Abstract
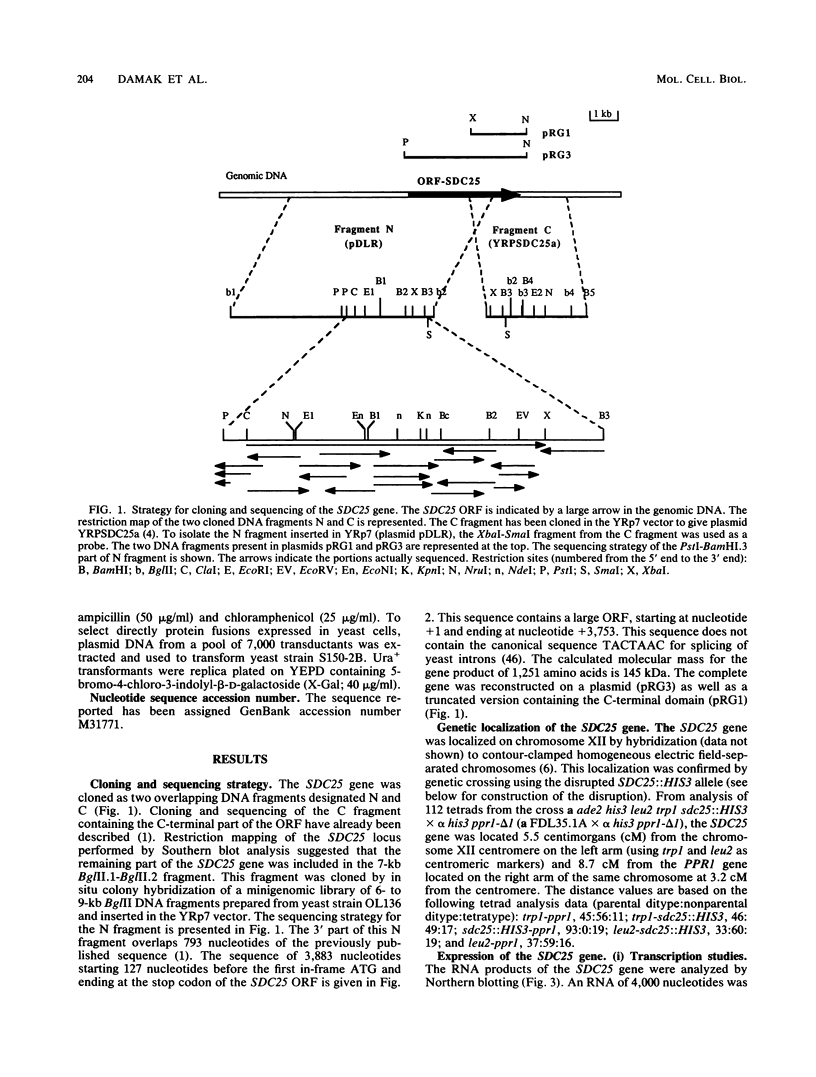
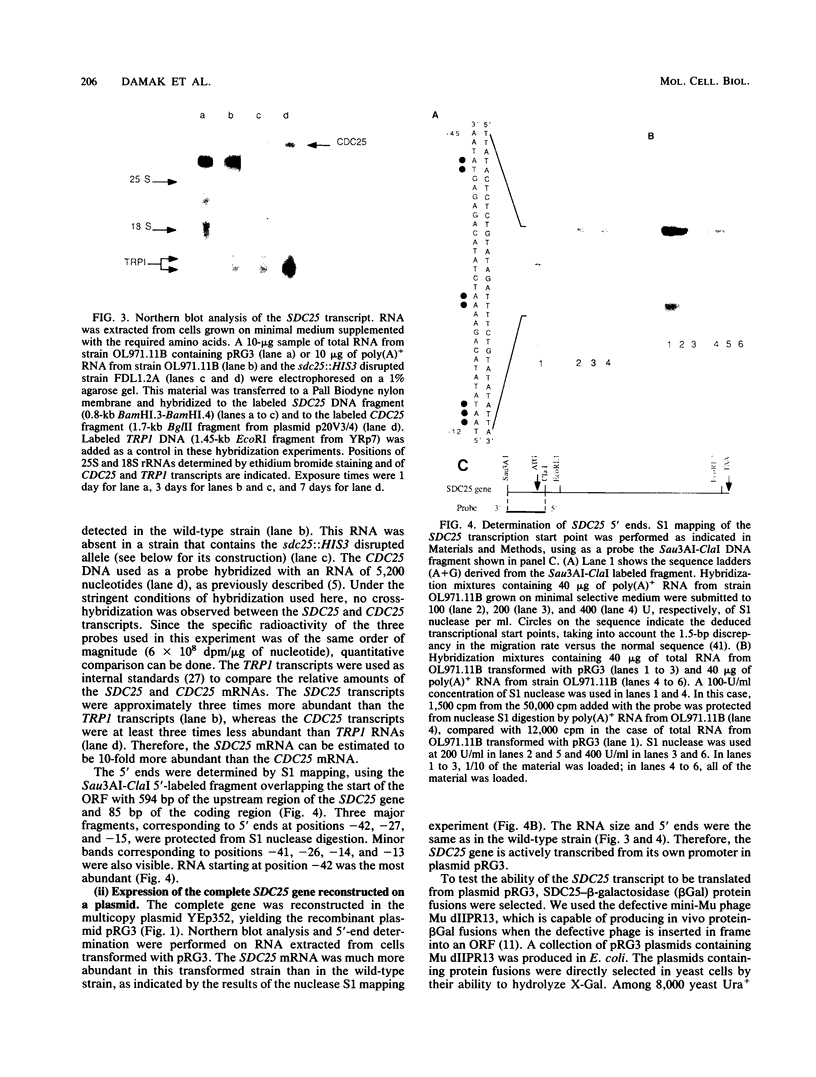
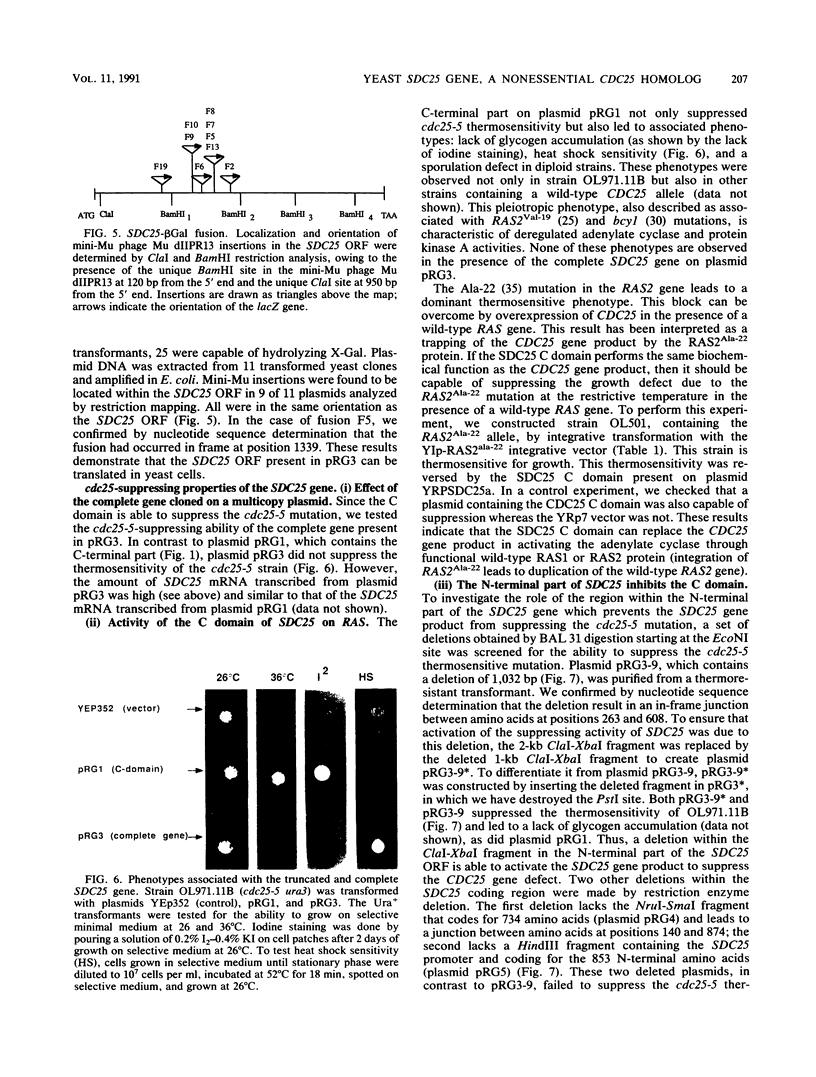
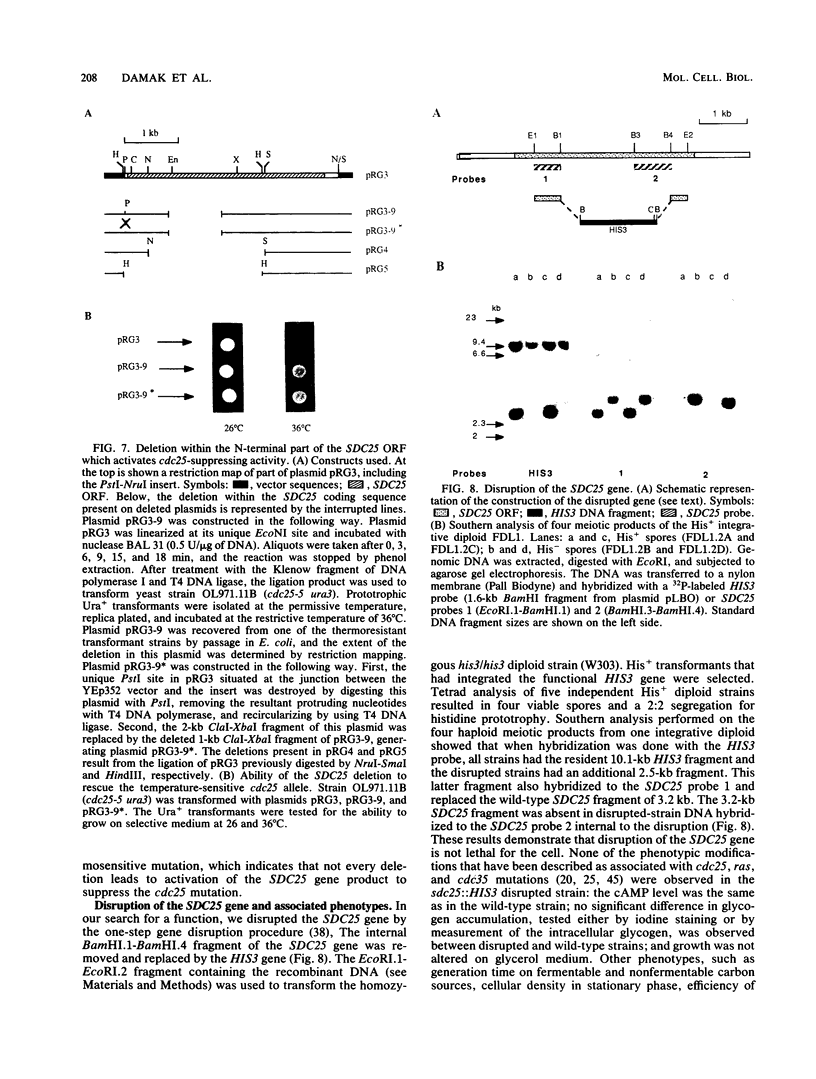
In the yeast Saccharomyces cerevisiae, the CDC25 gene product activates adenylate cyclase through RAS1 and RAS2 gene products. We have recently described the cloning of a DNA fragment which suppresses the cdc25 mutation but not ras1, ras2, or cdc35 mutations. This fragment contains a 5'-truncated open reading frame which shares 47% identity with the C-terminal part of the CDC25 gene. We named the entire gene SDC25. In this paper, we report the cloning, sequencing, and characterization of the complete SDC25 gene. The SDC25 gene is located on the chromosome XII close to the centromere. It is transcribed into a 4-kb-long mRNA that contains an open reading frame of 1,251 codons. Homology with the CDC25 gene extends in the N-terminal part, although the degree of similarity is lower than in the C-terminal part. In contrast with the C-terminal part, the complete SDC25 gene was found not to suppress the CDC25 gene defect. A deletion in the N-terminal part restored the suppressing activity, a result which suggests the existence of a regulatory domain. The SDC25 gene was found to be dispensable for cell growth under usual conditions. No noticeable phenotype was found in the deleted strain.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boy-Marcotte E., Damak F., Camonis J., Garreau H., Jacquet M. The C-terminal part of a gene partially homologous to CDC 25 gene suppresses the cdc25-5 mutation in Saccharomyces cerevisiae. Gene. 1989 Apr 15;77(1):21–30. doi: 10.1016/0378-1119(89)90355-7. