Abstract
Towards the development of a systems biology-based risk assessment approach for environmental toxicants, including tobacco products in a systems toxicology setting such as the “21st Century Toxicology”, we are building a series of computable biological network models specific to non-diseased pulmonary and cardiovascular cells/tissues which capture the molecular events that can be activated following exposure to environmental toxicants. Here we extend on previous work and report on the construction and evaluation of a mechanistic network model focused on DNA damage response and the four main cellular fates induced by stress: autophagy, apoptosis, necroptosis, and senescence. In total, the network consists of 34 sub-models containing 1052 unique nodes and 1538 unique edges which are supported by 1231 PubMed-referenced literature citations. Causal node-edge relationships are described using the Biological Expression Language (BEL), which allows for the semantic representation of life science relationships in a computable format. The Network is provided in .XGMML format and can be viewed using freely available network visualization software, such as Cytoscape.
Keywords: computable, network model, DNA damage, autophagy, apoptosis, necroptosis, senescence, Biological Expression Language (BEL)
Introduction
System-wide ‘omics’ data containing measurements of thousands of molecular species in a single experiment are increasingly being used to unravel the complex biological mechanisms contributing to pulmonary and cardiovascular diseases. Detailed mechanistic network models are needed to place the differential measurements obtained from molecular profiling data into the context of known biology. These mechanistic models can then be used to better understand the impact of biologically active substances/ toxicants and associated disease risks as outlined in systems toxicology settings such as the “21st Century Toxicology”.1,2
Previously, we have reported on the construction of network models describing cell proliferation and cellular stress.3,4 Extending on elements of these networks (eg, the Cellular Stress Network), which described the network perturbations occurring during cellular defense in response to acute exogenous or endogenous insults, we report here on the construction and evaluation of a third network model, describing the mechanisms that can be activated if these cellular defenses are overwhelmed.
The proper maintenance of homeostatic balance is essential for cell survival in a constantly changing environment. Human pulmonary tissue forms an interface between the external and internal microenvironments, and is therefore constantly exposed to both exogenous stressors including combustion products (diesel exhaust, carbon monoxide, cigarette smoke (CS)), particulate matter, ozone,5–7 and endogenous stressors (eg, mitochondrial-derived reactive oxygen species (ROS), unfolded proteins, nutrient deprivation), all of which can alter cellular homeostasis. Pulmonary cells are equipped with a variety of defense mechanisms to aid in the preservation of cellular homeostasis in the face of such harsh conditions8–10 as outlined in one of our previous network model describing the main CS-related cellular stress defense mechanisms in detail.3 However, these mechanisms can be overwhelmed by chronic stress, for example, ultimately culminating in the intracellular accrual of free radicals, oxidative damage to biomolecules including DNA, and the induction of the DNA damage response as a further protective mechanism. If all these responses to restore cellular homeostasis fail, compromised cells may commit to a terminal fate for the collective benefit of the surrounding tissue, adopting one of four main fates: apoptosis, necroptosis, autophagy, or senescence11 to prevent the nucleation of a potentially deleterious proinflammatory microenvironment.
The DNA damage response activates DNA repair enzymes and in cycling cells, halts cell division by activating G1/S or G2/M cell cycle checkpoints, allowing time for DNA repair.12 Apoptosis is initiated through two main pathways following appropriate extracellular or intracellular signals.13 It fragments a dying cell into apoptotic bodies, which are subsequently cleared from tissue by the phagocytic activity of neighboring or immune cells, minimizing local inflammation. Alternatively, cell clearance can occur through necrosis, a form of death that results in cell lysis and release of proinflammatory intracellular components into the surrounding milieu. Accumulating evidence indicates that at least some forms of necrotic cell death occur in a regulated manner, termed “necroptosis”.14,15 In contrast to apoptosis and necroptosis, which result in the removal of damaged cells, the induction of autophagy or senescence leaves cells surviving, but qualitatively changes their phenotype or function.16,17 During autophagy, lysosomal enzymes degrade and recycle damaged intracellular organelles and proteins in an effort to maintain nutrient and energy homeostasis.18,19 Cellular senescence is characterized by irreversible growth arrest,20,21 as response to a variety of external stimuli including DNA damage, oncogene amplification, and telomere dysfunction.
The DNA damage response, apoptosis, necroptosis, autophagy, and senescence are especially important in the context of CS, as smoke exposure in human pulmonary experimental systems has been shown to induce each, depending on the exposure or experimental context.22–29 Although these cellular fates generally serve in a protective capacity, emerging research also points to a prominent role for stress-induced cell fate choices in the pathogenesis of CS-related diseases, including lung cancer, chronic obstructive pulmonary disease (COPD), and cardiovascular disease.30–33 Understanding how this protective to pathogenic transition occurs requires both a thorough mechanistic understanding of the pathways involved and the appropriate input data.
Here we describe the construction and application of a literature-based network model depicting the DNA damage response, apoptosis, necroptosis, autophagy, and senescence, hereafter referred to by the acronym DACS (DNA damage, Autophagy, Cell death (apoptosis and necroptosis), and Senescence). The DACS Network is modular and computable with its edges supported by hundreds of scientific references. We applied the Network to an independent molecular profiling data set, verifying the content and computability of the network in the process. Together with our previously published network models, the Network will be an invaluable research tool to investigate the biological effects of environmental exposures including CS on human systems, both qualitatively and quantitatively, towards systems toxicology approaches.
Methods
Biological Expression Language (BEL)
The causal relationships in the model are expressed in the Biological Expression Language (BEL)44, which allows for the representation of biological processes in a computable format. BEL is designed to represent scientific findings by capturing causal and correlative relationships in context, where context can include information about the biological and experimental system in which the relationships were observed, the supporting publications cited and the curation process used.
Knowledgebase
The nodes and edges comprising the DACS Network were assembled from the Selventa Knowledgebase, a comprehensive repository containing over 1.5 million nodes (biological processes and entities) and over 7.5 million edges (assertions about causal and non-causal relationships between nodes). The assertions in the Selventa Knowledgebase are derived from peer-reviewed scientific literature as well as other public and proprietary databases. Specifically, each assertion describes an individual experimental observation from an experiment performed in a human, mouse or rat species context, either in vitro or in vivo. Assertions in one species (eg, human) are homologized to another species (eg, mouse) in cases where each element of an assertion has an orthologous counterpart in both species. Assertions also capture information about the referring source (eg, the PubMed ID (PMID) for journal articles listed in MEDLINE), as well as key contextual information including the species (human, mouse, or rat) and the tissue or cell line from which the experimental observation was derived. An example causal assertion is the increased transcriptional activity of TP53 (tumor protein p53) causes an increase in the mRNA expression of CDKN1A (cyclin-dependent kinase inhibitor 1A) [fibroblast; Human; PMID 15616590]. The Knowledgebase contains causal relationships derived from healthy tissues and disease areas such as inflammation, metabolic diseases, cardiovascular injury, liver injury and cancer.
Analysis of transcriptomic data sets
Four previously published data sets, GSE6206,34 E-MEXP-1968,35 GSE13330,36 and GSE19018 were used to construct the DACS Network. A fifth data set, GSE28464,37 was used to evaluate the DACS Network, with a specific focus on the relevant senescence sub-models (Supplementary Table 1). All data sets were downloaded either from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/gds) or from ArrayExpress (http://www.ebi.ac.uk/arrayexpress). RNA expression data were analyzed using the “affy”, “lumiHumanIDMapping”, and “limma” packages of the Bioconductor suite of microarray analysis tools available for the R statistical environment.38–42 Robust Microarray Analysis (RMA) background correction and quantile normalization were used to generate microarray expression values for the Affymetrix platform (CEL files from GSE6206, E-MEXP-1968, GSE13330, and GSE19018), while log2 transformation and quantile normalization were used to generate expression values for the Illumina platform (non-normalized data file from GSE28464). An overall linear model was fit to the data for all sample groups, and specific contrasts of interest were evaluated to generate raw P-values for each probe set on the expression array.43 The Benjamini-Hochberg False Discovery Rate (FDR) method was then used to correct for multiple testing effects.
For Affymetrix data sets, probe sets were considered to have statistically significant changed expression levels in a specific comparison if they had an adjusted P-value of less than 0.05, an absolute fold change greater than 1.3, and average expression intensity greater than 150 in either treatment group. NetAffx version na32 feature annotation files, available from Affymetrix (http://www.Affymetrix.com), were used to map probe sets to genes. For the Illumina platform, the criteria used for statistical significance in changed gene expression were if they had an adjusted P-value of less than 0.05 and an absolute fold change greater than 1.3. In our analysis, genes represented by multiple probe sets were considered to have changed if at least one probe set was observed to change. Gene expression changes that met these criteria are called ‘State Changes’ and have the directional qualities of ‘increased’ or ‘decreased’ ie, they were upregulated or downregulated, respectively, in response to the experimental condition. The number of State Changes for each data set is listed in Supplementary Table 1.
Reverse causal reasoning (RCR): automated hypothesis generation
Reverse causal reasoning (RCR) analysis of the five DNA damage and senescence transcriptomic data sets was used to generate lists of nodes that were predicted to be increased or decreased, and these lists of nodes were used to aid in the selection of nodes for inclusion in the DACS Network, as well as to evaluate the DACS Network using the data set. RCR interrogates the Selventa Knowledgebase to identify potential upstream controllers of entities observed to change significantly in an experiment (see Selventa 201044 and Additional File 1 for specific detail on RCR). Here we applied RCR to the mRNA State Changes in the five transcriptomic data sets to predict potential upstream controllers for the expression changes. These potential upstream controllers identified by RCR are called HYPs as they represent statistically significant hypotheses that are potential explanations for the observed downstream mRNA State Changes. Specifically, the upstream HYP is a potential explanation for the subset of State Changes that are causally downstream of the HYP in individual assertions in the Selventa Knowledgebase.
