Abstract
Sensitivity and specificity are two most important factors to take into account for molecule sensing, chemical detection and disease diagnosis. A perfect sensitivity is to reach the level where a single molecule can be detected. An ideal specificity is to reach the level where the substance can be detected in the presence of many contaminants. The rapidly progressing nanopore technology is approaching this threshold. A wide assortment of biomotors and cellular pores in living organisms perform diverse biological functions. The elegant design of these transportation machineries has inspired the development of single molecule detection based on modulations of the individual current blockage events. The dynamic growth of nanotechnology and nanobiotechnology has stimulated rapid advances in the study of nanopore based instrumentation over the last decade, and inspired great interest in sensing of single molecules including ions, nucleotides, enantiomers, drugs, and polymers such as PEG, RNA, DNA, and polypeptides. This sensing technology has been extended to medical diagnostics and third generation high throughput DNA sequencing. This review covers current nanopore detection platforms including both biological pores and solid state counterparts. Several biological nanopores have been studied over the years, but this review will focus on the three best characterized systems including α-hemolysin and MspA, both containing a smaller channel for the detection of single-strand DNA, as well as bacteriophage phi29 DNA packaging motor connector that contains a larger channel for the passing of double stranded DNA. The advantage and disadvantage of each system are compared; their current and potential applications in nanomedicine, biotechnology, and nanotechnology are discussed.
Keywords: bacteriophage phi29, α-hemolysin, MspA, solid state pore, synthetic nanopores, DNA packaging, nanomotor, connector, liposomes, ion channel, single channel conductance, membrane channel, viral assembly, stoichiometry quantification, nanostructure, bionanotechnology, nanobiotechnology, nanomedicine
Introduction
Translocation of ions, DNA, RNA, polypeptides and other macromolecules across the membrane within or between cells is a fundamental and ubiquitous process. The transportation process involves a wide assortment of passive pores, active ion channels, and viral motors with elegant and highly-ordered structures. The novel and sophisticated design of the transport machineries have inspired the development of nanopores for single molecule detection.
Nanopore based analysis is currently an area of great interest in many disciplines with the potential for incredibly versatile applications. These include sensing small molecules such as ions, nucleotides, enantiomers, and drugs, as well as larger polymers such as PEG, RNA, DNA, and polypeptides. Single pore sensing is a label-free single molecule recognition approach requiring very low sample volumes without sample preparations or amplifications. The detection can be carried out with high sensitivity in the presence of large number of contaminants. This review encompasses the concept of nanopores; types of nanopores along with their advantages and disadvantages; and their current and potential applications in nanomedicine and nanotechnology. The field of nanopore has skyrocketed over the last decade, as evidenced by 900+ publications from Pubmed. Thus a significant amount of nanopore literatures is not covered due to the limitations of space. For more in-depth analysis of past advances, interested readers are encouraged to read several excellent reviews published over the years[1–5]
Principles of nanopore detection
The stochastic nanopore technique is based on the working principle of the classical Coulter Counter or the ‘resistive-pulse’ routine[6], which demonstrated sizing of micron sized particles with a micron sized aperture. In the nanopore technique, charged polymers are electrophoretically driven though a nanometer sized aperture (typically a few nm to tens of nm) embedded in a thin membrane. The nanopore is located in an electrochemical chamber separated into cis- and trans-compartments, each containing conducting buffers. Under an applied voltage, electrolyte ions flow through the nanopore, which is measured as current in the electrical circuit. The current, typically in the pico-Ampere scale, is measured using a patch clamp setup along with associated ultra-sensitive electronics, housed inside a Faraday cage.
Polymer capture, entry and subsequent translocations are then characterized at the single molecule level by modulations of the individual current blockage events. The transient current is based on the volumetric exclusion of ions from the pore during transport of linearized polymers with current proportional to the cross-sectional area of the linearized polymer relative to the cross sectional area of the pore. Various parameters, such as the event duration, current amplitude, and unique electrical signature of the current blockages can be used either singly or in combination for single molecule fingerprinting.
