SUMMARY
SAMHD1 has recently been identified as an HIV-1 restriction factor operating in myeloid cells. As a countermeasure, the Vpx accessory protein from HIV-2 and certain lineages of SIV have evolved to antagonize SAMHD1 by inducing its ubiquitin-proteasome-dependent degradation. Here, we show that SAMHD1 experienced strong positive selection episodes during primate evolution that occurred in the Catarrhini ancestral branch prior to the separation between hominoids (gibbons and great apes) and Old World monkeys. The identification of SAMHD1 residues under positive selection led to mapping the Vpx-interaction domain of SAMHD1 to its C-terminal region. Importantly, we found that while SAMHD1 restriction activity toward HIV-1 is evolutionarily maintained, antagonism of SAMHD1 by Vpx is species-specific. The distinct evolutionary signature of SAMHD1 sheds light on the development of its antiviral specificity.
INTRODUCTION
Eukaryotic organisms have been exposed to viral infections for millions of years. This coevolutionary process has driven the development and adaptation of immune responses against invading viruses. In turn, viruses have evolved countermeasures to escape immune control. The human immunodeficiency virus (HIV) targets susceptible cells of the immune system that express the CD4 receptor and coreceptors (CCR5 or CXCR4) necessary for viral entry. However, certain cells of the immune system are nonpermissive to HIV-1 infection despite efficient entry. Indeed, dendritic cells (DCs) are largely refractory to HIV-1 infection (Coleman and Wu, 2009). This block to viral replication is linked to the presence of the cellular dominant negative factor SAMHD1 (Laguette et al., 2011). Three other cellular proteins have previously been shown to restrict HIV infection: tripartite motif 5 alpha (TRIM5α), APOBEC3G (apolipo-protein messenger RNA-editing enzyme catalytic polypeptide-like editing complex 3 [A3G]) and BST-2/tetherin (Neil et al., 2008; Sheehy et al., 2002; Stremlau et al., 2004). These restriction factors are all induced by interferon (IFN) treatment, act on discrete steps of the viral replication cycle, and are ubiquitously expressed in the organism (Douville and Hiscott, 2010). TRIM5α causes untimely uncoating after delivery of the viral core in the cytoplasm of target cells (Stremlau et al., 2006). A3G causes hypermutations of the viral genome, thus rendering it unstable, unable to integrate or to give rise to new functional viral particles (Harris et al., 2003). BST-2 tethers viral particles to the cell surface and therefore prevents viral budding (Neil et al., 2008). The precise step upon which SAMHD1 acts remains to be fully unraveled. Nonetheless, SAMHD1 silencing in DCs favors accumulation of full-length HIV-1 DNA, suggesting that it may affect reverse transcription (Laguette et al., 2011).
SAMHD1 belongs to a family of proteins that have been involved in a rare genetic disorder, the Aicardi-Goutières Syndrome (AGS) (Rice et al., 2009). Sequencing the genome of AGS patients revealed mutations in TREX1, RNASEH2A-C, and SAMHD1 (Crow et al., 2006a, 2006b; Crow and Rehwinkel, 2009). TREX1 and RNaseH2 both play important roles in nucleic acid metabolism (Mazur and Perrino, 1999), suggesting a similar mode of action for SAMHD1. Notwithstanding the involvement of these proteins in similar pathways, their impact on HIV replication is divergent. TREX1 and RNASH2 are both facilitating factors (Genovesio et al., 2011; Stetson et al., 2008; Yan et al., 2010), whereas SAMHD1 is a bona fide restriction factor (Hrecka et al., 2011; Laguette et al., 2011). AGS patients experience increased leukocytosis and IFN plasma levels (Dale et al., 2010; Rice et al., 2009). As it is the case for the TRIM family of proteins, SAMHD1 appears to be at the crossroads of inflammation and restriction.
Genetic conflict between hosts and lentiviruses leads to rapid selection of mutations that alter amino acid composition of both actors, especially at positions involved in protein-protein interaction (Emerman and Malik, 2010; Ortiz et al., 2009). This process of positive selection characterizes A3G interaction with Vif (Sawyer et al., 2004), TRIM5α interaction with CA (Sawyer et al., 2005), and BST-2 interaction with Vpu, Nef, or Env (McNatt et al., 2009; Sauter et al., 2009). While HIV-1 has no means to counteract SAMHD1 restriction, HIV-2 and certain SIV strains encode the auxiliary protein Vpx that potently overcomes the block to viral replication constituted by SAMHD1 by promoting its degradation by the proteasome machinery (Hrecka et al., 2011; Laguette et al., 2011). Thus, SAMHD1 is expected to be in a genetic conflict with the lentiviral protein Vpx.
Here, we show that SAMHD1 experienced strong positive selection episodes during primate evolution. The identification of SAMHD1 residues under positive selection allowed mapping of the interaction domain with Vpx to the C-terminal region. Furthermore, we demonstrate that SAMHD1 proteins of apes, monkeys, and lemurs are all active against HIV-1, whereas Vpx degrades and antagonizes SAMHD1 in a species-specific manner.
RESULTS
SAMHD1 Evolved under Strong Selective Pressures that Occurred along the Catarrhini Ancestral Branch
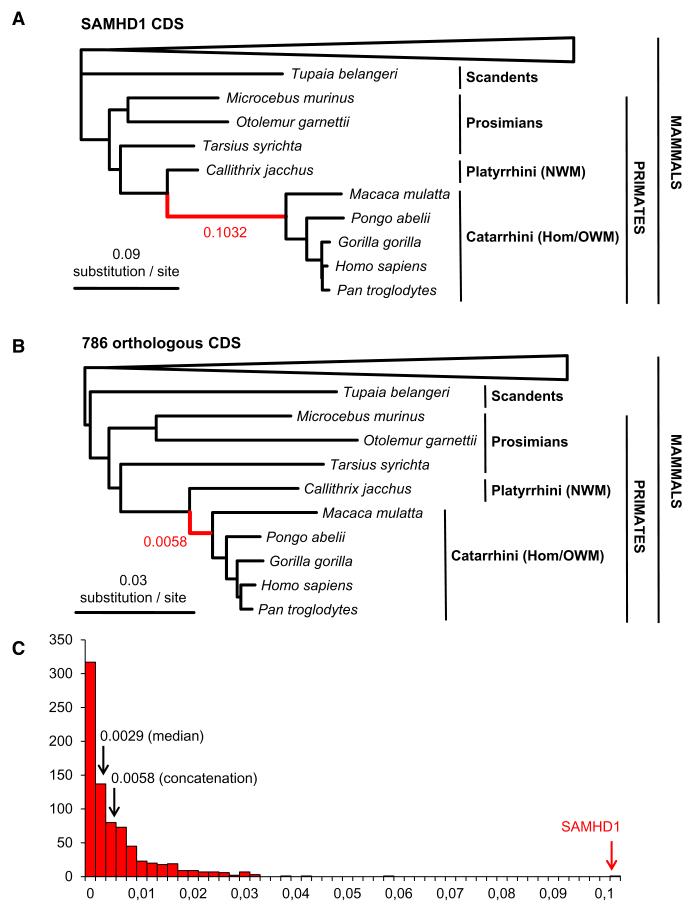
To determine whether selection pressure has shaped SAMHD1 evolution, we first constructed a phylogenetic tree using SAMHD1 coding sequences (CDSs) from 34 different mammals, including primates, available from ongoing sequencing projects and present in the database of Orthologous Mammalian Markers (OrthoMaM) (Ranwez et al., 2007). SAMHD1 showed an atypical gene tree relative to other 1:1 orthologous genes represented in the OrthoMaM database. Indeed, the maximum likelihood (ML) phylogenetic tree inferred from SAMHD1 amino acid sequences (Figure 1A) exhibited an exceptionally long ancestral branch for Catarrhini, including Old World monkeys (OWMs) and hominoids. This branch was more than 17 times as long in the SAMHD1 gene tree (0.1032 substitution/site) as in the ML phylogenetic tree (0.0058 substitution/site) inferred from the concatenation of the 786 1:1 orthologous CDS for which the same 34 mammalian taxa are available in OrthoMaM (Figure 1B). The distribution of the ancestral Catarrhini branch length across the same 786 ML gene trees confirms that SAMHD1 is an extreme outlier (Figure 1C and Figure S1 available online). The atypical branch length of the SAMHD1 gene tree underlines an exceptional accumulation of amino acid substitutions in this gene, suggesting the occurrence of an episode of adaptive evolution in the ancestral lineage leading to Catarrhini.