Each HYP is scored according to two probabilistic scoring metrics: richness and concordance. Richness is the probability that the number of observed mRNA State Changes connected to a given HYP could have occurred by chance alone, calculated using the hyper-geometric distribution. Concordance is the probability that the number of observed RNA State Changes that match the direction of the HYP (eg, increased or decreased activity or abundance of a node) could have occurred by chance alone, calculated using a binomial distribution. HYPs meeting both richness and concordance P-value cutoffs of 0.1 were considered to be statistically significant. When performing control analyses, applying these significance cutoffs to randomly generated data (with similar numbers of RNA State Changes as the experimental data) generally produces less than 5% of the number of HYPs meeting both significance criteria than are observed for experimental data (not shown). For the purposes of network model construction, top scoring HYPs meeting the minimum statistical cutoffs for richness and concordance were evaluated and selected for integration based on their biological plausibility and relevance to the perturbation and biological context (eg, cell type) of the experiment. For data set interrogation, scored HYPs meeting these same statistical cutoffs were considered, with the understanding that as potential explanations for a subset of State Changes, the connectivity and consistency of direction of individual HYPs needed to be considered within context of the models (Selventa 201044 and Additional File 1).
Results
Network structure and content
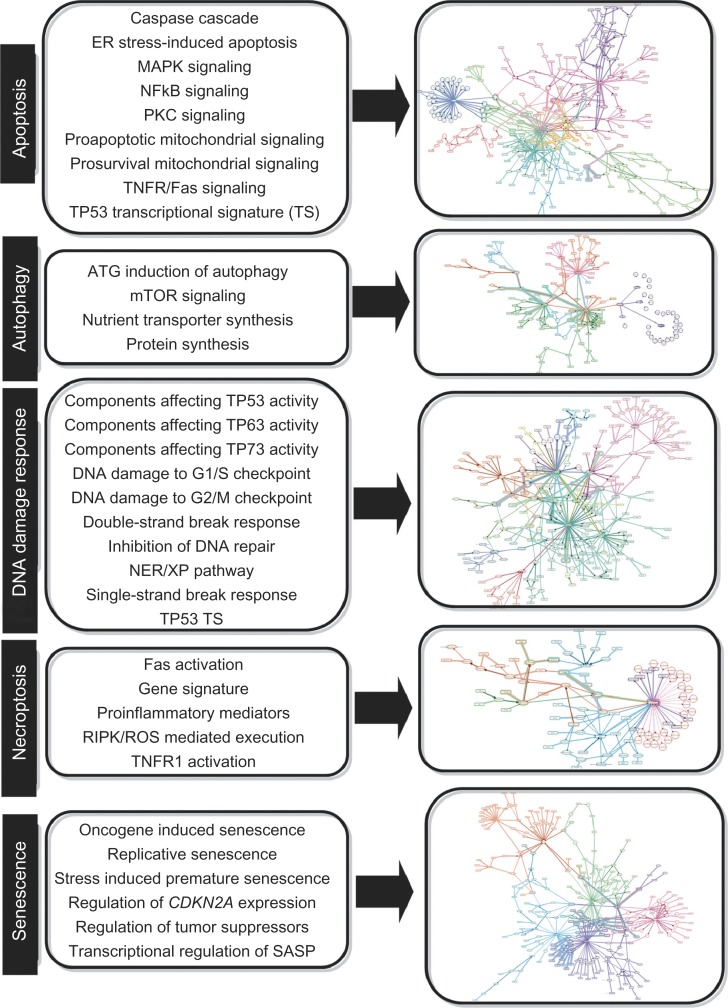
We constructed a network model focused on DNA damage response and the four main cellular fates induced by overwhelming stress: autophagy, apoptosis, necroptosis, and senescence (Fig. 1, Supplementary Fig. 1). The complete DACS Network is provided in Additional File 2 as an excel file and in Additional Files 3–7 in .XGMML format. The .XGMML format can be viewed using freely available network visualization software, such as Cytoscape.45 The DACS Network was constructed using a highly modular design, where the larger network is divided into sub-models. Discrete mechanisms affecting cell fate (eg, ‘NFKB signaling’ describing the prosurvival effects of NFKB-mediated transcriptional upregulation of anti-apoptotic genes) in the five DACS Network areas are described by 34 sub-models (Fig. 1).
Figure 1.
Overview of the DACS Subnetworks.
Notes: The DACS Network is comprised of 34 submodels that represent relevant signaling within five areas of biology – apoptosis, autophagy, DNA damage response, necroptosis, and senescence. Each of the 34 submodels describes the molecular signaling mechanisms shown to activate or inhibit the end process (eg, in the submodel ‘Replicative senescence,’ increased CDKN2A and CDKN1A protein abundances lead to the induction of replicative senescence, while increased abundance of WRN protein inhibits replicative senescence). The left panel lists the names of the submodels involved in each area (eg, ‘Replicative senescence’ under Senescence), and the right panel shows an agglomerated diagram of all submodels involved in each area, with different submodels highlighted in unique colors.
In total, the DACS Network contains 1052 unique nodes and 1538 unique edges (959 causal edges and 579 non-causal edges), which are supported by 1231 PubMed-referenced literature citations (Table 1, Additional Files 2–7). Nodes in the DACS Network are biological entities such as protein abundances, mRNA expression levels, and protein activities. In addition, nodes can also represent biological processes (eg, protein biosynthesis). Edges are relationships between the nodes, and are categorized as either causal or non-causal. Causal edges are directional cause-effect relationships between nodes (eg, NFKB directly increases the gene expression of BCL2), whereas non-causal edges connect different forms of a biological entity, such as gene expression to the related protein abundance. Node-edge relationships in the DACS Network are described using the BEL which allows for the semantic representation of life science relationships in a computable format (Selventa 201044 and Additional File 1). Overall, the DACS Network provides a comprehensive, detailed representation of the causal pathways involved in the DNA damage response, apoptosis, necroptosis, autophagy, and senescence.
Table 1.
DACS Network statistics.
| Nodes | 1052 |
| mRNAs | 138 |
| Proteins | 392 |
| Phosphoproteins | 105 |
| Activities | 224 |
| Complexes | 22 |
| Protein families | 25 |
| Biological processes/GO terms | 48 |
| Chemicals/small molecules | 22 |
| Other | 76 |
| Total edges | 1538 |
| Causal | 959 |
| Non-casual | 579 |
| Unique PMIDs | 1231 |
| Submodel name | Total nodes (predictable) |
| DNA Damage | 272 (72) |
| Autophagy | 161 (49) |
| Apoptosis | 280 (112) |
| Necroptosis | 94 (30) |
| Senescence | 365 (186) |
Notes: Summary of relevant statistics describing the contents of the DACS Network. For each DACS Network area, the total number of unique nodes in the agglomerated model is given, along with the number of those nodes that are capable of prediction by RCR (in parentheses).
Network construction
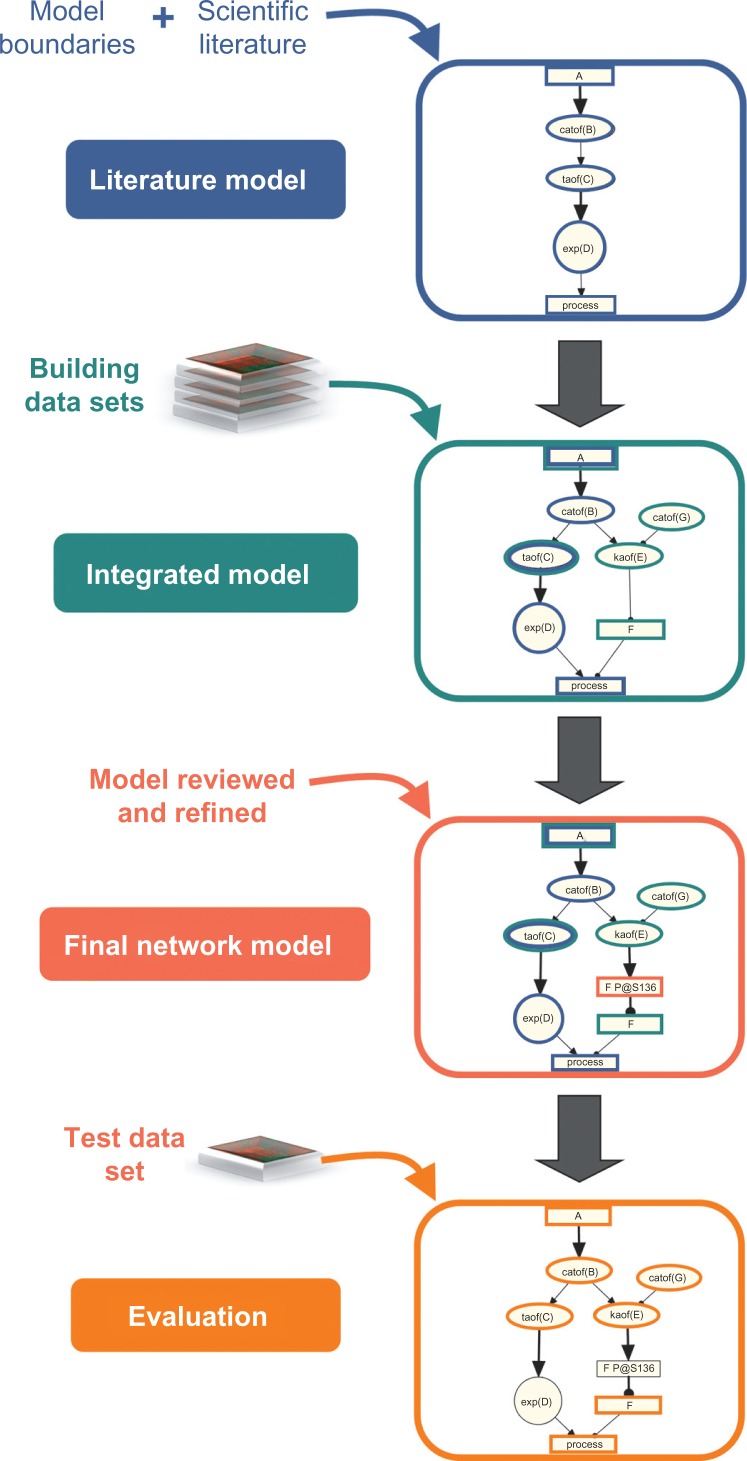
The DACS Network was constructed using the same iterative process used to create previously published network models.3,4 Using this strategy, the network is populated with nodes and edges from two main sources: prior knowledge described in the scientific literature, and results obtained from the computational analysis of transcriptomic profiling data via RCR (Selventa 201044 and Additional File 1) (Fig. 2).