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E., Garreau H., Jacquet M. Cyclic AMP controls the switch between division cycle and resting state programs in response to ammonium availability in Saccharomyces cerevisiae. Yeast. 1987 Jun;3(2):85–93. doi: 10.1002/yea.320030205. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Jacquet M. A new RAS mutation that suppresses the CDC25 gene requirement for growth of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2980–2983. doi: 10.1128/mcb.8.7.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camonis J. H., Kalékine M., Gondré B., Garreau H., Boy-Marcotte E., Jacquet M. Characterization, cloning and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 1986 Feb;5(2):375–380. doi: 10.1002/j.1460-2075.1986.tb04222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Cohen P. The role of protein phosphorylation in the hormonal control of enzyme activity. Eur J Biochem. 1985 Sep 16;151(3):439–448. doi: 10.1111/j.1432-1033.1985.tb09121.x. [DOI] [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Camonis J., Jacquet M., Parmeggiani A. Different kinetic properties of the two mutants, RAS2Ile152 and RAS2Val19, that suppress the CDC25 requirement in RAS/adenylate cyclase pathway in Saccharomyces cerevisiae. J Biol Chem. 1990 Jan 25;265(3):1563–1568. [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Mistou M. Y., Parmeggiani A., Camonis J., Boy-Marcotte E., Damak F., Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990 May 18;248(4957):866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B., Bolotin-Fukuhara M. In vivo functional characterization of a yeast nucleotide sequence: construction of a mini-Mu derivative adapted to yeast. Gene. 1988;62(1):45–54. doi: 10.1016/0378-1119(88)90578-1. [DOI] [PubMed] [Google Scholar]