Figure 1. Characterization of the Atypical Catarrhini Ancestral Branch Length in the SAMHD1 Gene Tree.
(A) Amino acid branch lengths inferred for SAMHD1 sequences available for 34 mammals in the OrthoMaM database (Ranwez et al., 2007) as inferred by maximum likelihood under the LG+G+F model implemented in RAxML (Stamatakis, 2006). Only the Scandentia+primates sub-tree is presented. Note the extremely long ancestral branch (0.1032 substitutions per site) leading to Old World monkeys (OWMs, Catarrhini) represented in red.
(B) Amino acid branch lengths inferred from a concatenation of 786 1:1 orthologous CDSs available for the same 34 taxa in the OrthoMaM database. Note the much shorter Catarrhini ancestral branch (0.0058 substitutions per site) represented in red.
(C) Distribution of the ancestral Catarrhini amino-acid branch length inferred across the 786 1:1 orthologous gene trees showing that SAMHD1 is an extreme outlier with a branch length about 17 times longer than the one inferred from the concatenation.
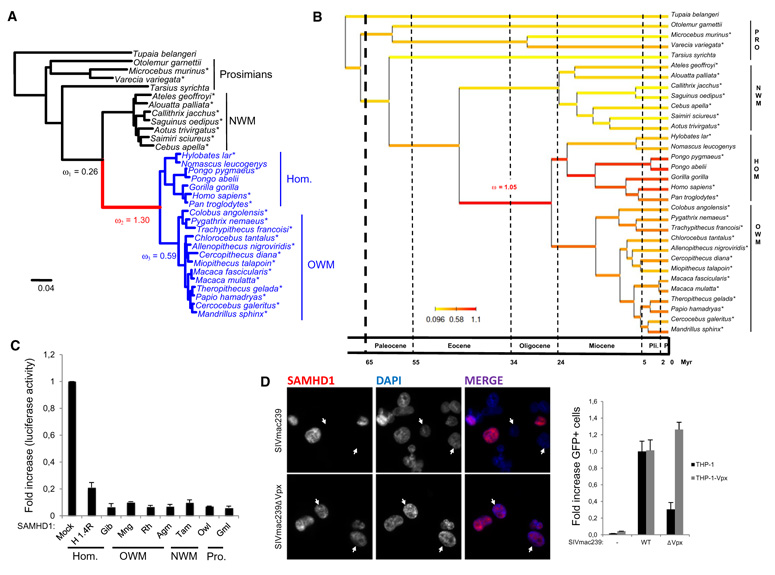
To test this hypothesis, we sequenced the complete SAMHD1 open reading frame (ORF; approximately 1880 bp) from 25 primate species (Table S1 and Figure S1). ML analysis of a 32-taxa data set allowed reconstruction of a well-resolved phylogeny of primates including seven New World Monkeys (NWMs, Platyrrhini) and 20 OWMs and hominoids (Catarrhini) (Figure 2A). The inferred phylogeny is almost fully compatible with the reference topology obtained from the most recent primate multigene phylogeny (Perelman et al., 2011), except for some difficulties to resolve phylogenetic relationships between short internal branches within OWMs and NWMs. Using the ML topology, we performed a number of statistical tests based on the ratio of nonsynonymous (dN) to synonymous (dS) substitutions (dN/dS) to define the selective pressures acting on the SAMHD1 molecule in primates (Table 1). Estimation of the global dN/dS ratio across the whole tree using the one-ratio branch model (M0) resulted in a ω value of 0.36, showing that SAMHD1 is globally under purifying selection in primates. However, hierarchical likelihood ratio tests (LRTs) performed between different branch models allowing ω to vary among branches of the phylogeny (Yang, 1998) reveals that strong positive selection occurred in the Catarrhini ancestral branch (Table 1). Indeed, the alternative two-ratio model (M2ω), that allows the single ancestral Catarrhini branch to have its own ω, results in a significant increase in likelihood over the one-ratio M0 model (p < 0.001). Under this two-ratio model, ω is estimated to be 1.27 along the ancestral branch leading to Catarrhini, indicating episodes of positive selection. Overall, the best-fitting branch model distinguishes three categories for ω distributed across the phylogeny. This model confirms that positive selection occurred along the Catarrhini ancestral branch (ω = 1.30), followed by a decrease in Old World monkeys and hominoids (ω = 0.59) whereas New World Monkeys and other primates present a dN/dS ratio characteristic of purifying selection (ω = 0.26) (Figure 2A). Such branch models nevertheless require a priori choice of the branches or group of branches of the phylogenetic tree among which ω might differ. To relax this assumption, we used a recently developed alternative approach that consists in an integrated Bayesian framework that allows joint reconstruction of variations in the molecular evolutionary process, including dN/dS ratios, life-history traits and divergence times (Lartillot and Poujol, 2011). We used this approach to reconstruct the variation of the dN/dS ratio in SAMHD1 along primate phylogeny while controlling divergence times and the effect of three life-history traits: body mass, longevity, and maturity (Figure 2B). The Bayesian reconstruction shows a clear increase of the dN/dS ratio in the Catarrhini ancestral branch length (mean dN/dS = 1.05) with elevated values persisting in Hominidae, especially in orangutans, while Platyrrhini show uniformly low dN/dS ratios. Moreover, the Bayesian chronogram infers that positive selection episodes experienced by SAMHD1 during primate evolution occurred approximately 45 to 25 million years ago, just before diversification of Catarrhini and their separation into hominoids and cercopithecoids.
Figure 2. Evolutionary Conservation of SAMHD1 Restriction Activity against HIV-1.
(A) SAMHD1 phylogenetic tree for 31 primate species rooted by the treeshrew Tupaia belangeri. The topology was inferred by maximum likelihood from the nucleotide sequences under a GTR+G model using RAxML. This topology is fully congruent with the most recent multigene phylogeny obtained by Perelman et al. (2011). Branch lengths are those inferred by Codeml under the best fitting branch model (Yang and Nielsen, 1998). Note the episode of positive selection along the Catarrhini ancestral branch (ω = 1.30), followed by a decrease in OWMs and hominids (ω = 0.59), whereas New World monkeys (NWMs, Platyrrhini) and other primates show a dN/dS characteristic of purifying selection (ω = 0.26). * indicates sequenced SAMHD1 in this study. Scale is in number of substitutions per codon. (B) The variation of ω in SAMHD1 along the primate phylogeny was jointly reconstructed with divergence times while controlling the effect of three life-history traits (body mass, longevity, and maturity) using the Bayesian framework recently proposed by Lartillot and Poujol (2011). The Bayesian chronogram obtained shows that the positive selection episode experienced by SAMHD1 during primate evolution occurred along the ancestral Catarrihini approximately between 45 and 25 million years ago. Time scale is expressed in million years with vertical lines delimitating the main geological periods (Pli., Pliocene; P., Pleistocene). (C) SAMHD1 expression was induced through retroviral transduction of SAMHD1-silenced THP-1 myeloid cells. Forty-eight hours after transduction, cells were differentiated and infected with a HIV-LUC-G. Luciferase activity was measured 24 hr after infection, normalized for protein levels and represented as relative infection compared to untransduced cells. Mock, untransduced; H1.4R, shRNA-resistant huSAMHD1; Gib, gibbon; Mng, mangabey; Rh, rhesus macaque; Agm, African green monkey; Tam, tamarin; Owl, owl monkey; Gml, gray mouse lemur; OWM, Old World monkeys; NWM, New World monkeys. (D) THP-1 and THP-1 stably expressing Vpx were differentiated on coverslips and infected with a SIVmac239-IRES-eGFP molecular clone with an IRES-eGFP sequence as a reporter (SIVmac239) or with the same virus with a stop codon in the vpx ORF (SIVmac239ΔVpx). Cells were analyzed by immunofluorescence 48 hr after infection for SAMHD1 expression. Nuclei were stained with DAPI. Cells were further analyzed by flow cytometry for the expression of GFP. Results are presented as the percent of GFP-positive cells, with the percent of GFP+ THP-1 cells infected with SIVmac239 set to 1. Error bars represent the standard deviation from the mean.