Figure 2.
Workflow used to construct and evaluate the DACS Network.
Notes: The DACS Network is a literature based model containing content derived from two main sources. The literature model was constructed from causal relationships extracted from relevant scientific literature following the definition of network boundaries. The literature model was then augmented with additional nodes derived from Reverse Causal Reasoning (RCR) analysis of transcriptomic data sets, forming the integrated model. In this step, RCR analysis was also used to verify the placement of existing nodes in the literature model. Manual review and refinement of the integrated model resulted in the final network model. The final network model was evaluated using RCR analysis of an independent test transcriptomic data set.
In order to build a network model that describes the DACS-related biological mechanisms in non-diseased pulmonary and cardiovascular cells/tissues, we first defined and applied a set of criteria for selecting network content similar to those used in previously published network models.3,4 Starting with a list of nodes identified by a survey of published literature in the five DACS Network areas, we searched for causal relationships describing the mechanistic relationships between these nodes with literature support from normal lung and cardiovascular cell types. In cases where the relevant experiments have not been published in these contexts, relationships derived from non-lung contexts using cell types found in normal lung (fibroblasts, epithelial cells, endothelial cells, etc.) were used. Canonical mechanisms that are well-known in the literature were also included in the network model even if literature support explicitly demonstrating the presence of the mechanism in normal lung or cardiovascular tissues was not found (eg, the catalytic activity of the FAS receptor increasing the catalytic activity of FADD in the activation of TNFR signaling). For direct and proximal connections such as a kinase phosphorylating a residue on a target or protein-protein interactions, evidence from cell free in vitro systems, which lack a single specified tissue context, were also used when normal lung or cardiovascular tissues were not available. Lastly, relationships derived from human and rodent (specifically mouse and rat) systems were included and homologized, with human contexts prioritized (see Methods).
Using these network boundaries, a literature model was created by compiling causal relationships extracted from the Selventa Knowledgebase, a unified collection of over 1.5 million elements of biological knowledge captured from public literature and other resources (see Methods). When critical causal connections did not exist in the Knowledgebase, they were identified and manually curated from literature into the Knowledgebase. During the course of model building, over 7,500 new causal relationships related to DNA damage, cell death, and senescence from 685 unique literature references were added to the Knowledgebase to support the biology reflected in the DACS Network. Following this effort, the literature model encompassed experimentally proven and well-established mechanistic signaling within the five DACS areas.
Next, the literature model was augmented with additional nodes derived from the computational analysis of molecular profiling data using RCR. RCR-derived HYPs were included as new nodes in the DACS Network model if they had literature support for a mechanistic role in the process of interest. RCR analysis was done to confirm the relevance of nodes already present in the literature model, and to uncover relevant nodes that were not identified during the construction of the literature model. RCR-based augmentation of the DACS Network was performed using four transcriptomic data sets (two for DNA damage and two for senescence), referred to as ‘building’ data sets (Supplementary Table 1). Ideally, transcriptomic data sets addressing all five DACS areas would be used in order to maximize network coverage. However, because three of the DACS Network areas (apoptosis, autophagy and necroptosis) have not been classically described as driven by or executed through transcriptomic changes, we focused our efforts on transcriptomic data from experiments describing DNA damage response and the induction of senescence. Candidate data sets for RCR analysis were selected from public gene expression data repositories GEO and ArrayExpress. We prioritized data sets according to three main criteria: (1) whether the biological process relevant to the DACS Network was induced in non-diseased cell types found in normal lung, (2) whether phenotypic endpoint data was available to provide additional verification of the experimental setup/transcriptomic data, and (3) the statistical rigor of the design of transcriptomic profiling experiments. The four building data sets (Supplementary Table 1) were all derived from in vitro experiments done in human or mouse fibroblasts, and represent the response to DNA damage, induction of replicative senescence (RS) and stress-induced premature senescence (SIPS). Applying RCR to the four network building data sets, 575 HYPs were evaluated for biological plausibility. From this initial list of 575 HYPs, 63 were considered biologically plausible in the context of previous literature reports, and were placed into the appropriate sub-model(s) based on their mechanistic connections to the DACS areas (Supplementary Table 2). The literature model augmented with the data-driven nodes formed the integrated model. As a final step in the construction of the DACS Network, the nodes and edges were manually reviewed and refined (eg, by additional specific literature curation), producing the final DACS Network model (Fig. 2).
Application of the DACS Network to an independent data set
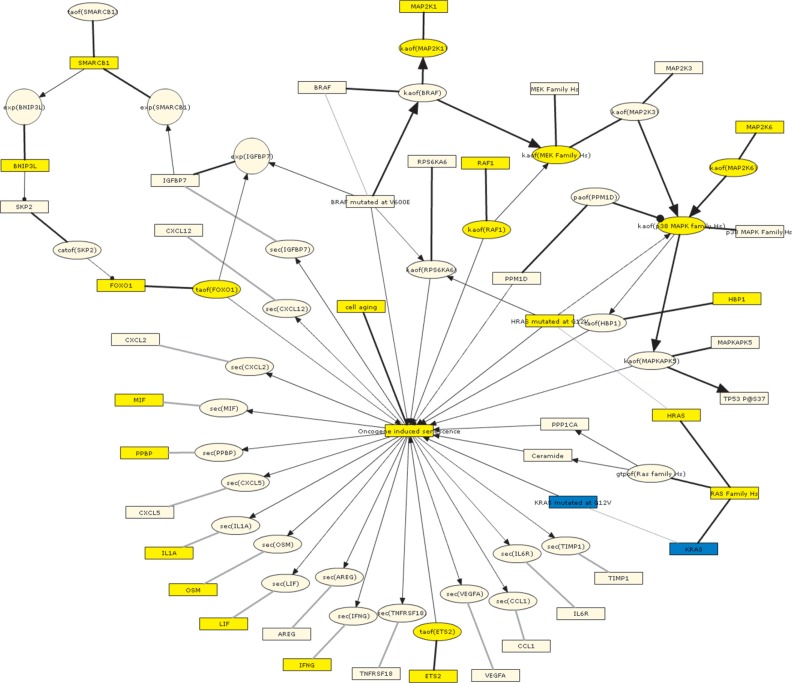
Following network finalization, the DACS Network was applied to investigate a transcriptomic test data set, not included in the construction process, from a well-accepted model of senescence induction ie, oncogene-induced senescence through tamoxifen-inducible HRAS G12V expression in lung fibroblasts (GSE28464)37 (Supplementary Table 1).46–48 This data set also met the boundary criteria for data set selection described above. Although the test data set did not reflect biological activity occurring in all areas of the DACS Network, it enabled a detailed proof-of-principle evaluation of a specific portion of the network (ie, relevant senescence sub-models) as a means to ensure that the nodes and edges placed into the network through manual curation provided an accurate reflection of currently known biology. The four senescence sub-models representing the biology most closely related to the experimental perturbation (constitutively active HRAS by G12V mutation) were selected for investigation using this data set: oncogene-induced senescence (OIS), regulation of CDKN2A expression, regulation by tumor suppressors, and transcriptional regulation of the senescence-associated secretory phenotype (SASP). The OIS sub-model directly reflects the mechanism expected to be seen given the GSE28464 experimental perturbation. The other three sub-models describe mechanisms that are generally applicable to all modes of cellular senescence.
In total, the four senescence sub-models used for evaluation contain 259 unique nodes, 126 (49%) of which were eligible for prediction (meaning that they contain four or more downstream gene expression relationships and thus are capable of prediction as a hypothesis) by RCR. Eighty three of the 126 RCR-capable nodes (66%) are predicted as HYPs in the test set, 79 of which (95%) are predicted in directions consistent with increased oncogene-induced senescence that was experimentally observed.
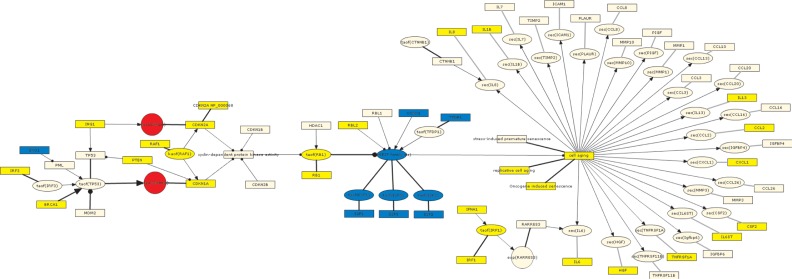
In particular, the oncogene-induced senescence sub-model describes the upstream signaling pathways associated with the induction of OIS as well as the unique SASP proteins produced by cells following OIS.49 When GSE28464 was used to interrogate this sub-model, 30 of the 43 RCR-capable nodes (70%) comprising this sub-model were predicted as HYPs, with 28 of the 30 (93%) predicted in directions consistent with increased OIS (Table 2). These directionally consistent HYP predictions include increased HRAS mutated at G12V, oncogene-induced senescence, and cell aging, all of which match the experimental perturbation from the test data set.37 Increased p38 MAPK activity, FOXO1 activity, RAF1 activity, and HBP1 abundance are all involved in known pathways leading to OIS (Fig. 3).8,46,50,51 Several SASP proteins were also predicted increased in abundance, including OSM, MIF, VEGFA, IL1A, LIF, PPBP and IFNG, consistent with what has been observed following OIS.49 The two directionally inconsistent predictions are for KRAS abundance and KRAS mutated at G12V (which can lead to OIS, but are predicted to be decreased). These inconsistencies were further clarified by reviewing the underlying State Change support for the KRAS HYPs. First, we performed a Gene Ontology (GO) biological process enrichment query on the State Changes supporting both the HRAS mutated at G12V and KRAS mutated at G12V HYPs using the Database for Annotation, Visualization and Integrated Discovery (DAVID). While the State Changes supporting the HRAS mutated at G12V HYP converged on GO biological processes indicative of cell cycle modulation known to be affected during cellular senescence, the State Changes supporting the KRAS at G12V HYP did not converge on any specific biological process (data not shown). In addition, underlying evidence for the KRAS HYP comes, at least in part, from transformed cells that have already bypassed senescence during the transformation process, thus excluding the KRAS HYP from further consideration in the OIS sub-model.