Types and attributes of various nanopores
Nanopores are classified into three classes: (1) Biological pores embedded in a lipid bilayer; (2) Synthetic nanopores fabricated in solid substrates, such as Si3N4, Al2O3, TiO2, and graphene; and (3) Hybrid of biological and synthetic nanopores.
(A) Biological nanopores
Over the years, relatively larger channels compared to ion channels have been explored for the purposes of nanopore detection. The substrate of choice for all biological pores is planar lipid membranes, liposomes or polymer membranes housed inside an electrochemical chamber. Large scale production and purification of various channel proteins are possible by employing standard molecular biology techniques. In majority of the cases, the purified channel pores are homogeneous from different batches. In addition, explicit engineering of the channel pores via site-directed mutagenesis is possible due to available crystal structure of several channel proteins. The defining aspects of three well studied biological pores are discussed briefly below:
(1) α-Hemolysin channel
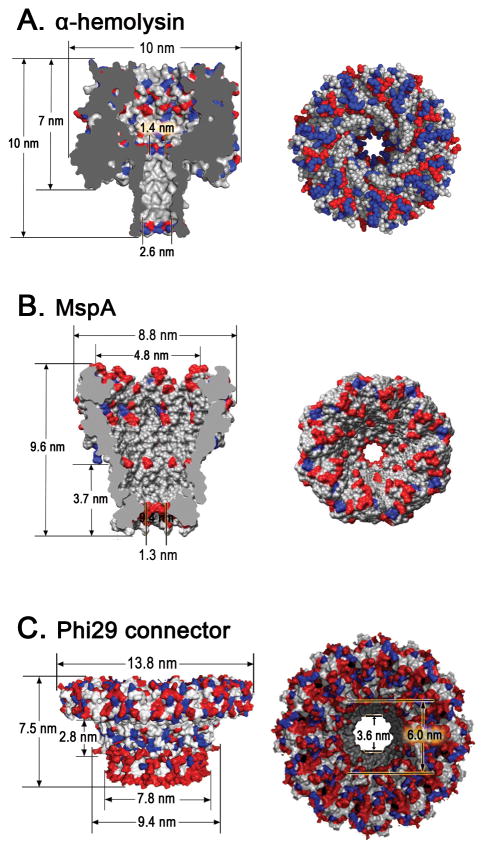
α-Hemolysin is an exotoxin secreted by the human pathogen Staphylococcus aureus bacterium. It’s a 232.4 kDa mushroom-like heptameric transmembrane pore, consisting of a vestibule (3.6 nm in diameter; ~5 nm in length) connected to a transmembrane β-barrel (~2.6 nm in diameter; ~5 nm in length) (Fig. 1A)[7]. The pore is narrowest at the vestibule - transmembrane domain junction with a diameter of ~1.4 nm. Owing to its intrinsic nanopore structure, α-hemolysin has shown great potential in stochastic sensing of various analytes including metal ions[8,9], small organic molecules[10–12], DNA[13], RNA[14], proteins[15,16], and so forth. Due to the limitation of the pore size, the use of the α-hemolysin channel is restricted to the translocation of ssDNA (~1-nm in diameter).
Figure 1. Structure of three biological nanopores.
Side and top views of (A) heptameric α-hemolysin toxin from Staphylococcus aureus (PDB ID: 3ANZ)[7]; (B) octameric MspA porin from Mycobacterium smegmatis[18,20]; (C) dodecameric connector channel from bacteriophage phi29 DNA packaging motor[33]. In the figures, acidic (red), basic (blue), and other (white) amino acids are shown. Figures reproduced with permissions from: (B) Ref.[20], © The National Academy of Sciences of the USA; (C) Ref. [33], © Nature Publishing Group.
α-Hemolysin channel proteins can insert into the lipid bilayer spontaneously. It is thermally stable, functioning at temperatures close to 100°C[17]. α-Hemolysin can withstand a wide pH range (pH 2–12) while maintaining the nanopore structure. The α-hemolysin channels (in particular the β-barrel portion) are amenable to rational modifications by genetic engineering or chemical means for introducing specific binding elements.