See also Figure S2.
Table 1.
Results of Likelihood Ratio Tests for Positive Selection in SAMHD1
| Hypotheses | LRT | |||
|---|---|---|---|---|
| Null Hypothesis | Alternative Hypothesis | −2ΔLnL | df | p Value |
| Branch Models | ||||
| M0: one-ratio model | M2ω: two-ratio modela | 20.46 | 1 | p < 0.001*** |
| M0: one-ratio model | M3ω: three-ratio modelb | 46.92 | 2 | p< 0.001*** |
| M0: one-ratio model | M4ω: four-ratio modelc | 48.76 | 3 | p< 0.001*** |
| M0: one-ratio model | M5ω: five-ratio modeld | 49.13 | 4 | p< 0.001*** |
| M2ω: two-ratio model | M3ω: three-ratio model | 26.46 | 1 | p< 0.001*** |
| M2ω: two-ratio model | M4ω: four-ratio model | 28.31 | 2 | p< 0.001*** |
| M2ω: two-ratio model | M5ω: five-ratio model | 28.67 | 3 | p< 0.001*** |
| M3ω: three-ratio model | M4ω: four-ratio model | 1.85 | 1 | ns |
| M3ω: three-ratio model | M5ω: five-ratio model | 2.21 | 2 | ns |
| M4ω: four-ratio model | M5ω: five-ratio model | 0.36 | 1 | ns |
| Branch-Site Models | ||||
| M1a (nearly neutral) | Model A (ω2 ≥ 1) | 3.26 | 1 | ns |
| Null model A(ω2 = 1) | Model A (ω2 ≥ 1) | 2.78 | 1 | p < 0.05* |
| Site Models | ||||
| M7: beta | M8: beta and ω | 40.41 | 2 | p < 0.001*** |
| M1a: nearly neutral | M2a: positive selection | 50.92 | 2 | p< 0.001*** |
highly significant;
significant; ns, not significant.
Ancestral Catarrhini branch versus all other branches.
Ancestral Catarrhini branch, Catarrhini versus all other branches.
Ancestral Catarrhini branch, Catarrhini, Platyrrhini versus all other branches.
Ancestral Catarrhini branch, Hominoidea, Cercopithecoidea, Platyrrhini versus all other branches.
These observations lead us to explore SAMHD1 restriction activity across evolution. For this purpose, we analyzed the anti-HIV-1 activity of a panel of SAMHD1 orthologs from hominoids, OWMs, NWMs, and prosimians (Figure 2C). SAMHD1-silenced THP-1 myeloid cells or U937 promyeloid cells—that express no detectable levels of endogenous SAMHD1—were transduced with retroviral vectors to express human (hu) short hairpin RNA (shRNA) resistant SAMHD1 (H1.4R) or a panel of primate SAMHD1. Cells were differentiated and subsequently infected with a vesicular stomatitis virus G protein pseudotyped HIV-1 molecular clone that harbors the luciferase gene in place of nef as a reporter—HIV-LUC-G. Measuring the luciferase activity shows that all tested SAMHD1 alleles were able to restrict HIV-1 infection (Figure 2C and Figure S2). Next, we asked whether SIVmac239 is restricted by huSAMHD1. We infected differentiated THP-1 with either a wild-type (WT) SIVmac239 molecular clone that possesses an IRES-eGFP sequence as a reporter or its ΔVpx counterpart. Infection with SIVmac239 causes a net decrease of SAMHD1 levels as observed by immunofluorescence, whereas infection with SIVmac239ΔVpx causes no significant variation in SAMHD1 levels (Figure 2D and Figures S2D and S2E). Moreover, infection of parental THP-1 cells or THP-1 cells stably expressing Vpxmac251 (THP-1-Vpx) with the same molecular clones of SIVmac239 shows, in a flow cytometry assay, that expression of Vpxmac251 restores SIVmac239ΔVpx infection to levels similar to those of SIVmac239 (Figure 2D). This suggests that optimal SIVmac239 infection of THP-1 cells requires the presence of a functional Vpx. Taken together, these experiments show that SAMHD1 restriction activity has been conserved throughout the evolutionary history of primates.
Identification of Positive Selected Sites Clustered in SAMHD1 C-Terminal Domain
In order to further characterize episodes of molecular adaptation that occurred in SAMHD1 and identify putative Vpx-interacting domains, we explored branch-site models for detection of positive selection on SAMHD1 domains. These models, that allow ω variation among sites in the protein and across branches on the tree, aim at detecting positive selection that affects a few sites along particular lineages (Yang and Nielsen, 2002). The branch-site tests 1 and 2 (Zhang et al., 2005) were applied to the Catarrhini ancestral branch, which was set as the foreground branch. The conservative branch-site test 1 was not significant, whereas the branch-site test 2 was significant (p < 0.05). Under branch-site model A, the empirical Bayes procedure did not identify positively selected sites with significant posterior probability along the Catarrhini ancestral branch (data not shown).
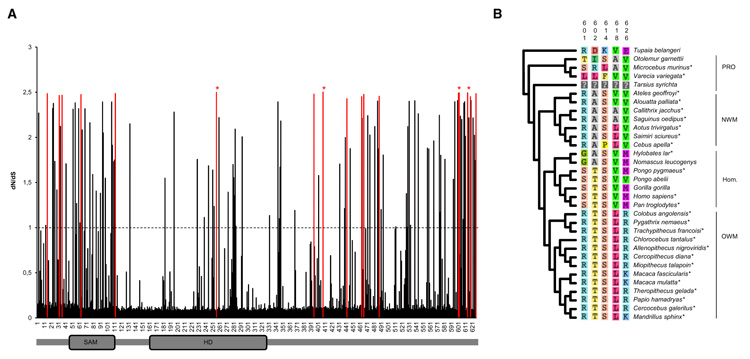
We investigated the among-site dN/dS ratio variation along the SAMHD1 molecule using site-specific codon models for detection of positively selected sites (Nielsen and Yang, 1998; Yang et al., 2000). Both LRT computed between nearly neutral models and models allowing for site-specific positive selection rejected the null hypothesis of nearly neutral evolution of SAMHD1 (p < 0.001) with the identification of sites under positive selection (Table 1). The site-specific selection profile of SAMHD1 inferred under the M8 model revealed 17 positively selected codons with a posterior probability of more than 0.95 (Figure 3A). The more conservative but less powerful M2a model identified four of these codons at positions 256, 408, 601, and 614 to be under positive selection. One of these codons (position 256) falls within the functional HD domain of the molecule. Strikingly, we identified a cluster of positively selected sites in the C-terminal part of the protein, comprising five sites (positions 601, 602, 614, 618, and 626) that localized within the last 25 amino acids of SAMHD1 (Figure 3B).
Figure 3. Site-Specific Positive Selection Profile of SAMHD1 in 31 Primates.
(A) The dN/dS ratio (ω) of the 626 codons of the human SAMHD1 sequence was inferred under the M8 site model (Nielsen and Yang, 1998) implemented in Codeml (Yang, 2007) from an alignment of 31 primates species with Tupaia belangeri as an outgroup. The dash line represents the dN/dS = 1 limit, above which codons are inferred to be under positive selection. The 17 codons shown in red are those identified as being under positive selection with posterior probability PP > 0.95 under M8 model. The four codons indicated by red stars are those identified using the more conservative M2a site model with PP > 0.95 (Zhang et al., 2005). The position of the two functional SAM and HD domains is indicated.