Table 2.
Nodes from the oncogene-induced senescence submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Oncogene Induced Senescence HYPs | Expected Direction | Test Data set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| BNIP3L | ||
| cell aging | ||
| ETS2 | ||
| FOXO1 | ||
| gtpof(Ras family Hs) | ||
| HBP1 | ||
| HRAS | ||
| HRAS mutated at G12V | ||
| IFNG | ||
| IL1A | ||
| kaof(MAP2K1) | ||
| kaof(MAP2K6) | ||
| kaof(MEK Family Hs) | ||
| kaof(p38 MAPK family Hs) | ||
| kaof(RAF1) | ||
| LIF | ||
| MAP2K1 | ||
| MAP2K6 | ||
| MIF | ||
| Oncogene induced senescence | ||
| OSM | ||
| PPBP | ||
| RAF1 | ||
| RAS Family Hs | ||
| SMARCB1 | ||
| taof(ETS2) | ||
| taof(FOXO1) | ||
| VEGFA | ||
| Predicted in inconsistent directions | ||
| KRAS | ||
| KRAS mutated at G12V | ||
Notes: Expected direction is based on internal causality of the oncogene-induced senescence submodel. Yellow = predicted increase in abundance or activity; blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
Abbreviations: gtpof(X), GTP-bound activity of X; kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Figure 3.
Graph showing the oncogene-induced senescence submodel as depicted using the BEL framework and colored according to the GSE28464 test data set.
Notes: Yellow = predicted increase in abundance or activity; blue = predicted decrease in abundance or activity.
Abbreviations: catof(X), catalytic activity of X; exp(X), mRNA expression of X; gtpof(X), GTP-bound activity of X; kaof(X), kinase activity of X; paof(X), phosphatase activity of X; sec(X), cell secretion of X; taof(X), transcriptional activity of X.
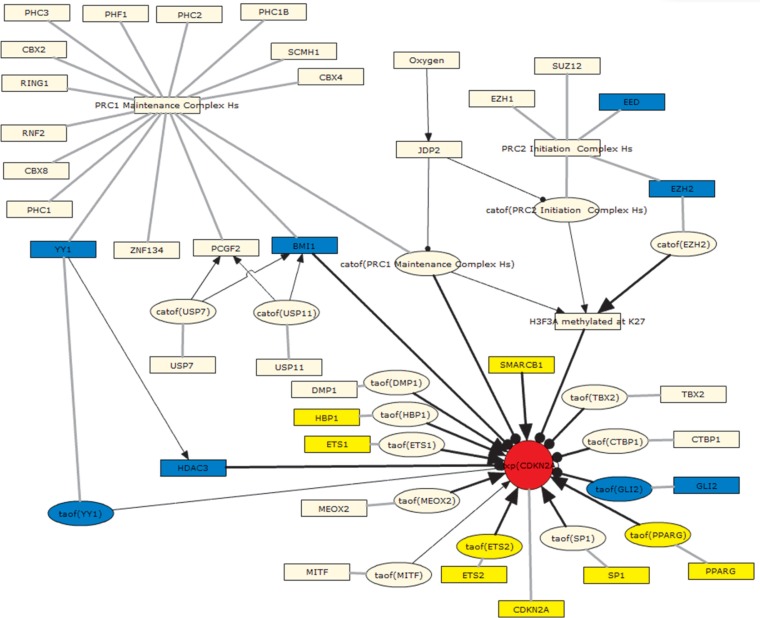
The regulation of CDKN2A expression sub-model includes direct transcriptional regulators of CDKN2A, a cyclin-dependent kinase inhibitor whose increased expression at the gene and protein levels are hallmarks of cellular senescence.52 When interrogated using the test data set, 17 of the 33 RCR-capable nodes (52%) in this sub-model were predicted as HYPs, all in directions consistent with increased CDKN2A expression (Supplementary Table 3). Notably, the prediction for increased CDKN2A protein abundance was also supported by the observed increase in CDKN2A mRNA levels in the test data set. Both positive (SMARCB1, HBP1, ETS1, ETS2, SP1, and PPARG) and negative (HDAC3 and GLI2) regulators of CDKN2A expression were predicted in directions consistent with their previously reported roles.51,53–57 Finally, members of both the Polycomb Repressive Complexes 1 and 2 (PRC1/2) were predicted decreased (YY1 and BMI1 for PRC1, EED and EZH2 for PRC2), consistent with their known role as negative regulators of CDKN2A expression (Fig. 4).58,59
Figure 4.
Graph showing the regulation of CDKN2A expression submodel as depicted using the BEL Framework and colored according to the GSE28464 test data set.
Notes: Yellow = predicted increase in abundance or activity; blue = predicted decrease in abundance or activity; red = observed increase in mRNA expression.
Abbreviations: catof(X), catalytic activity of X; exp(X), mRNA expression of X; taof(X), transcriptional activity of X.
Next, we interrogated the regulation by tumor suppressors sub-model, which describes the cell cycle exit characteristic of senescence regulated by the E2F/Rb axis and CDK inhibitors.60 Thirty-eight of the 49 RCR-capable nodes (78%) in this sub-model were predicted as HYPs in the test data set, all but two (95%) in directions consistent with cell cycle exit and increased senescence (Supplementary Table 4). The consistent HYPs include predictions for decreased abundance/activity of cell cycle activators (E2F family members and CCND1) and conversely, increased abundance/activity of cell cycle inhibitors (RB1, CDKN1A, and CDKN2A). The content of the regulation by tumor suppressors sub-model also included SASP proteins that are shared between multiple modes of senescence, and several of these are predicted increased at the HYP level as well, including CCL2, CSF2, CXCL1, HGF, IL1B, IL6ST, IL8, IL13, and TNFRSF1A (Fig. 5).49 The inconsistent HYPs are IL6, which is predicted to be decreased, and PTEN, which is predicted to be increased (Supplementary Table 4). Upon further exploration of the directionality of the IL6 HYP, we noted that a large fraction of the supporting State Changes (74 out of 173) were IL6 targets related to cell proliferation in multiple myeloma, which falls outside of the network boundaries.61,62 Because many of these proliferative genes were observed to be downregulated (presumably as a consequence of senescent cells exiting the cell cycle), this set of genes could account for the RCR prediction of decreased IL6. When the directionality of the IL6 HYP was re-evaluated excluding this set of genes, it was predicted increased in abundance (Supplementary Fig. 2), consistent with its role as a proinflammatory mediator. Due to those findings, the related literature evidence (Supplementary Fig. 2) will be excluded from future RCR analysis in this model. PTEN is a multifunctional protein and the prediction for increased abundance may be reflective of its role in areas outside of senescence.
Figure 5.
Graph showing the regulation by tumor suppressors submodel as depicted using the BEL framework and colored according to the GSE28464 test data set.
Notes: Yellow = predicted increase in abundance or activity; blue = predicted decrease in abundance or activity; red = observed increase in mRNA expression. IL6 is shown as predicted increased in this figure, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: exp(X), mRNA expression of X; kaof(X), kinase activity of X; sec(X), cell secretion of X; taof(X), transcriptional activity of X.
To further evaluate the mechanisms responsible for the expression of the SASP proteins, we interrogated the transcriptional regulation of the senescence-associated secretory phenotype sub-model, which centers on the transcriptional activities of NFKB and CEBPB upstream of the mRNA expression of SASP proteins. Of the 23 RCR-capable nodes in this sub-model, 16 (70%) are predicted as HYPs in GSE28464, and 15 (94%) are predicted in directions consistent with an increased SASP (Supplementary Table 5). In addition, the transcriptional activity of both NFKB and CEBPB are predicted to be increased, consistent with their roles as central transcriptional mediators of the SASP.63,64 SASP proteins such as CCL2, CCL5, CXCL1, IFNG, IL1A, IL13, IL8, IL6, and VEGFA are all predicted increased. Complementing the RCR predictions, six of these proteins (CCL2, CXCL1, IL1A, IL8, IL6 and VEGFA) are also observed increased at the mRNA level in GSE28464 (Supplementary Fig. 3).
In summary, the evaluation of the transcriptomic profiling data set from human lung cells expressing oncogenic HRAS (GSE28464) using four relevant sub-networks from the DACS Network reveals molecular processes known to be involved in the major hallmarks of oncogene-induced senescence, eg, decreased abundance/activity of cell cycle activators (E2F family members and CCND1), increased abundance/activity of cell cycle inhibitors (RB1, CDKN1A, and CDKN2A), and induction of SASP proteins via activation of the transcription factors NFKB and CEBPB.
Discussion
Comparison with other DACS-related computational networks
Several different modeling approaches have been used to build models of biological systems depending on the biological complexity being captured, the specific goals of the study, and the experimental details involved. The DACS Network was constructed using a prior knowledge of causal relationships from literature, and augmented with nodes derived from RCR, a data-driven method that infers pathway activity based on differentially expressed entities and knowledge of their upstream regulators. Here, we compare and contrast three previously published networks that share features with the DACS Network.65–67
Behrends et al performed a systematic proteomic analysis and utilized existing protein interaction databases to construct an autophagy interaction network (AIN).65 Like the DACS Network, the AIN consists of functional sub-networks, representing unique biological areas of autophagy. In contrast to the protein-protein interactions of the Behrends AIN, the DACS Network shows directionality through mechanistic causal relationships between proteins and other entities including genes, protein activities, biological processes, complexes, etc. Additionally, the DACS Network incorporates transcriptomic data through the integration of computationally derived nodes to infer pathway activity.