(2) MspA channel
MspA (Mycobacterium smegmatis porin A) is a funnel shaped octameric channel pore which allows the transport of water soluble molecules across bacterial cell membranes. It contains a single constriction ~1.2 nm wide and 0.6 nm long (Fig. 1B), as revealed by the crystal structure published by the Neiderweis lab[18]. This fundamental finding has enabled all subsequent work with MspA including the site specific mutagenesis that has enabled nucleotide differentiation using MspA[19].
MspA channels can spontaneously insert into a planar bilayer to form a nanopore[20], similar to α-hemolysin. MspA is very robust and retains channel-forming activity at pH 0 – 14; after extraction at 100°C for 30 min; or even incubation at 80°C in presence of 2% SDS[20,21]. Since the crystal structure of is available[18], site-directed mutagenesis can be carried out to chemically reengineer mutant channels for desired applications[19,22,23].
(3) Phi29 Connector channel
The first demonstration of a nanochannel that is neither a membrane protein nor an ion channel inserted into a lipid bilayer was the phi29 connector protein, by the Guo lab[32]. The bacterial virus phi29 DNA-packaging nanomotor contains an elegant and elaborate channel composed of twelve copies of the protein gp10, which encircle to form a dodecamer channel[24–30] that acts as a path for the translocation of double-stranded DNA. The length of the connector is ~7 nm, while the cross-sectional area of the channel is 10 nm2 (3.6 nm in diameter) at the narrow end and 28 nm2 (6 nm in diameter) at its wider end (Fig. 1C)[24–26]. The mode of connector insertion and anchoring within the viral capsid is mediated via protein-protein interactions[26,31,32]. The connector has been inserted into a lipid bilayer and the resulting system has been shown to exhibit robust properties and generate extremely reliable, precise and sensitive conductance signatures when ions or DNA pass through the channel, as revealed by single channel conductance measurements (Fig. 2A–F) [33–38].
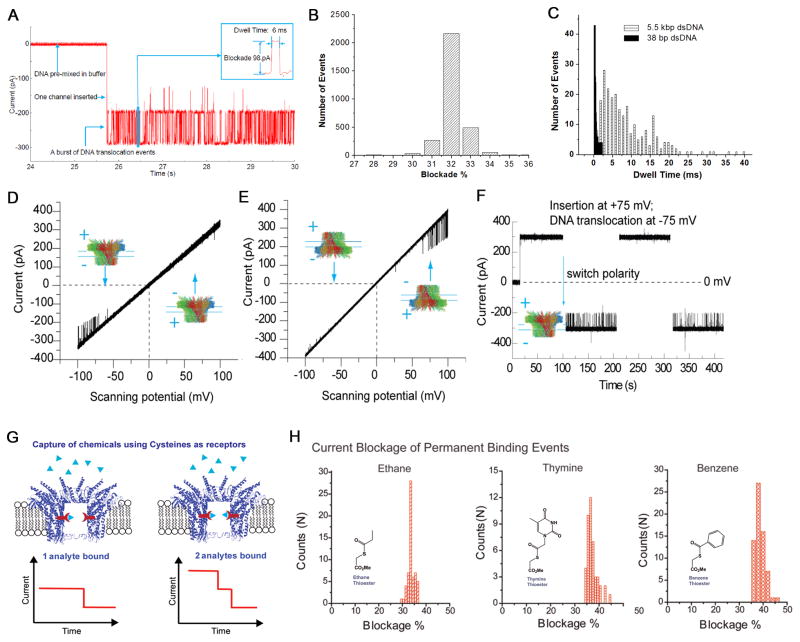
Figure 2. Application of phi29 connector channel for translocation of dsDNA and sensing of single chemicals.