(B) Amino acids in the C-terminal domain of SAMHD1 that are subject to strong positive selection and their distribution among primates is represented.
See also Figure S3.
Vpx/SAMHD1 Interaction Is Mediated by the C-Terminal Domain of SAMHD1
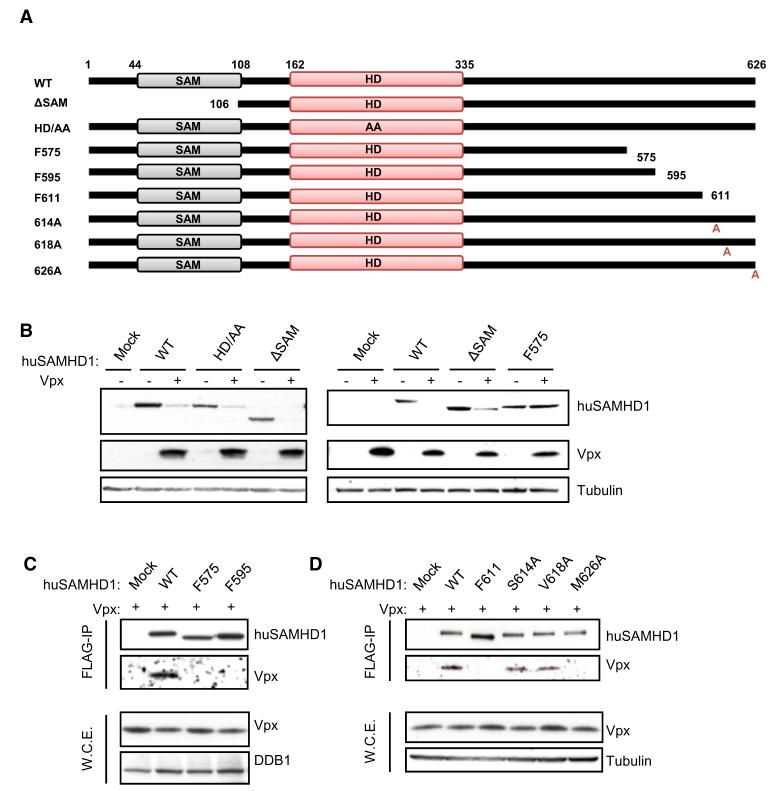
Based on the assumption that genetic conflict between viral proteins and restriction factors leads to the rapid fixation of mutations at the site of protein-protein interaction, we hypothesized that the C-terminal domain of SAMHD1 might be involved in interaction with Vpx. To test this hypothesis, we generated constructs where human FLAG-tagged wild-type SAMHD1 (WT-SAMHD1) is truncated for either the N-terminal SAM domain (ΔSAM) or portions of the C-terminal tail (SAMHD1-F575, SAMHD1-F595, SAMHD1-F611; Figure 4A). The mutant SAMHD1-HD/AA, that has no restriction activity toward HIV-1 (Laguette et al., 2011), was used as a control. Like WT-SAMHD1, all truncated SAMHD1 mutants localized to the nucleus as demonstrated by immunofluorescence in HeLa cells (Figure S4).
Figure 4. Interaction between Vpxmac251 and huSAMHD1 through the C-Terminal Domain of SAMHD1.
(A) Schematic representation of truncations and point mutants of huSAMHD1 that were engineered in the pOZ retroviral vector. All constructs were C-terminally FLAG-tagged. The limits of SAM and HD domains are indicated.
(B) Amino acids 575 to 626 of SAMHD1 are required for Vpxmac251-induced hSAMHD1 degradation. FLAG-tagged WT-SAMHD1, SAMHD1-HD/AA, SAMHD1-ΔSAM, and SAMHD1-F575 were transfected in 293T cells together or not with HA-tagged Vpxmac251. Whole-cell extracts were analyzed 48 hr later by immunoblot using anti-FLAG, anti-HA, and anti-tubulin antibodies.
(C) Amino acids 595 to 626 of huSAMHD1 are involved in the interaction between Vpxmac251 and huSAMHD1. Whole-cell extracts from 293T cells transfected to express HA-Vpxmac251 together with WT-SAMHD1, SAMHD1-F575, or SAMHD1-F595 were FLAG immunoprecipitated. After peptide elution, immunoprecipitates and whole-cell extracts were analyzed by immunoblot with anti-FLAG, anti-HA, and anti-DDB1 antibodies.
(D) FLAG-tagged WT-SAMHD1, SAMHD1-F611, SAMHD1-S614A, SAMHD1-V618A, and SAMHD1-M626A were FLAG immunoprecipitated from 293T cells expressing HA-Vpxmac251. Peptide-eluted immunoprecipitates were analyzed by western blot with anti-HA, anti-FLAG, and anti-tubulin antibodies.
See also Figure S4.
We tested the ability of HA-tagged Vpx from SIVmac251 (Vpxmac251) to degrade ΔSAM, HD/AA and F575 as compared to WT-SAMHD1, in 293T cells (Figure 4B). Vpxmac251 expression correlated with decreased levels of WT-SAMHD1, SAMHD1-ΔSAM, and SAMHD1-HD/AA, whereas no significant decrease of SAMHD1-F575 levels was observed. We next tested the ability of Vpxmac251 to interact with SAMHD1 mutants. FLAG-tagged WT-SAMHD1, SAMHD1-F575, SAMHD1-F595 were coexpressed with HA-tagged Vpxmac251 in 293T cells. FLAG immunoprecipitation of whole-cell extracts reveals the interaction of Vpxmac251 with WT-SAMHD1 whereas no interaction with SAMHD1-F575 and SAMHD1-F595 was detected (Figure 4C). This indicates that the interaction of SAMHD1 with Vpxmac251 is mediated by the last 31 amino acids of SAMHD1 that comprises amino acids 601, 602, 614, 618, and 626, which are under significant positive selection. To further map this interaction, we generated a fragment of SAMHD1 (SAMHD1-F611) that allows discriminating between amino acids 601/602 and 614/618/626. The fact that SAMHD1-F611 fails to interact with Vpx indicates that the last 11 residues of SAMHD1 (Figure 4D) comprising amino acids 614, 618, and 626 are involved in the SAMHD1/Vpx interaction. We next generated point mutants of SAMHD1 where amino acids S614, V618, and M626 are substituted by alanines and tested their ability to interact with Vpxmac251. Whole-cell extracts of 293T cells coexpressing FLAG-SAMHD1-S614A, -V618A, or -M626A together with HA-Vpxmac251 were subjected to FLAG immunoprecipitation. Analysis by immunoblot revealed that SAMHD1-S614A, SAMHD1-V618A, both interacted with Vpxmac251 at levels similar to WT-SAMHD1, whereas SAMHD1-M626A failed to interact with Vpxmac251 (Figure 4D). Taken together our results demonstrate that the C-terminal region of huSAMHD1 is required for its interaction with Vpxmac251 and is therefore critical for Vpxmac251-induced degradation of SAMHD1. We further map amino acid M626 of huSAMHD1 as required for this interaction.