Caron et al manually constructed a comprehensive, detailed network of mTOR signaling based on 522 published articles and a protein interaction network (PIN) using 85 key mTOR proteins and protein-protein interactions from multiple databases.66 Comparable to the DACS Network, the mTOR network represents biochemical modifications, directionality, biological entities, and annotations (cell lines, cited literature references). While the integrated mTOR network provides a highly granular view of mTOR signaling, the DACS Network covers a wider range in addition to basic mTOR signaling.
Finally, Passos et al utilized several ‘omics’ approaches to investigate cellular senescence.67 Using target gene inhibition, in silico interactome analysis based on the BioGrid database, and statistical inference, they identified a signaling pathway involving TP53, CDKN1A, GADD45A, MAPK14, GRB2, SRC, DAB2, TGFRB2, and TGFβ. Overlaying these results with those from previous gene expression analysis, they were able to confirm the upregulation of these pathway genes in senescent MRC5 fibroblasts. Similarly, the DACS Network uses transcriptomic data, but applies a computational approach to infer the activity of upstream controllers that fall in a pathway rather than overlaying the genes onto the network itself. Although the Passos network depicts the interconnections between senescence-related entities, it is undirected, as BioGrid interactions lack inherent directionality.
Thus, although the DACS Network shares many features with other previously published networks, we believe the inherent computability conferred upon it by the BEL Framework and the ability to evaluate biological mechanisms by RCR (as opposed to direct mapping of differentially expressed genes onto pathways) differentiates the DACS Network from previously existing resources. In addition, the broad scientific coverage of five distinct yet overlapping biological areas makes the DACS Network a unique resource for the scientific community.
The knowledgebase used to build the network model contains information curated from published literature. We concede that the peer review process is far from perfect and any errors that exist in the public literature could be translated to the knowledgebase. However, the prior knowledge encoded in the knowledgebase has been subject to two additional layers of peer review by PhD level curators. We believe that any inaccuracies that exist in the knowledgebase constitute a minor fraction and occur without a systematic bias that would profoundly affect the results presented here.
While the results shown here indicate that network models have utility in evaluating ‘omics’ data, there are some elements that could be improved in the future. The methodology depends on up-to-date prior knowledge of both the signaling pathways that are represented by the network models and the genes that are regulated by network components. As new discoveries in these areas are made and published, a process for maintaining the connectivity of the network models will need to be put in place to ensure the networks constantly reflect the current state of the field; being dynamic and updatable, any new knowledge can be added to the existing DACS network.
Future application in systems biology-based risk assessment
Understanding how exposure to chemical products affects biological systems is a key first step in the development of effective risk assessment programs. Historically, chemical mechanism-of-action (MOA) studies used simple in vitro or in vivo models and measured a relatively limited number of biological entities. Modern toxicological assessment using system-wide ‘omics’ approaches can now generate thousands of biological data points for a single experiment, and the field of systems toxicology has evolved in order to distil discrete MOA information from this sea of data.68,69 Detailed mechanistic network models are needed to place the differential measurements obtained from molecular profiling data into the context of known biology. These mechanistic models can then be used to better understand the impact of biologically active substances/toxicants and associated disease risks. We are currently developing an application of these network models to derive quantitative measures of network perturbations to compare the impact of biologically active substances, including CS, on human systems in order to assess relative disease risk.
The biological mechanisms represented in the DACS Network, combined with its inherent computability, make it an ideal resource in systems toxicology approaches. For example, the DACS Network could be used in combination with molecular profiling data from human in vitro toxicological studies to characterize the degree to which a simple chemical entity induces a DNA damage response or initiates cell death pathways. In addition, the DACS Network could be used with molecular profiling data from rodents exposed to environmental toxicants in vivo in order to identify the mechanisms whose activation or suppression precedes the development of known genotoxic markers. In each case, the information obtained by combining systems-level data with network-level analyses would provide invaluable mechanistic insight into the biological effects of potentially harmful exposures, and would serve to aid in the development of risk assessment pipelines.
Conclusions
We have presented here a network model that broadly covers the biology within five distinct yet overlapping cellular processes: DNA damage and the main cell fates resulting from cellular stress. The computability enabled by BEL and the broad coverage of toxicologically relevant biology make the DACS Network an exceptional, open-source tool for evaluating modern ‘omics’ data.
Acknowledgments
We would like to acknowledge Michael J. Maria for project management and support with preparation of this manuscript, Stephanie Boue and Vincenzo Belcastro for reviewing the manuscript, and Natalie Catlett for methodological assistance.
Footnotes
Author Contributions
Conceived and designed the experiments: SG, RBL, WKS, MT, AH, CM, EV, MP, MCP, JH. Analysed the data: SG, RBL, WKS, VH, MT, AH, CM, EV, MP, JWW. Wrote the first draft of the manuscript: JWW. Contributed to the writing of the manuscript: SG, BF, WKS, VH, JWW. Agree with manuscript results and conclusions: All authors. Jointly developed the structure and arguments for the paper: SG, RD, JWW. Made critical revisions and approved final version: All authors. All authors reviewed and approved of the final manuscript.
Funding
Selventa and PMI authors performed this work under a joint research collaboration funded by PMI.
Competing Interests
Authors disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Supplementary Materials
Table S1.
Data sets analyzed by RCR for model augmentation and evaluation.
| Model building data sets | Model evaluation data set | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Process | DNA damage | Senescence | Senescence | ||
| Data set ID | GSE6206 | E-MEXP-1968 | GSE13330 | GSE19018 | GSE28464 |
| PubMed ID | 19584263 | 19363488 | 19155301 | Unpublished | 21512002 |
| Species | Mouse | Mouse | Human | Human | Human |
| Context | In vitro | In vitro | In vitro | In vitro | In vitro |
| Cell type | Embryonic fibroblasts | Dermal fibroblasts | Foreskin BJ fibroblasts | IMR90 lung fibroblasts | IMR90 lung fibroblasts |
| Perturbation | Cisplatin (16 μM) | UV irradiation (4 J/m2) | Bleomycin (100 μg/mL) Long term culture | Long term culture in 20% oxygen | Tamoxifen-inducible HRAS G12V expression |
| Timepoint(s) | 24 hr | 6 hr after UV exposure | 24 hr Late passage | 48 population doublings (old) | Day 4 post HRAS G12V induction |
| Control | Untreated | Non-irradiated (0 J/m2) | Early passage (young) | 30 population doublings (young) | Day 0 |
| # State changes | 3684 | 472 | 3355 2799 |
2257 | 3691 |
Notes: Four data sets relating to two different DACS Network areas (DNA damage and Senescence) were analyzed by RCR for the data-driven phase of model construction (Model Building Data Sets). One senescence data set was analyzed by RCR for model evaluation. This table provides a summary of the experimental details and comparisons used for each data set as well as the number of gene expression State Changes observed in each data set.
The DACS Network.
Notes: Graphical representation of the five central biological areas covered by the DACS Network. An agglomerated view of all subnetwork nodes and edges relating the mechanisms involved in the cellular DNA damage response (green) and the induction of senescence (teal), necroptosis (red), autophagy (magenta), and apoptosis (purple), forming an interconnected network of shared and non-overlapping biology.
Table S2.
Data-driven nodes that were predicted as HYPs by RCR on the GSE6206, E-MEXP-1968, GSE13330 RS, GSE13330 SIPS, and GSE19018 building data sets.
| Data-Driven Nodes Added to DACS Network | Expected Direction | DNA Damage Data Sets | Senescence Data Sets | Submode I | |||
|---|---|---|---|---|---|---|---|
| GSE6206 | E-MEXP-1968 | GSE13330 RS | GSE13330 SIPS | GSE19018 | |||
| ATF2 | DNA Damage—Double—strand break response | ||||||
| BACH1 | Senescence—Stress—induced premature senescence | ||||||
| BIRC5 | Apoptosis—NFKB signaling | ||||||
| BNIP3L | Senescence—Oncogene—induced senescence | ||||||
| catof(proteasome complex (sensu Eukarya) Hs) | Senescence—Stress—induced premature senescence | ||||||
| catof(PTGS2) | Senescence—Stress—induced premature senescence | ||||||
| CCL5 | Senescence—Transcriptional regulation of the SASP | ||||||
| CCND1 | DNA Damage—Double—strand break response | ||||||
| CTNNB1 | Senescence—Regulation by tumor suppressors | ||||||
| DDIT3 | Apoptosis—ER stress—induced apoptosis | ||||||
| DNMT3A | Senescence—Replicative senescence | ||||||
| ENO1 | Senescence—Regulation by tumor suppressors | ||||||
| EP300 | Apoptosis—MAPK signaling | ||||||
| ETS2 | Senescence—Oncogene—induced senescence | ||||||
| FHIT | DNA Damagee—single—strand break response | ||||||
| gtpof(RAC1) | Apoptosis—MAPK signaling | ||||||
| gtpof(RHOA) | DNA Damagee—single—strand break response | ||||||
| gtpof(RHOB) | DNA Damagee—single—strand break response | ||||||
| HDAC1 | Senescence—Regulation by tumor suppressors | ||||||
| HDAC3 | Senescence—Regulation of p16INK expression | ||||||
| IFNA1 | Senescence—Regulation by tumor suppressors | ||||||
| IL1A | Senescence—Transcriptional regulation of the SASP | ||||||
| IRF1 | Senescence—Regulation by tumor suppressors | ||||||
| IRF3 | Senescence—Regulation by tumor suppressors | ||||||
| IRF5 | Senescence—Replicative senescence | ||||||
| kaof(MAP2K1) | Senescence—Oncogene—induced senescence | ||||||
| kaof(PKC Family Hs) | Apoptosis—PKC signaling | ||||||
| kaof(RAF1) | Senescence—Oncogene—induced senescence | ||||||
| MAP2K1 | Senescence—Oncogene—induced senescence | ||||||
| MYC | DNA Damage—Double—strand break response | ||||||
| PKC Family Hs | Apoptosis—PKC signaling | ||||||
| PPARG | Senescence—Regulation of p16INK expression | ||||||
| proteasome complex (sensu Eukarya) Hs | Senescence—Stress—induced premature senescence | ||||||
| PTGS2 | Senescence—Stress—induced premature senescence | ||||||
| RAC1 | Apoptosis—MAPK signaling | ||||||
| RAF1 | Senescence—Oncogene—induced senescence | ||||||
| RASSF1 | DNA Damage—Double—strand break response | ||||||
| RHOA | DNA Damage—Double—strand break response | ||||||
| RHOB | DNA Damage—Double—strand break response | ||||||
| SMARCB1 | Senescence—Regulation of p16INK expression | ||||||
| SP1 | Senescence—Regulation of p16INK expression | ||||||
| taof(ATF2) | DNA Damage—Double—strand break response | ||||||
| taof(BACH1) | Senescence—Stress—induced premature senescence | ||||||
| taof(CTNNB1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(EP300) | Apoptosis—MAPK signaling | ||||||
| taof(ETS2) | Senescence—Oncogene—induced senescence | ||||||
| taof(IRF1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(IRF3) | Senescence—Regulation by tumor suppressors | ||||||
| taof(IRF5) | Senescence—Replicative senescence | ||||||
| taof(PPARG) | Senescence—Regulation of p16INK expression | ||||||
| taof(SP1) | Senescence—Regulation of p16INK expression | ||||||
| taof(TFDP1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(TP63) | DNA Damage—Componenets affecting TP63 activity | ||||||
| taof(TP73) | DNA Damage—Componenets affecting TP73 activity | ||||||
| taof(XBP1) | Apoptosis—ER stress—induced apoptosis | ||||||
| taof(YY1) | DNA Damage—Componenets affecting TP53 activity | ||||||
| TFDP1 | Senescence—Regulation by tumor suppressors | ||||||
| TP63 | DNA Damage—Componenets affecting TP63 activity | ||||||
| TP73 | DNA Damage—Componenets affecting TP73 activity | ||||||
| TWIST1 | Senescence—Stress—induced premature senescence | ||||||
| VHL | Senescence—Stress—induced premature senescence | ||||||
| XBP1 | Apoptosis—ER stress—induced apoptosis | ||||||
| YY1 | DNA Damage—Componenets affecting TP53 activity | ||||||
Notes: These data-driven nodes were added to the indicated submodels of the DACS Network based on their mechanistic connections to the processes reflected by the submodels. Expected direction is based on internal causality of the indicated submodels. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity.
