338–348. doi: 10.1016/j.ydbio.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 50.Mitarai N, Dodd IB, Crooks MT, Sneppen K. The generation of promoter-mediated transcriptional noise in bacteria. PLoS Comput Biol. 2008;4:e1000109. doi: 10.1371/journal.pcbi.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nature Cell Biology. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 52.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 53.Ostermayer FW, Allen RB, Dierschke EG. Room Temperature cw Operation of a GaAs1−xPx Diode Pumped YAG:Nd Laser. Applied Physics Letters. 1971;19:289–292. [Google Scholar]
- 54.Paige JS, Wu KY, Jaffrey SR. RNA Mimics of Green Fluorescent Protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedraza JM, van Oudenaarden A. Noise Propagation in Gene Networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 56.Pedraza JM, Paulsson J. Effects of Molecular Memory and Bursting on Fluctuations in Gene Expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- 57.Pinaud F, Dahan M. Targeting and imaging single biomolecules in living cells by complementation-activated light microscopy with split-fluorescent proteins. PNAS. 2011;108:E201–E210. doi: 10.1073/pnas.1101929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potrykus K, Cashel M. (p)ppGpp: Still Magical?*. Annual Review of Microbiology. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 59.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raser JM, O'Shea EK. Control of Stochasticity in Eukaryotic Gene Expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene Regulation at the Single-Cell Level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 64.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saffer AM, Kim DH, van Oudenaarden A, Horvitz HR. The Caenorhabditis elegans Synthetic Multivulva Genes Prevent Ras Pathway Activation by Tightly Repressing Global Ectopic Expression of lin-3 EGF. PLoS Genet. 2011;7:e1002418. doi: 10.1371/journal.pgen.1002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santangelo P, Nitin N, Bao G. Nanostructured Probes for RNA Detection in Living Cells. Annals of Biomedical Engineering. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 67.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-Activated Localization Microscopy of DNA Structures. Nano Lett. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 68.Schonhoft JD, Stivers JT. Timing facilitated site transfer of an enzyme on DNA. Nature Chemical Biology. 2012;8:205–210. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Selvin PR, Ha T. Single-molecule Techniques: A Laboratory Manual. CSHL Press; 2008. [Google Scholar]
- 70.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 71.Silverman AP, Kool ET. Quenched autoligation probes allow discrimination of live bacterial species by single nucleotide differences in rRNA. Nucl. Acids Res. 2005;33:4978–4986. doi: 10.1093/nar/gki814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nature Reviews Molecular Cell Biology. 2011;12:119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- 73.So L, Ghosh A, Zong C, Sepulveda LA, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stephen LZ-D, Webb ED. Multicolour single molecule imaging on cells using a supercontinuum source. Biomedical Optics Express. 2012;3:400–6. doi: 10.1364/BOE.3.000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Current Biology. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 76.Tan RZ, van Oudenaarden A. Transcript counting in single cells reveals dynamics of rDNA transcription. Mol. Syst. Biol. 2010;6:358. doi: 10.1038/msb.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, et al. Quantifying E. coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Testa I, Wurm CA, Medda R, Rothermel E, von Middendorf C, Fölling J, et al. Multicolor Fluorescence Nanoscopy in Fixed and Living Cells by Exciting Conventional Fluorophores with a Single Wavelength. Biophysical Journal. 2010;99:2686–2694. doi: 10.1016/j.bpj.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trcek T, Larson DR, Moldón A, Query CC, Singer RH. Single-Molecule mRNA Decay Measurements Reveal Promoter-Regulated mRNA Stability in Yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Umbarger MA, Toro E, Wright MA, Porreca GJ, Baù D, Hong S-H, et al. The Three-Dimensional Architecture of a Bacterial Genome and Its Alteration by Genetic Perturbation. Molecular Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nature Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 83.Vargas DY, Raj A, Marras SAE, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SAE, et al. Single-Molecule Imaging of Transcriptionally Coupled and Uncoupled Splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–51. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 86.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. PNAS. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–864. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Li G-W, Chen C, Xie XS, Zhuang X. Chromosome Organization by a Nucleoid-Associated Protein in Live Bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. PNAS. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, Heilemann M, Cornish VW, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nature Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 91.Wu B, Piatkevich KD, Lionnet T, Singer RH, Verkhusha VV. Modern fluorescent proteins and imaging technologies to study gene expression, nuclear localization, and dynamics. Current Opinion in Cell Biology. 2011;23:310–317. doi: 10.1016/j.ceb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys. 2008;37:417–44. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 93.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing Gene Expression in Live Cells, One Protein Molecule at a Time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 94.Zeng L, Skinner SO, Zong C, Sippy J, Feiss M, Golding I. Decision Making at a Subcellular Level Determines the Outcome of Bacteriophage Infection. Cell. 2010;141:682–691. doi: 10.1016/j.cell.2010.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 1.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 2.English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. PNAS. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flors C. DNA and chromatin imaging with super-resolution fluorescence microscopy based on single-molecule localization. Biopolymers. 2011;95:290–297. doi: 10.1002/bip.21574. [DOI] [PubMed] [Google Scholar]
- 4.Golding I, Cox EC. RNA dynamics in live Escherichia coli cells. Proc Natl Acad Sci U S A. 2004;101:11310–5. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-Time Kinetics of Gene Activity in Individual Bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Hammar P, Leroy P, Mahmutovic A, Marklund EG, Berg OG, Elf J. The lac Repressor Displays Facilitated Diffusion in Living Cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 7.Hensel Z, Feng H, Han B, Hatem C, Wang J, Xiao J. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nature Structural & Molecular Biology. 2012 doi: 10.1038/nsmb.2336. [DOI] [PubMed] [Google Scholar]
- 8.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Current Biology. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Li G-W, Chen C, Xie XS, Zhuang X. Chromosome Organization by a Nucleoid-Associated Protein in Live Bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]