Vpx-Induced huSAMHD1 Degradation Depends on Vpx/SAMHD1 Interaction and Involves Relocalization of SAMHD1 to the Cytoplasm
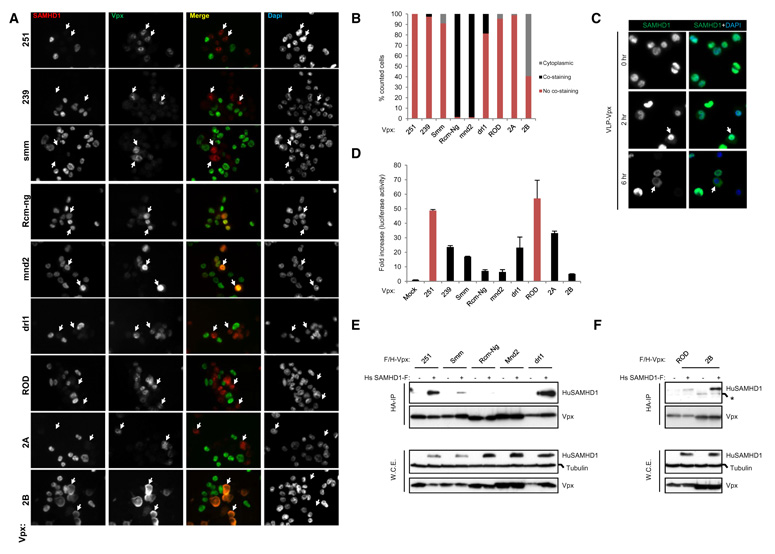
We previously observed that, when expressed in THP-1 cells, Vpx from SIVmac251, and HIV-2ROD (VpxROD) potently induced huSAMHD1 degradation whereas Vpx from SIVRcm-ng (VpxRcm-ng) failed to affect huSAMHD1 levels (Laguette et al., 2011). To extend this observation, we analyzed the ability of Vpx from additional HIV-2 and SIV strains to cause huSAMHD1 degradation in THP-1 cells, using Vpxmac251, VpxROD, and VpxRcm-ng as controls. In the tested panel of Vpx alleles, we included both alleles from laboratory-adapted lentiviral strains—Vpx from SIVmac239 (Vpxmac239), Vpxmac251, and VpxROD—and from “primary” viral strains—SIV sooty mangabey (VpxSmm), mandrill (Vpxmnd2), drill (Vpxdrl1), HIV-2 A (Vpx2A), and HIV-2 B (Vpx2B) (Figure 5). These alleles were all engineered to be FLAG and HA tagged and inserted in a retroviral vector allowing for transduction of proliferating THP-1 cells. Immunostaining of SAMHD1 and Vpx showed no significant costaining of Vpxmac251, Vpxmac239, VpxSmm, Vpxdrl1, VpxROD, and Vpx2A with huSAMHD1 (Figures 5A and 5B), suggesting a degradation of huSAMHD1 in the presence of these Vpx alleles. To the contrary, significant costaining between VpxRcm-Ng and Vpxmnd2 with huSAMHD1 was observed (Figure 5B), suggesting that no significant decrease of huSAMHD1 levels was induced by these Vpx alleles. Intriguingly, Vpx/huSAMHD1 costaining was also observed when Vpx2B was expressed, but this costaining was cytoplasmic (Figures 5A and 5B), indicating a relocalization of huSAMHD1 to the cytoplasm in the presence of Vpx2B. To investigate whether this cytoplasmic localization of SAMHD1 might be an intermediate step en route toward degradation, we treated differentiated THP-1 cells with pseudoparticles expressing Vpxmac251 (VLP-Vpx) and followed SAMHD1 staining over time by immunofluorescence (Figure 5C). Two hours after VLP-Vpx treatment, a decrease of huSAMHD1 staining could be observed, whereas 6 hr after VLP-Vpx treatment, the cells displayed a heterogeneous SAMHD1 staining pattern, including cells where no SAMHD1 signal could be detected and cells where the SAMHD1 staining was cytoplasmic. Additionally, Vpx was unable to degrade SAMHD1 when differentiated THP-1 cells were treated with leptomycin B an inhibitor of protein export (Figure S5). This strongly suggests that relocalization of SAMHD1 from the nucleus to the cytoplasm might be an intermediate step toward its final degradation.
Figure 5. Ability of Vpx Alleles from SIV and HIV-2 Strains to Degrade huSAMHD1.
(A) THP-1 cells were transduced with retroviral vectors allowing expression of FLAG- and HA-tagged Vpxmac251, Vpxmac239, VpxSmm, VpxRcm-Ng, Vpxmnd2, Vpxdrl1, VpxROD, Vpx2A, and Vpx2B. Forty-eight hours later, transduced cells were differentiated overnight on polylysine treated coverslips prior to immunostaining with anti-SAMHD1 and anti-HA antibodies. Nuclei are stained in mounting media with DAPI. Arrows point at examples of cells displaying a representative staining.
(B) Vpx-positive cells from (A) were counted. Results are expressed as the percent of Vpx-positive cells where Vpx/SAMHD1 costaining, Vpx/SAMHD1 exclusion and cytoplasmic SAMHD1 is observed.
(C) THP-1 cells differentiated on coverslips were treated with VLP-Vpx. Cells were fixed and stained with anti-SAMHD1 antibody at 0, 2, and 6 hr after VLP-Vpx exposure. Nuclei are stained in mounting media with DAPI. Arrows point at examples of cells displaying a representative staining.
(D) Cells treated as in (A) were infected with HIV-LUC-G. Luciferase activity was measured 24 hr after infection and is expressed as fold increase luciferase activity over untransduced parental THP-1 cells. Error bars represent the standard deviation from the mean.
(E) FLAG- and HA-tagged Vpx alleles from selected SIV strains were coexpressed with FLAG-tagged huSAMHD1 or empty vector in 293T cells. Whole-cell extracts were subjected to HA immunoprecipitation prior to peptide elution and analysis by immunoblot with anti-FLAG, anti-HA, and anti-tubulin antibodies.
(F) FLAG- and HA-tagged Vpx alleles from HIV-2ROD and HIV-2B strains were coexpressed with FLAG-tagged huSAMHD1 or empty vector in 293T cells. Whole-cell extracts were subjected to HA immunoprecipitation prior to peptide elution and analysis by immunoblot using anti-FLAG, anti-HA and anti-tubulin antibodies.
See also Figure S5.
We next analyzed the ability of these Vpx alleles to overcome huSAMHD1 restriction activity. Differentiated THP-1 cells were transduced so as to express Vpx alleles. Cells were infected with HIV-LUC-G. Luciferase activity shows a significant increase of infection by HIV-LUC-G after Vpxmac251, Vpxmac239, VpxSmm, Vpxdrl1, VpxROD, and Vpx2A expression (Figure 5D). In this context, VpxRcm-Ng, Vpxmnd2, and Vpx2B poorly affected huSAMHD1 restriction. These experiments show a correlation between Vpx ability to degrade huSAMHD1 and to overcome its restriction activity.
To further investigate the importance of Vpx/SAMHD1 interaction in this phenotype, we performed HA-immunoprecipitation on whole-cell extracts from 293T cells coexpressing FLAG- and HA-tagged Vpx and FLAG-huSAMHD1. We thereby show that Vpxmac251, VpxSmm, Vpxdrl1, VpxROD, and Vpx2B interact with huSAMHD1, whereas only weak interaction was observed with Vpx from SIVRcm-ng and SIVmnd2 (Figures 5E and 5F). Of note, the interaction of SAMHD1 with VpxSmm was weaker than with Vpxmac251. It should be noted however that Vpx2B which interacts with huSAMHD1 will ultimately induce its degradation and enhance HIV-1 infectivity but with slower kinetics compared to Vpx2A. Indeed, when Vpx2B-expressing THP-1 cells are harvested 96 hr after transduction, or when HeLa cells are transiently transfected with huSAMHD1 and Vpx2B, protein levels of huSAMHD1 are decreased at similar levels as when cells express VpxROD or Vpx2A (data not shown).