Abbreviations: catof(X), catalytic activity of X; gtpof(X), GTP-bound activity of X; kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Table S3.
Nodes from the regulation of CDKN2A expression submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Regulation of CDKN2A HYPs | Expected Direction | Test Data set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| CDKN2A | ||
| ETS1 | ||
| ETS2 | ||
| HBP1 | ||
| PPARG | ||
| SMARCB1 | ||
| SP1 | ||
| taof(ETS2) | ||
| taof(PPARG) | ||
| taof(YY1) | ||
| YY1 | ||
| BMI1 | ||
| EED | ||
| EZH2 | ||
| GLI2 | ||
| HDAC3 | ||
| taof(GLI2) | ||
Notes: Expected direction is based on internal causality of the regulation of CDKN2A expression submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
Abbreviation: taof(X), transcriptional activity of X.
Table S4.
Nodes from the regulation by tumor suppressors submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Regulation by Tumor Suppressors HYPs | Expected Direction | Test Data set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| BRCA1 | ||
| CCL2 | ||
| CDKN1A | ||
| CDKN2A | ||
| CDKN2A NP_000068 | ||
| Cell aging | ||
| CSF2 | ||
| CXCL1 | ||
| HGF | ||
| IFNA1 | ||
| IL6 * | ||
| IL13 | ||
| IL1B | ||
| IL6ST | ||
| IL8 | ||
| ING1 | ||
| IRF1 | ||
| IRF3 | ||
| Kaof(RAF1) | ||
| Oncogene induced senescence | ||
| RAF1 | ||
| RB1 | ||
| RBL2 | ||
| replicative cell aging | ||
| taof(IRF1) | ||
| taof(RB1) | ||
| TNFRSF1A | ||
| CCND1 | ||
| E2F1 | ||
| E2F2 | ||
| E2F3 | ||
| ENO1 | ||
| taof(E2F family Hs) | ||
| taof(E2F1) | ||
| taof(E2F2) | ||
| taof(E2F3) | ||
| TFDP1 | ||
| Predicted in inconsistent directions | ||
| PTEN | ||
Notes: Expected direction is based on internal causality of the regulation by tumor suppressors submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
IL6 is shown as predicted increased in this table, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Directionality investigation for the IL6 HYP in the DACS Network Model test data set.
Notes: The scored IL6 HYP is shown for the DACS Network evaluation data set GSE28464 before (left) and after (right) removal of proliferation-related genes in multiple myeloma as reported in two previously published studies. Circles represent observed gene expression State Changes (red for increased, green for decreased), and edges connecting the central IL6 HYP node to State Changes represent literature-derived causal relationships between IL6 and its downstream target genes. Solid lines indicate relationships that support the HYP prediction (counted as “Score”) and dotted lines indicate relationships that are contradictory to the HYP prediction (counted as “Contra”). When proliferation-related genes are retained in the IL6 HYP, RCR produces a prediction for decreased IL6 abundance. Excluding this set of proliferation-related genes results in an RCR prediction for increased IL6 abundance.
Table S5.
Nodes from the transcriptional regulation of the senescence-associated secretory phenotype (SASP) submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Transcriptional Regulation of the SASP HYPs | Expected Direction | Test Data Set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| CCL2 | ||
| CCL5 | ||
| CEBPB | ||
| CXCL1 | ||
| IFNG | ||
| IL1 Family Hs | ||
| IL6 * | ||
| IL13 | ||
| IL1A | ||
| IL8 | ||
| kaof(p38 MAPK family Hs) | ||
| NFKB Complex Hs | ||
| RELA | ||
| taof(CEBPB) | ||
| taof(NFKB Complex Hs) | ||
| VEGFA | ||
Notes: Expected direction is based on internal causality of the transcriptional regulation of the SASP submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
IL6 is shown as predicted increased in this table, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Graph showing the transcriptional regulation of the senescence-associated secretory phenotype (SASP) submodel as depicted using the BEL framework and colored according to the GSE28464 test data set.
Notes: Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity, red = observed increase in mRNA expression. IL6 is shown as predicted increased in this figure, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: exp(X), mRNA expression of X; kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Additional Files
Additional File 1 Reverse Causal Reasoning.pdf
Additional File 2 DACS Network.xlsx
Additional File 3 DNA Damage-Agglomerated.cy.xgmml
Additional File 4_Autophagy-Agglomerated.cy.xgmml
Additional File 5_Apoptosis-Agglomerated.cy.xgmml
Additional File 6_Necroptosis-Agglomerated.cy.xgmml
Additional File 7_Senescence-Agglomerated.cy.xgmml
References
- 1.Mahadevan B, Snyder RD, Waters MD, et al. Genetic toxicology in the 21st century: reflections and future directions. Environ Mol Mutagen. 2011 Jun;52(5):339–54. doi: 10.1002/em.20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stephens ML, Barrow C, Andersen ME, et al. Accelerating the development of 21st-century toxicology: outcome of a Human Toxicology Project Consortium workshop. Toxicol Sci. 2012;125(2):327–34. doi: 10.1093/toxsci/kfr248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlage WK, Westra JW, Gebel S, et al. A computable cellular stress network model for non-diseased pulmonary and cardiovascular tissue. BMC Syst Biol. 2011;5:168. doi: 10.1186/1752-0509-5-168. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westra JW, Schlage WK, Frushour BP, et al. Construction of a computable cell proliferation network focused on non-diseased lung cells. BMC Syst Biol. 2011;5:105. doi: 10.1186/1752-0509-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuper H, Adami HO, Boffetta P. Tobacco use, cancer causation and public health impact. J Intern Med. 2002;251(6):455–66. doi: 10.1046/j.1365-2796.2002.00993.x. [DOI] [PubMed] [Google Scholar]
- 6.Ris C. US EPA health assessment for diesel engine exhaust: a review. Inhal Toxicol. 2007;19(Suppl 1):229–39. doi: 10.1080/08958370701497960. [DOI] [PubMed] [Google Scholar]
- 7.Budinger GR, Mutlu GM. Update in environmental and occupational medicine 2010. Am J Respir Crit Care Med. 2011;183(12):1614–9. doi: 10.1164/rccm.201103-0467UP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtois-Cox S, Genther Williams SM, Reczek EE, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10(6):459–72. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JY, Wang Y, Prakash C. Xenobiotic-metabolizing enzymes in human lung. Curr Drug Metab. 2006;7(8):939–48. doi: 10.2174/138920006779010575. [DOI] [PubMed] [Google Scholar]
- 10.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 11.Vicencio JM, Galluzzi L, Tajeddine N, et al. Senescence, apoptosis or autophagy? When a damaged cell must decide its path—a mini-review. Gerontology. 2008;54(2):92–9. doi: 10.1159/000129697. [DOI] [PubMed] [Google Scholar]
- 12.Langerak P, Russell P. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3562–71. doi: 10.1098/rstb.2011.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulukaya E, Acilan C, Yilmaz Y. Apoptosis: why and how does it occur in biology? Cell Biochem Funct. 2011;29(6):468–80. doi: 10.1002/cbf.1774. [DOI] [PubMed] [Google Scholar]
- 14.Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16(1):3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11(10):700–14. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 16.Cho S, Hwang ES. Fluorescence-based detection and quantification of features of cellular senescence. Methods Cell Biol. 2011;103:149–88. doi: 10.1016/B978-0-12-385493-3.00007-3. [DOI] [PubMed] [Google Scholar]
- 17.Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol. 2012;74:377–401. doi: 10.1146/annurev-physiol-020911-153348. [DOI] [PubMed] [Google Scholar]
- 18.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2011;23(2):198–206. doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 21.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192(4):547–56. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyunoya T, Monick MM, Klingelhutz A, Yarovinsky TO, Cagley JR, Hunninghake GW. Cigarette smoke induces cellular senescence. Am J Respir Cell Mol Biol. 2006;35(6):681–8. doi: 10.1165/rcmb.2006-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HP, Wang X, Chen ZH, et al. Autophagic proteins regulate cigarette smoke-induced apoptosis: protective role of heme oxygenase-1. Autophagy. 2008;4(7):887–95. doi: 10.4161/auto.6767. [DOI] [PubMed] [Google Scholar]