- Daniel J. H. The CDC25 "Start" gene of Saccharomyces cerevisiae: sequencing of the active C-terminal fragment and regional homologies with rhodopsin and cytochrome P450. Curr Genet. 1986;10(12):879–885. doi: 10.1007/BF00398284. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Mulholland J., Zhu Z. M., Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990 Jan 18;343(6255):288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Field J., Broek D., Kataoka T., Wigler M. Guanine nucleotide activation of, and competition between, RAS proteins from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2128–2133. doi: 10.1128/mcb.7.6.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau H., Camonis J. H., Guitton C., Jacquet M. The Saccharomyces cerevisiae CDC25 gene product is a 180 kDa polypeptide and is associated with a membrane fraction. FEBS Lett. 1990 Aug 20;269(1):53–59. doi: 10.1016/0014-5793(90)81117-7. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Iida H., Yahara I. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J Cell Biol. 1984 Apr;98(4):1185–1193. doi: 10.1083/jcb.98.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P., Baltimore D. N-terminal mutations activate the leukemogenic potential of the myristoylated form of c-abl. EMBO J. 1989 Feb;8(2):449–456. doi: 10.1002/j.1460-2075.1989.tb03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Boy-Marcotte E., Rossier C., Kessin R. H. A fragment of Dictyostelium discoideum genomic DNA that complements the URA1 mutation of Saccharomyces cerivisiae. J Mol Appl Genet. 1982;1(6):513–525. [PubMed] [Google Scholar]