Species-Specific Degradation of Primate SAMHD1 by Vpx
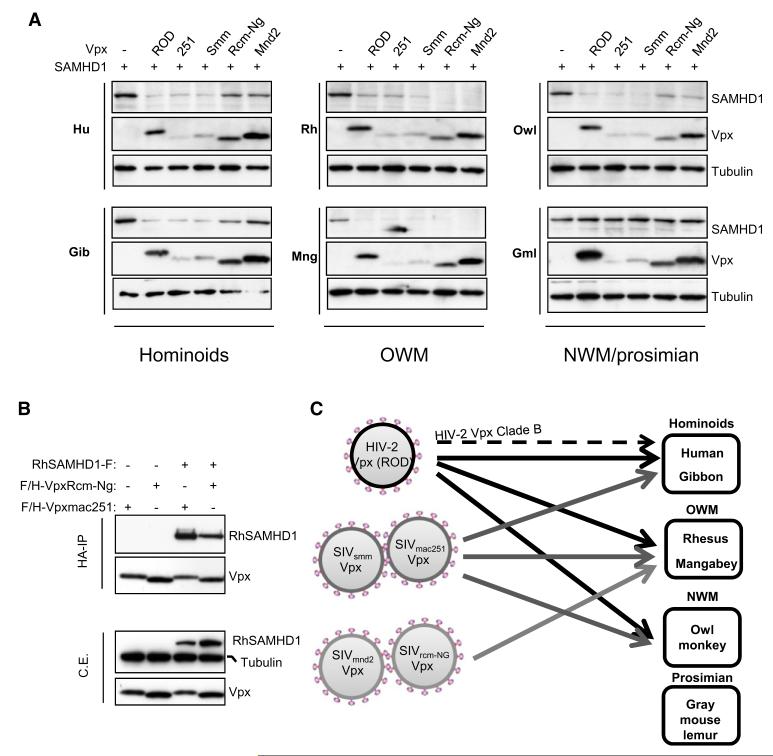
Given the differential activity of Vpx alleles to counteract huSAMHD1 restriction activity, we finally tested the ability of Vpx from different lentiviral strains to degrade SAMHD1 from various primates by cotransfection of Vpx and SAMHD1 expressing plasmids in HeLa cells (Figure 6A). In agreement with previous degradation assays performed in THP-1 cells (Laguette et al., 2011), human SAMHD1 was significantly degraded in the presence of VpxROD, Vpxmac251, and VpxSmm but not in the presence of VpxRcm-Ng and Vpxmnd2. A similar degradation pattern was observed for SAMHD1 from another hominoid, the gibbon, which is not naturally infected by SIV. Efficient degradation of rhesus macaque SAMHD1 was observed when coexpressed with Vpx from HIV-2ROD, and Vpx from two SIV strains able to infect rhesus macaques (SIVmac251 and SIVSmm). Additionally, protein levels of rhesus SAMHD1 were strongly decreased in the presence of Vpx from SIV strains infecting mangabeys (VpxRcm-ng) and mandrills (SIVmnd2). Consistently, Vpxmac251 and VpxRcm-Ng interacted with rhesus SAMHD1 in an immunoprecipitation assay (Figure 6B). Similar to rhesus SAMHD1, mangabey SAMHD1 was strongly degraded when coexpressed with all Vpx proteins, including VpxRcm-ng and Vpxmnd2. These results show that VpxRcm-Ng and Vpxmnd2 can induce the degradation of SAMHD1 proteins from Old World monkeys (rhesus and mangabeys) and not from hominoids (human and gibbon). Levels of SAMHD1 from the owl monkey, a NWM that has not been exposed to SIV, were significantly decreased in the presence of VpxROD, Vpxmac251, and VpxSmm. Interestingly, SAMHD1 from the gray mouse lemur, the most distant primate as compared to humans, was resistant to degradation in the presence of all tested Vpx alleles. These data highlight that Vpx alleles mediate the degradation of primate SAMHD1 in a species-specific manner (Figure 6C).
Figure 6. Vpx-Induced SAMHD1 Degradation Is Species Specific.
(A) SAMHD1 expression patterns in the presence of Vpx from HIV-2 and SIV strains. FLAG- and HA-tagged SAMHD1 proteins from a selected panel of primates were coexpressed with various FLAG- and HA-tagged Vpx proteins in HeLa cells. Whole-cell extracts were analyzed 30 hr after transfection by immunoblot with anti-HA and anti-tubulin antibodies. Hu, human; Gib, gibbon; Rh, rhesus macaque; Mng, mangabey; OWM, Old World monkeys; Owl, owl monkey; NWM, New World monkeys; Gml, gray mouse lemur.
(B) VpxRcm-Ng and Vpxmac251 interact with rhesus SAMHD1. FLAG- and HA-tagged Vpx alleles were coexpressed with FLAG-tagged rhesus SAMHD1 or empty vector in 293T cells. Whole-cell extracts were subjected to HA immunoprecipitation prior to peptide elution and analysis by immunoblot with anti-FLAG, anti-HA, and anti-tubulin antibodies.
(C) Schematic representation summarizing the data obtained showing that Vpx targeting of SAMHD1 is species specific. HIV-2 Clade B Vpx (discontinuous arrow) degrades huSAMHD1 but with slower kinetics compared to HIV-2ROD Vpx.
DISCUSSION
Evolutionary analysis of host restriction factors (Strebel et al., 2009) such as TRIM5α, APOBEC3G, and BST-2 highlight episodes of selective pressure during primate evolution that likely result from genetic conflict with retroviral elements (Ortiz et al., 2009; Patel et al., 2011). Likewise, by analyzing the molecular evolution of SAMHD1 in primates, and consistent with Lim et al. (2012), we present evidence that SAMHD1 was subjected to strong positive selection since catarrhines diverged from platyrrhines. Unlike the above-cited host restriction factors, the episodes of selection did not abrogate SAMHD1 restriction activity toward HIV-1, since SAMHD1 orthologs from very distant primates exhibit potent anti-HIV-1 activity similar to that of huSAMHD1. However, certain lentiviruses have evolved means of counteracting SAMHD1 restriction through the viral auxiliary protein Vpx. Here, we harnessed positive selection analysis as a tool to identify the C-terminal domain of huSAMHD1 as required for interaction with Vpxmac251 and subsequent targeting of SAMHD1 to proteasomal degradation. We further mapped the amino acid M626 of SAMHD1 as required for the Vpx/SAMHD1 interaction. Finally, we show that, in contrast to SAMHD1 broad range activity, the ability of Vpx to induce the degradation of primate SAMHD1 is species specific.
Inference of Catarhinni, Platyrhinni, Simiiformes, and primate ancestral SAMHD1 (Figure S3) illustrates the accumulation of amino acid changes along SAMHD1 sequence over 60 million years of evolution. Adaptation of the SAMHD1 sequence prior to diversification of the Catarrhini coincides with multiple events of invasion of the primate lineage by endogenous retroviruses such as HERV-K, HERV-H, HERV-E, HERV-W, and ERV-9 (Bannert and Kurth, 2006). In this perspective, it will be important to investigate whether endogenous retroviruses played a role in SAMHD1 adaptation and whether SAMHD1 may affect other retroviruses. In agreement, Vpx may alleviate a monocyte-specific restriction of murine leukemia virus infection (Jarrosson-Wuilleme et al., 2006; Kaushik et al., 2009). This notwithstanding, the accumulation of positively selected sites clustered in the N-terminal and C-terminal of the protein possibly results from lineage-specific adaptation of SAMHD1, after the split between hominoids and OWMs that occurred around 18 million years ago and may therefore result from genetic conflict between SAMHD1 and more recent viruses such as lentiviruses. Indeed, the most recent dating estimates the age of lentiviruses to a minimum of 12 million years (Katzourakis et al., 2007; Keckesova et al., 2009).
One of the hallmarks of positively selected restriction factors in primates is their species-specific antiretroviral activity (Mariani et al., 2003; McNatt et al., 2009; Song et al., 2005). However, we observed that primate SAMHD1 orthologs distant by 60 million years (divergence human/gray mouse lemur) were able to block HIV-1. Thus, despite selective pressure occurring over millions of years, SAMHD1 restrictive potential against HIV-1 appears to be evolutionarily conserved.