- 24.Nyunoya T, Monick MM, Klingelhutz AL, et al. Cigarette smoke induces cellular senescence via Werner’s syndrome protein down-regulation. Am J Respir Crit Care Med. 2009;179(4):279–87. doi: 10.1164/rccm.200802-320OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sdralia ND, Patmanidi AL, Velentzas AD, et al. The mode of lymphoblastoid cell death in response to gas phase cigarette smoke is dose-dependent. Respir Res. 2009;10(1):82. doi: 10.1186/1465-9921-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H, Albino AP, Jorgensen E, Traganos F, Darzynkiewicz Z. DNA damage response induced by tobacco smoke in normal human bronchial epithelial and A549 pulmonary adenocarcinoma cells assessed by laser scanning cytometry. Cytometry A. 2009;75(10):840–7. doi: 10.1002/cyto.a.20778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen ZH, Lam HC, Jin Y, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010;107(44):18880–5. doi: 10.1073/pnas.1005574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias ST, Diniz J, Almeida RS, et al. Cytotoxic effect of tobacco extracts on human oral squamous cell carcinoma cell-line. Oral Oncol. 2010;46(12):869–73. doi: 10.1016/j.oraloncology.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen ED, Zhao H, Traganos F, Albino AP, Darzynkiewicz Z. DNA damage response induced by exposure of human lung adenocarcinoma cells to smoke from tobacco- and nicotine-free cigarettes. Cell Cycle. 2010;9(11):2170–6. doi: 10.4161/cc.9.11.11842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leone A, Landini L, Leone A. What is tobacco smoke? Sociocultural dimensions of the association with cardiovascular risk. Curr Pharm Des. 2010;16(23):2510–7. doi: 10.2174/138161210792062948. [DOI] [PubMed] [Google Scholar]
- 31.Ryter SW, Choi AM. Autophagy in the lung. Proc Am Thorac Soc. 2010;7(1):13–21. doi: 10.1513/pats.200909-101JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haspel JA, Choi AM. Autophagy: A Core Cellular Process with Emerging Links to Pulmonary Disease. Am J Respir Crit Care Med. 2011;184(11):1237–46. doi: 10.1164/rccm.201106-0966CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro SD. Merging personalized medicine and biology of aging in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;184(8):864–6. doi: 10.1164/rccm.201108-1486ED. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Knabb JR, Spike BT, Macleod KF. Elevated poly-(ADP-ribose)-polymerase activity sensitizes retinoblastoma-deficient cells to DNA damage-induced necrosis. Mol Cancer Res. 2009;7(7):1099–109. doi: 10.1158/1541-7786.MCR-08-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garinis GA, Uittenboogaard LM, Stachelscheid H, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11(5):604–15. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pazolli E, Luo X, Brehm S, et al. Senescent stromal-derived osteopontin promotes preneoplastic cell growth. Cancer Res. 2009;69(3):1230–9. doi: 10.1158/0008-5472.CAN-08-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narita M, Young AR, Arakawa S, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332(6032):966–70. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpentier J, Luyckx AS, Lefebvre PJ. Influence of metformin on arginine-induced glucagon secretion in human diabetes. Diabete Metab. 1975;1:23–8. [PubMed] [Google Scholar]
- 39.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du P, Kibbe WA, Lin SM. nuID: a universal naming scheme of oligonucleotides for illumina, affymetrix, and other microarrays. Biol Direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team 2007. R: A Language and Environment for Statistical Computing;
- 43.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 44.2010. Selventa. Reverse Causal Reasoning Methods Whitepaper;
- 45.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12(19):2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei S, Wei S, Sedivy JM. Expression of catalytically active telomerase does not prevent premature senescence caused by overexpression of oncogenic Ha-Ras in normal human fibroblasts. Cancer Res. 1999;59(7):1539–43. [PubMed] [Google Scholar]
- 48.Batsi C, Markopoulou S, Vartholomatos G, et al. Chronic NF-kappaB activation delays RasV12-induced premature senescence of human fibroblasts by suppressing the DNA damage checkpoint response. Mech Ageing Dev. 2009;130(7):409–19. doi: 10.1016/j.mad.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16(5):238–46. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun P, Yoshizuka N, New L, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128(2):295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Wang W, Liu X, Paulson KE, Yee AS, Zhang X. Transcriptional factor HBP1 targets P16(INK4A), upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene. 2010;29(36):5083–94. doi: 10.1038/onc.2010.252. [DOI] [PubMed] [Google Scholar]
- 52.Sharpless NE. Ink4a/Arf links senescence and aging. Exp Gerontol. 2004;39(11–12):1751–9. doi: 10.1016/j.exger.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 53.Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65(22):10192–8. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 54.Wu J, Xue L, Weng M, et al. Sp1 is essential for p16 expression in human diploid fibroblasts during senescence. PLoS One. 2007;2(1):e164. doi: 10.1371/journal.pone.0000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gan Q, Huang J, Zhou R, et al. PPAR{gamma} accelerates cellular senescence by inducing p16INK4{alpha} expression in human diploid fibroblasts. J Cell Sci. 2008;121(Pt 13):2235–45. doi: 10.1242/jcs.026633. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Feng Y, Xu L, et al. YY1 restrained cell senescence through repressing the transcription of p16. Biochim Biophys Acta. 2008;(10):1783, 1876–83. doi: 10.1016/j.bbamcr.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 57.Lanigan F, Geraghty JG, Bracken AP. Transcriptional regulation of cellular senescence. Oncogene. 2011;30(26):2901–11. doi: 10.1038/onc.2011.34. [DOI] [PubMed] [Google Scholar]
- 58.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang SW, Lee JK, Ku CC, et al. Jun Dimerization Protein 2 in Oxygen Restriction; Control of Senescence. Curr Pharm Des. 2011;17(22):2278–89. doi: 10.2174/138161211797052394. [DOI] [PubMed] [Google Scholar]
- 60.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 61.Croonquist PA, Linden MA, Zhao F, Van Ness BG. Gene profiling of a myeloma cell line reveals similarities and unique signatures among IL-6 response, N-ras-activating mutations, and coculture with bone marrow stromal cells. Blood. 2003;102(7):2581–92. doi: 10.1182/blood-2003-04-1227. [DOI] [PubMed] [Google Scholar]
- 62.Tsuyama N, Danjoh I, Otsuyama K, et al. IL-6-induced Bcl6 variant 2 supports IL-6-dependent myeloma cell proliferation and survival through STAT3. Biochem Biophys Res Commun. 2005;337(1):201–8. doi: 10.1016/j.bbrc.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 63.Coppe JP, Patil CK, Rodier F, et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One. 2010;5(2):e9188. doi: 10.1371/journal.pone.0009188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Freund A, Patil CK, Campisi J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011;30(8):1536–48. doi: 10.1038/emboj.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caron E, Ghosh S, Matsuoka Y, et al. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Passos JF, Nelson G, Wang C, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plant N. Can systems toxicology identify common biomarkers of non-genotoxic carcinogenesis? Toxicology. 2008;254(3):164–9. doi: 10.1016/j.tox.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Kienhuis AS, Bessems JG, Pennings JL, et al. Application of toxicogenomics in hepatic systems toxicology for risk assessment: acetaminophen as a case study. Toxicol Appl Pharmacol. 2011;250(2):96–107. doi: 10.1016/j.taap.2010.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Data sets analyzed by RCR for model augmentation and evaluation.
| Model building data sets | Model evaluation data set | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Process | DNA damage | Senescence | Senescence | ||
| Data set ID | GSE6206 | E-MEXP-1968 | GSE13330 | GSE19018 | GSE28464 |
| PubMed ID | 19584263 | 19363488 | 19155301 | Unpublished | 21512002 |
| Species | Mouse | Mouse | Human | Human | Human |
| Context | In vitro | In vitro | In vitro | In vitro | In vitro |
| Cell type | Embryonic fibroblasts | Dermal fibroblasts | Foreskin BJ fibroblasts | IMR90 lung fibroblasts | IMR90 lung fibroblasts |
| Perturbation | Cisplatin (16 μM) | UV irradiation (4 J/m2) | Bleomycin (100 μg/mL) Long term culture | Long term culture in 20% oxygen | Tamoxifen-inducible HRAS G12V expression |
| Timepoint(s) | 24 hr | 6 hr after UV exposure | 24 hr Late passage | 48 population doublings (old) | Day 4 post HRAS G12V induction |
| Control | Untreated | Non-irradiated (0 J/m2) | Early passage (young) | 30 population doublings (young) | Day 0 |
| # State changes | 3684 | 472 | 3355 2799 |
2257 | 3691 |
Notes: Four data sets relating to two different DACS Network areas (DNA damage and Senescence) were analyzed by RCR for the data-driven phase of model construction (Model Building Data Sets). One senescence data set was analyzed by RCR for model evaluation. This table provides a summary of the experimental details and comparisons used for each data set as well as the number of gene expression State Changes observed in each data set.
The DACS Network.
Notes: Graphical representation of the five central biological areas covered by the DACS Network. An agglomerated view of all subnetwork nodes and edges relating the mechanisms involved in the cellular DNA damage response (green) and the induction of senescence (teal), necroptosis (red), autophagy (magenta), and apoptosis (purple), forming an interconnected network of shared and non-overlapping biology.
Table S2.