- Johnson D. I., Jacobs C. W., Pringle J. R., Robinson L. C., Carle G. F., Olson M. V. Mapping of the Saccharomyces cerevisiae CDC3, CDC25, and CDC42 genes to chromosome XII by chromosome blotting and tetrad analysis. Yeast. 1987 Dec;3(4):243–253. doi: 10.1002/yea.320030405. [DOI] [PubMed] [Google Scholar]
- Jung G., Saxe C. L., 3rd, Kimmel A. R., Hammer J. A., 3rd Dictyostelium discoideum contains a gene encoding a myosin I heavy chain. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6186–6190. doi: 10.1073/pnas.86.16.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kato J. Y., Takeya T., Grandori C., Iba H., Levy J. B., Hanafusa H. Amino acid substitutions sufficient to convert the nontransforming p60c-src protein to a transforming protein. Mol Cell Biol. 1986 Dec;6(12):4155–4160. doi: 10.1128/mcb.6.12.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Multiple control elements in the TRP1 promoter of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Dec;6(12):4251–4258. doi: 10.1128/mcb.6.12.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto V. P., Wasenius V. M., Salvén P., Saraste M. Transforming and membrane proteins. Nature. 1988 Aug 4;334(6181):388–388. doi: 10.1038/334388a0. [DOI] [PubMed] [Google Scholar]
- Liljelund P., Losson R., Kammerer B., Lacroute F. Yeast regulatory gene PPR1. II. Chromosomal localization, meiotic map, suppressibility, dominance/recessivity and dosage effect. J Mol Biol. 1984 Dec 5;180(2):251–265. doi: 10.1016/s0022-2836(84)80003-0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Initiation of meiosis in yeast mutants defective in adenylate cyclase and cyclic AMP-dependent protein kinase. Cell. 1983 Feb;32(2):417–423. doi: 10.1016/0092-8674(83)90461-0. [DOI] [PubMed] [Google Scholar]