Genetic conflict between host restriction factors and viruses is predicted to lead to rapid selection of mutations that alter amino acid composition of both actors, especially at positions that affect protein-protein interaction. This type of genetic conflict has been shown to occur between APOBEC3G and Vif (Sawyer et al., 2004), TRIM5α and the viral capsid (Stremlau et al., 2005), BST-2 and Vpu, Nef, or Env (McNatt et al., 2009). In the case of APOBEC3G, TRIM5α, and BST-2, domains involved in direct interaction with retroviral factors are enriched in positively selected sites (Ortiz et al., 2009). Similarly, analysis of positive selection on SAMHD1 sequence reveals a cluster of sites in the C-terminal domain under selective pressure. Deletion of the C terminus of huSAMHD1 results in loss of interaction with Vpxmac251 and subsequent loss of SAMHD1 degradation. This suggests that Vpx may have dictated the evolution of the C-terminal domain of SAMHD1. Mutational analysis of the sites under selection in C terminus reveals that at least M626 of huSAMHD1 is important for the interaction with Vpxmac251. M626 is one of the few amino acids of SAMHD1 that vary in a lineage-specific manner in primates. Position 626 may be important for the interaction of Vpx with cognate SAMHD1, considering the species-specificity of Vpx-induced SAMHD1 degradation. However, both owl monkey and gray mouse lemur SAMHD1 harbor a V626, but only owl monkey SAMHD1 is efficiently degraded by Vpxmac251. This raises the possibility that position 626 is not the only site recognized by Vpx. Indeed, Lim et al. (2012) show that positively selected residues within the N-terminal domain of SAMHD1 determine both binding and susceptibility to Vpx. Additional traces of positive selection are seen in the N-terminal domain of SAMHD1, comprising the SAM domain predicted to mediate SAM/SAM, protein/protein, or protein/RNA interaction. It is therefore possible that SAMHD1 does not solely interact with Vpx and suggests that SAMHD1 may target another viral component to exert its restriction activity. Interestingly, position 256, within the HD domain, was also detected to be under positive selection. It will be of importance to determine whether this residue is involved in SAMHD1 function.
Additionally, we also show that Vpx2A and Vpx2B differ in their ability to degrade SAMHD1. Indeed, Vpx2B causes SAMHD1 degradation at a slower rate than Vpx2A. This feature allows for a mechanistic insight into Vpx-induced SAMHD1 degradation: Vpx seems to cause relocalization of SAMHD1 to the cytoplasm prior to routing toward the proteasome machinery. Since Vpx2B efficiently interacts with SAMHD1, one may speculate that the interaction of Vpx2B with the E3-ligase complex involved in SAMHD1 degradation and its export from the nucleus are less efficient than those of Vpx2A. The different clades of HIV-2 may have originated from independent cross-species transmission event (Lemey et al., 2003). Whether the differential kinetics of Vpx-mediated enhancement of DC infection after infection with HIV-2 from clades A or B affects pathogeny, remains to be explored.
Since the viral auxiliary protein Vpx is not shared by all lentiviruses, this raises concerns of the necessity to bypass this restriction for efficient viral spread. In particular, pandemic HIV-1 strains have not evolved means to induce SAMHD1 degradation, whereas nonpandemic HIV-2 induces SAMHD1 degradation through its Vpx. A consequence of HIV-1 not possessing Vpx is the inability of this virus to infect DCs, particularly at an early stage of viral transmission when viral loads are very low. Whether DCs from HIV-2 patients are productively infected in the context of a natural infection is unknown. Interestingly, phylogenetic studies have traced the origin of pandemic and nonpandemic HIV-1 (group M and groups N, O, and P, respectively) infections to cross-species transmissions from chimpanzees and gorillas (Keele et al., 2006). SIVcpz has been shown to originate from a probable recombination of lentiviruses from red-capped mangabeys (SIVRcm) and greater spot-nosed monkeys (SIVgsn) (Bailes et al., 2003). In this process, it appears that the vpx gene of SIVRcm was lost. Infection by SIVcpz in chimpanzees causes symptoms closely related to AIDS (Keele et al., 2009). Whether this ORF was lost before the transmission to chimpanzees is unknown. This observation is to be examined in the light of previous reports hypothesizing that vpx would have originated from gene duplication of an ancestral vpr gene or by acquisition of a heterolougous vpr allele (Tristem et al., 1992; Tristem et al., 1990). HIV-2 infection originated from an independent cross-species transmission of SIVSmm from Sooty mangabeys (Hirsch et al., 1989; Wertheim and Worobey, 2009) that possess a vpx gene. One may question whether the presence of Vpx may have represented an advantage favoring the cross-species transmission. However, HIV-2 infection is characterized by its nonpandemicity and the decline in its prevalence as compared to the increasing prevalence of HIV-1 (Tebit and Arts, 2011). Nonetheless, Vpx seems to be dispensable for persistence and spread in humans. The restriction factor SAMHD1 appears to bridge anti-viral responses and innate immune responses. The interplay between SAMHD1 and Vpx reflects how the immune system adapts to the reciprocal constraints exerted by viruses and the host, especially in the light of restriction factors being involved in the triggering of innate immune responses.
EXPERIMENTAL PROCEDURES
Evolutionary Analyses and Ancestral Sequence Reconstruction
The CDS amino acid alignment containing the 34 sequences available for SAMHD1 was retrieved from the OrthoMaM database (Ranwez et al., 2007). Other 786 1:1 orthologous CDS amino acid alignments for which the same 34 taxa were available were also downloaded from OrthoMaM. Ambiguously aligned sites were excluded using Gblocks default parameters (Castresana, 2000). Maximum likelihood branch lengths were then optimized for each alignment by using maximum likelihood under the LG+G8+F model on a fixed topology with PhyML 3 (Guindon et al., 2010). The same branch length optimization protocol was applied to the concatenation of the 786 genes but using RAxML v7.2.8 (Stamatakis, 2006).
The 32-taxa data set was constructed from the 25 newly obtained SAMHD1 sequences and seven sequences retrieved from EnsEMBL (Table S1). The alignment length was restricted to the length of the human SAMHD1 sequence (626 codons) by excluding all sites containing gaps in nonhuman sequences. Maximum likelihood phylogenetic reconstruction was performed from this nucleotide alignment under the GTR+GAMMA model with RAxML.
Most of the analyses aiming at describing the selective constraints acting on SAMHD1 in primates were conducted using the site-specific, branch-specific, and branch-site models for detecting positive selection implemented in the CODEML program of the PAML 4.4 package (Yang, 2007). All calculations were performed using the ML topology previously inferred by RAxML.
The Bayesian reconstruction of dN/dS variation along the primate SAMHD1 ML phylogeny was conducted using the dsom procedure implemented in the CoEvol program (Lartillot and Poujol, 2011). We used the seven calibrations proposed by Perelman et al. (2011) that were compatible with our taxon sampling to infer divergence times. The values of the three life-history traits incorporated in the analysis (body mass, longevity, and maturity) were obtained from the PanTheria database. Two independent MCMC were run for a total of 100,000 cycles sampling points every ten cycles. The first 1,000 points were then excluded as the burnin and inferences were made from the remaining 9,000 sampled points.
Expression Constructs and Plasmids
All SAMHD1 coding sequences were C-terminally tagged with FLAG and HA by cloning into the previously described MMLV-based retroviral vector (Kumar et al., 2009). For immunoprecipitation experiments, selected SAMHD1 alleles were subcloned into a similar vector except that the HA tag was removed. Constructs expressing FLAG- and HA-tagged Vpxmac251, VpxROD, VpxRCM-NG, and VpxRCM-GAB were previously described (Laguette et al., 2011). Vpx alleles from SIVmac239, SIVsm, SIVmnd2, SIVdrl1, HIV-2A, and HIV-2B were synthesized by GenScript (Table S2) and subcloned in pOZ vector (Nakatani and Ogryzko, 2003). For immunoprecipitation experiments, Vpxmac251 was further subcloned in pOZvector where the FLAG tag was deleted.