Data-driven nodes that were predicted as HYPs by RCR on the GSE6206, E-MEXP-1968, GSE13330 RS, GSE13330 SIPS, and GSE19018 building data sets.
| Data-Driven Nodes Added to DACS Network | Expected Direction | DNA Damage Data Sets | Senescence Data Sets | Submode I | |||
|---|---|---|---|---|---|---|---|
| GSE6206 | E-MEXP-1968 | GSE13330 RS | GSE13330 SIPS | GSE19018 | |||
| ATF2 | DNA Damage—Double—strand break response | ||||||
| BACH1 | Senescence—Stress—induced premature senescence | ||||||
| BIRC5 | Apoptosis—NFKB signaling | ||||||
| BNIP3L | Senescence—Oncogene—induced senescence | ||||||
| catof(proteasome complex (sensu Eukarya) Hs) | Senescence—Stress—induced premature senescence | ||||||
| catof(PTGS2) | Senescence—Stress—induced premature senescence | ||||||
| CCL5 | Senescence—Transcriptional regulation of the SASP | ||||||
| CCND1 | DNA Damage—Double—strand break response | ||||||
| CTNNB1 | Senescence—Regulation by tumor suppressors | ||||||
| DDIT3 | Apoptosis—ER stress—induced apoptosis | ||||||
| DNMT3A | Senescence—Replicative senescence | ||||||
| ENO1 | Senescence—Regulation by tumor suppressors | ||||||
| EP300 | Apoptosis—MAPK signaling | ||||||
| ETS2 | Senescence—Oncogene—induced senescence | ||||||
| FHIT | DNA Damagee—single—strand break response | ||||||
| gtpof(RAC1) | Apoptosis—MAPK signaling | ||||||
| gtpof(RHOA) | DNA Damagee—single—strand break response | ||||||
| gtpof(RHOB) | DNA Damagee—single—strand break response | ||||||
| HDAC1 | Senescence—Regulation by tumor suppressors | ||||||
| HDAC3 | Senescence—Regulation of p16INK expression | ||||||
| IFNA1 | Senescence—Regulation by tumor suppressors | ||||||
| IL1A | Senescence—Transcriptional regulation of the SASP | ||||||
| IRF1 | Senescence—Regulation by tumor suppressors | ||||||
| IRF3 | Senescence—Regulation by tumor suppressors | ||||||
| IRF5 | Senescence—Replicative senescence | ||||||
| kaof(MAP2K1) | Senescence—Oncogene—induced senescence | ||||||
| kaof(PKC Family Hs) | Apoptosis—PKC signaling | ||||||
| kaof(RAF1) | Senescence—Oncogene—induced senescence | ||||||
| MAP2K1 | Senescence—Oncogene—induced senescence | ||||||
| MYC | DNA Damage—Double—strand break response | ||||||
| PKC Family Hs | Apoptosis—PKC signaling | ||||||
| PPARG | Senescence—Regulation of p16INK expression | ||||||
| proteasome complex (sensu Eukarya) Hs | Senescence—Stress—induced premature senescence | ||||||
| PTGS2 | Senescence—Stress—induced premature senescence | ||||||
| RAC1 | Apoptosis—MAPK signaling | ||||||
| RAF1 | Senescence—Oncogene—induced senescence | ||||||
| RASSF1 | DNA Damage—Double—strand break response | ||||||
| RHOA | DNA Damage—Double—strand break response | ||||||
| RHOB | DNA Damage—Double—strand break response | ||||||
| SMARCB1 | Senescence—Regulation of p16INK expression | ||||||
| SP1 | Senescence—Regulation of p16INK expression | ||||||
| taof(ATF2) | DNA Damage—Double—strand break response | ||||||
| taof(BACH1) | Senescence—Stress—induced premature senescence | ||||||
| taof(CTNNB1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(EP300) | Apoptosis—MAPK signaling | ||||||
| taof(ETS2) | Senescence—Oncogene—induced senescence | ||||||
| taof(IRF1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(IRF3) | Senescence—Regulation by tumor suppressors | ||||||
| taof(IRF5) | Senescence—Replicative senescence | ||||||
| taof(PPARG) | Senescence—Regulation of p16INK expression | ||||||
| taof(SP1) | Senescence—Regulation of p16INK expression | ||||||
| taof(TFDP1) | Senescence—Regulation by tumor suppressors | ||||||
| taof(TP63) | DNA Damage—Componenets affecting TP63 activity | ||||||
| taof(TP73) | DNA Damage—Componenets affecting TP73 activity | ||||||
| taof(XBP1) | Apoptosis—ER stress—induced apoptosis | ||||||
| taof(YY1) | DNA Damage—Componenets affecting TP53 activity | ||||||
| TFDP1 | Senescence—Regulation by tumor suppressors | ||||||
| TP63 | DNA Damage—Componenets affecting TP63 activity | ||||||
| TP73 | DNA Damage—Componenets affecting TP73 activity | ||||||
| TWIST1 | Senescence—Stress—induced premature senescence | ||||||
| VHL | Senescence—Stress—induced premature senescence | ||||||
| XBP1 | Apoptosis—ER stress—induced apoptosis | ||||||
| YY1 | DNA Damage—Componenets affecting TP53 activity | ||||||
Notes: These data-driven nodes were added to the indicated submodels of the DACS Network based on their mechanistic connections to the processes reflected by the submodels. Expected direction is based on internal causality of the indicated submodels. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity.
Abbreviations: catof(X), catalytic activity of X; gtpof(X), GTP-bound activity of X; kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Table S3.
Nodes from the regulation of CDKN2A expression submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Regulation of CDKN2A HYPs | Expected Direction | Test Data set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| CDKN2A | ||
| ETS1 | ||
| ETS2 | ||
| HBP1 | ||
| PPARG | ||
| SMARCB1 | ||
| SP1 | ||
| taof(ETS2) | ||
| taof(PPARG) | ||
| taof(YY1) | ||
| YY1 | ||
| BMI1 | ||
| EED | ||
| EZH2 | ||
| GLI2 | ||
| HDAC3 | ||
| taof(GLI2) | ||
Notes: Expected direction is based on internal causality of the regulation of CDKN2A expression submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
Abbreviation: taof(X), transcriptional activity of X.
Table S4.
Nodes from the regulation by tumor suppressors submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Regulation by Tumor Suppressors HYPs | Expected Direction | Test Data set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| BRCA1 | ||
| CCL2 | ||
| CDKN1A | ||
| CDKN2A | ||
| CDKN2A NP_000068 | ||
| Cell aging | ||
| CSF2 | ||
| CXCL1 | ||
| HGF | ||
| IFNA1 | ||
| IL6 * | ||
| IL13 | ||
| IL1B | ||
| IL6ST | ||
| IL8 | ||
| ING1 | ||
| IRF1 | ||
| IRF3 | ||
| Kaof(RAF1) | ||
| Oncogene induced senescence | ||
| RAF1 | ||
| RB1 | ||
| RBL2 | ||
| replicative cell aging | ||
| taof(IRF1) | ||
| taof(RB1) | ||
| TNFRSF1A | ||
| CCND1 | ||
| E2F1 | ||
| E2F2 | ||
| E2F3 | ||
| ENO1 | ||
| taof(E2F family Hs) | ||
| taof(E2F1) | ||
| taof(E2F2) | ||
| taof(E2F3) | ||
| TFDP1 | ||
| Predicted in inconsistent directions | ||
| PTEN | ||
Notes: Expected direction is based on internal causality of the regulation by tumor suppressors submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
IL6 is shown as predicted increased in this table, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Directionality investigation for the IL6 HYP in the DACS Network Model test data set.
Notes: The scored IL6 HYP is shown for the DACS Network evaluation data set GSE28464 before (left) and after (right) removal of proliferation-related genes in multiple myeloma as reported in two previously published studies. Circles represent observed gene expression State Changes (red for increased, green for decreased), and edges connecting the central IL6 HYP node to State Changes represent literature-derived causal relationships between IL6 and its downstream target genes. Solid lines indicate relationships that support the HYP prediction (counted as “Score”) and dotted lines indicate relationships that are contradictory to the HYP prediction (counted as “Contra”). When proliferation-related genes are retained in the IL6 HYP, RCR produces a prediction for decreased IL6 abundance. Excluding this set of proliferation-related genes results in an RCR prediction for increased IL6 abundance.
Table S5.
Nodes from the transcriptional regulation of the senescence-associated secretory phenotype (SASP) submodel of the DACS Network that are predicted as HYPs by RCR on the GSE28464 test data set.
| Transcriptional Regulation of the SASP HYPs | Expected Direction | Test Data Set |
|---|---|---|
| GSE28464 | ||
| Predicted in consistent directions | ||
| CCL2 | ||
| CCL5 | ||
| CEBPB | ||
| CXCL1 | ||
| IFNG | ||
| IL1 Family Hs | ||
| IL6 * | ||
| IL13 | ||
| IL1A | ||
| IL8 | ||
| kaof(p38 MAPK family Hs) | ||
| NFKB Complex Hs | ||
| RELA | ||
| taof(CEBPB) | ||
| taof(NFKB Complex Hs) | ||
| VEGFA | ||
Notes: Expected direction is based on internal causality of the transcriptional regulation of the SASP submodel. Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity. Submodel nodes that are shared with other senescence models are bolded.
IL6 is shown as predicted increased in this table, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Graph showing the transcriptional regulation of the senescence-associated secretory phenotype (SASP) submodel as depicted using the BEL framework and colored according to the GSE28464 test data set.
Notes: Yellow = predicted increase in abundance or activity, blue = predicted decrease in abundance or activity, red = observed increase in mRNA expression. IL6 is shown as predicted increased in this figure, in contrast to the initial prediction by RCR (See Section 3.3 Application of the DACS Network to an Independent Data Set for additional detail and Supplementary Fig. 2).
Abbreviations: exp(X), mRNA expression of X; kaof(X), kinase activity of X; taof(X), transcriptional activity of X.
Additional File 1 Reverse Causal Reasoning.pdf
Additional File 2 DACS Network.xlsx
Additional File 3 DNA Damage-Agglomerated.cy.xgmml
Additional File 4_Autophagy-Agglomerated.cy.xgmml
Additional File 5_Apoptosis-Agglomerated.cy.xgmml
Additional File 6_Necroptosis-Agglomerated.cy.xgmml
Additional File 7_Senescence-Agglomerated.cy.xgmml