- Mayer B. J., Hamaguchi M., Hanafusa H. A novel viral oncogene with structural similarity to phospholipase C. Nature. 1988 Mar 17;332(6161):272–275. doi: 10.1038/332272a0. [DOI] [PubMed] [Google Scholar]
- Munder T., Mink M., Küntzel H. Domains of the Saccharomyces cerevisiae CDC25 gene controlling mitosis and meiosis. Mol Gen Genet. 1988 Oct;214(2):271–277. doi: 10.1007/BF00337721. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. At least 1400 base pairs of 5'-flanking DNA is required for the correct expression of the HO gene in yeast. Cell. 1985 Aug;42(1):213–223. doi: 10.1016/s0092-8674(85)80117-3. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Sass P., Wigler M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3629–3636. doi: 10.1128/mcb.7.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Ferenz C. R., Kelleher K. L., Kriz R. W., Knopf J. L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988 Mar 17;332(6161):269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Tamanoi F. Yeast RAS genes. Biochim Biophys Acta. 1988 Aug 3;948(1):1–15. doi: 10.1016/0304-419x(88)90002-9. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., McMullen B., Hurwitz M., Krebs E. G., Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Apr;7(4):1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987 Jul 17;50(2):277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. 1984 May 31-Jun 6Nature. 309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]