N-terminal (SAMHD1DSAM) and C-terminal (SAMHD1-F575, SAMHD1-F595, SAMHD1-F611) truncations in SAMHD1 were generated with the phusion enzyme (Finnzym). Point mutations in the SAMHD1 (SAMHD1-S614A, V618A, and M626A) sequence were introduced using the Quickchange lightning kit (Agilent Technologies) according to the manufacturer’s recommendations. SAMHD1-HD/AA and SAMHD1-R mutants, HIV-LUC, VSV-G, MMLV packaging, A-MLV envelope, and shRNA constructs were previously described (Laguette et al., 2011).
pBRSIVmac239_nef+_IRES_eGFPd5 was generated by introducing a MluI restriction site just upstream of the unique SmaI site located downstream of the nef ORF in pBRSIVmac239nef+TPId5 (Münch et al., 2001). Next, a SmaI-IRES-eGFP-MluI fragment was inserted. SIVmac vpx STOP was generated by mutating the start codon of vpx to ACG and introducing a stop codon (TGA) at position 2 without changing the reading frame of vif. These mutations were introduced by PCR site-directed mutagenesis using the Phire polymerase (Finnzymes) and cloned into the SIVmac proviral contruct via PacI/SphI.
Immunopurification
For immunoprecipitation experiments, whole-cell extracts were prepared with 10% glycerol, 0.5% triton, 150 mM NaCl, 10 mM KCL, 1.5 mM MgCl2, 0.5 mM EDTA, 10 mM β-mercaptoethanol, and 0.5 mM PMSF. When FLAG-tagged SAMHD1 was immunoprecipitated, extracts were incubated with anti-FLAG antibody conjugated agarose beads (Sigma), and the bound polypeptides were eluted with Flag peptide (Sigma). When HA-tagged Vpx was immunoprecipitated, extracts were incubated with anti-HA antibody conjugated agarose beads (Santa Cruz), and the bound polypeptides were eluted with Flag peptide (Roche) under native conditions.
Immunofluoresence
THP-1 cells were transduced with Vpx expressing retroviral constructs 48 hr prior to 16 hr differentiation on polylysine treated coverslips. Alternatively, THP-1 cells were differentiated on polylysine treated coverslips prior to VLP-Vpx treatment. HeLa cells were differentiated on coverslips prior to transfection with SAMHD1 constructs. Fixation was performed using PBS with 3% paraformaldehyde and 2% sucrose. Permeabilization was achieved with 0.5% Triton X-100, 20 mM Tris (pH 7.6), 50 mM Nacl, 3 mM MgCl2, and 300 mM sucrose. Wash steps and antibody incubation steps were performed in PBS-0.1%Tween. For SAMHD1 visualization, anti-SAMHD1 (Abcam) was used (1:1000 dilution). For Vpx visualization, anti-HA from Covance was used (1:1000 dilution). Secondary antibodies were purchased from Invitrogen. Nuclei were stained with DAPI in mounting media (Vectashield; Vector Labs) and images were collected on a Leica DM6000 microscope.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Christine Goffinet for critical reading of the manuscript, Sabine Chabalier-Laurent, Xuehua Li, Susanne Engelhart, Raquel Martinez, and Simon Meister for excellent assistance. We are also thankful to the many zoos and primate centers for providing the simian specimens, including the Duke Lemur Center (DLC). N.L. is recipient of SIDACTION fellowship. Work in M.B.’s laboratory was supported by ERC (250333), ANRS, SIDACTION, and FRM “Equipe labéllisée FRM” to M.B. J.S. was supported by the Research Foundation Flanders (FWO, 1.2.627.07.N.01.). N.R. and A.T. are supported by the Swiss National Science Foundation (grant 31003A_132863/1). Work in F.K.’s laboratory was supported by the Deutsche Forschungsgemeinschaft. Use of trade names is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. This is contribution ISEM 2012-015 of the Institut des Sciences de l’Evolution de Montpellier. We are grateful to Montpellier RIO Imaging for help with the microscopy analyses. This is DLC publication #1214.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at doi:10.1016/j.chom.2012.01.007.
REFERENCES
- Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, Hahn BH, Sharp PM. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Coleman CM, Wu L. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology. 2009;6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum. Mol. Genet. 2009;18(R2):R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 2006a;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 2006b;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Dale RC, Gornall H, Singh-Grewal D, Alcausin M, Rice GI, Crow YJ. Familial Aicardi-Goutières syndrome due to SAMHD1 mutations is associated with chronic arthropathy and contractures. Am. J. Med. Genet. A. 2010;152A:938–942. doi: 10.1002/ajmg.a.33359. [DOI] [PubMed] [Google Scholar]
- Douville RN, Hiscott J. The interface between the innate interferon response and expression of host retroviral restriction factors. Cytokine. 2010;52:108–115. doi: 10.1016/j.cyto.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Emerman M, Malik HS. Paleovirology—modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovesio A, Kwon YJ, Windisch MP, Kim NY, Choi SY, Kim HC, Jung S, Mammano F, Perrin V, Boese AS, et al. Automated genome-wide visual profiling of cellular proteins involved in HIV infection. J. Biomol. Screen. 2011;16:945–958. doi: 10.1177/1087057111415521. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hirsch VM, Edmondson P, Murphey-Corb M, Arbeille B, Johnson PR, Mullins JI. SIV adaptation to human cells. Nature. 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix JL, Cimarelli A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J. Virol. 2006;80:1152–1159. doi: 10.1128/JVI.80.3.1152-1159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik R, Zhu X, Stranska R, Wu Y, Stevenson M. A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe. 2009;6:68–80. doi: 10.1016/j.chom.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Jones JH, Terio KA, Estes JD, Rudicell RS, Wilson ML, Li Y, Learn GH, Beasley TM, Schumacher-Stankey J, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol. Biol. Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 estriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Poujol R. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol. Biol. Evol. 2011;28:729–744. doi: 10.1093/molbev/msq244. [DOI] [PubMed] [Google Scholar]
- Lemey P, Pybus OG, Wang B, Saksena NK, Salemi M, Vandamme AM. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. USA. 2003;100:6588–6592. doi: 10.1073/pnas.0936469100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Fregoso OI, McCoy CO, Matsen FA, Malik HS, Emerman M. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11 doi: 10.1016/j.chom.2012.01.004. in press. Published online January 26, 2012. 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′—>5′ exonucleases. J. Biol. Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch J, Adam N, Finze N, Stolte N, Stahl-Hennig C, Fuchs D, Ten Haaft P, Heeney JL, Kirchhoff F. Simian immunodeficiency virus in which nef and U3 sequences do not overlap replicates efficiently in vitro and in vivo in rhesus macaques. J. Virol. 2001;75:8137–8146. doi: 10.1128/JVI.75.17.8137-8146.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M, Guex N, Patin E, Martin O, Xenarios I, Ciuffi A, Quintana-Murci L, Telenti A. Evolutionary trajectories of primate genes involved in HIV pathogenesis. Mol. Biol. Evol. 2009;26:2865–2875. doi: 10.1093/molbev/msp197. [DOI] [PubMed] [Google Scholar]
- Patel MR, Emerman M, Malik HS. Paleovirology - Ghosts and gifts of viruses past. Curr Opin Virol. 2011;1:304–309. doi: 10.1016/j.coviro.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V, Delsuc F, Ranwez S, Belkhir K, Tilak MK, Douzery EJ. OrthoMaM: a database of orthologous genomic markers for placental mammal phylogenetics. BMC Evol. Biol. 2007;7:241. doi: 10.1186/1471-2148-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Münch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Song B, Gold B, O’Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J. Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J. Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 2011;11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- Tristem M, Marshall C, Karpas A, Petrik J, Hill F. Origin of vpx in lentiviruses. Nature. 1990;347:341–342. doi: 10.1038/347341b0. [DOI] [PubMed] [Google Scholar]
- Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 2009;5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.