(A) DsDNA translocation through membrane-embedded phi29 connector induced numerous current blockades. Insert: magnified image of a single translocation event. (B) Histogram of current blockade percentage induced by linear 2 kbp dsDNA. (C) Comparison of dwell times for translocation of 38 bp and 5.5 kbp dsDNA. One-way traffic in DNA translocation through a single connector channel in a lipid bilayer, examined under a ramping potential (D–E); and switching polarity (F). DNA is present in both cis- and trans-chambers. (G) Illustration of conjugation of chemical ligands to channel wall resulted in the reduction of channel size as indicated by uniform stepwise blockage of channel current. (H) Analysis of current blockage events induced by thioesters. Histogram of permanent binding events for the binding of thioesters groups containing ethane, thymine, and benzene respectively. Figures reproduced with permissions from: (A) Ref. [33], © Nature Publishing Group; (B) Ref. [35], © The Royal Society of Chemistry; (C) Ref. [33], © Nature Publishing Group; (D–F) Ref. [34], © American Chemical Society; (G–H) Ref. [38], © American Chemical Society.
The connector channel does not insert into the lipid bilayer by itself. A two step approach is required, whereby the connector is first reconstituted into lipid vesicles during the rehydration step, followed by vesicle fusion with a planar bilayer[33]. The conductance of each pore is almost identical and is perfectly linear with respect to the applied voltage. The connector channel is stable under a wide range of experimental conditions, including high salt and extreme pH [35]. The most significant advantage of the phi29 system, different from other well-studied systems, is that the phi29 connector has a larger channel allowing for the passage of ssDNA, dsDNA, peptides and possibly small proteins. The larger pore size is also advantageous in that it makes it easier for channel modifications to either create a sharper detection region for attaining single nucleotide resolution or for the insertion or conjugation of chemical groups for sensing and diagnostic applications (Fig. 2G–H) [38].
(B) Synthetic solid-state nanopores
Solid-state nanopores have emerged as a versatile alternative to biological nanopores because of their unique properties including well-defined geometries and dimensions, mechanical robustness, ease of modifications, and compatibility with various electronic or optical measurement techniques. The diameter of the solid-state nanopores can be precisely controlled ranging from sub-nanometers to hundreds of nanometers according to the experimental requirements. In general, solid-state nanopores in dielectric materials, such as SiN exhibit superior chemical and thermal stability over lipid membranes. However, their stability is dependent on the conditions used to form these pores[39]. As for graphene based nanopores, even though their chemical and thermal stability has not been demonstrated conclusively[40], its unique electrical properties is a big advantage over its biological counterparts. In recent years, solid-state nanopores have opened the door to a wide range of potential applications in DNA sequencing[41], monitoring protein interactions[42], controlling molecular transport, and fabrication of nanofluidic devices[43]. To date, various kinds of solid-state nanopore fabrication technologies have been developed, such as anodic oxidation method on the metal aluminum[44], ion-track-etching technology[45], ion beam sculpting[46] and electron beam fabrication[47–49] of nanopores in the thin synthetic membranes, and laser etching glass nanopores[50]. Detailed fabrication methods have been reviewed elsewhere[51].
(1) Silicon based substrates
Silicon nitride (SiN) and silicon dioxide (SiO2) membranes have over the years been used as a substrate due to its low mechanical stress and high chemical resistance. They are often fabricated by low-pressure chemical vapor deposition at high temperatures (800°C). Photolithography and standard wet-etching techniques are used to form a 100-μm × 100-μm window on the Si side, and focused electron beam is used to sputter atoms away from the SiN membrane to drill a hole in the center. The diameter of the nanopore can be precisely controlled according to the demands of the sensing system[47,52]. Conventional SiN membranes are typically ~30 nm thick. High salt containing conducting buffer is necessary to screen the negatively charged surface of the SiN or SiO2[53]. Linear I–V relationship are observed even at extremely high voltages. SiN pores have found use primarily in DNA translocation studies. They are potentially applicable to DNA sequencing but their application to sequencing has not yet been demonstrated.
(2) Aluminum Oxide (Al2O3) membranes
Using atomic layer deposition (ALD), ultra-thin pores can be fabricated in Al2O3 membranes with angstrom level control of membrane thickness (typically ~45–60 nm). Compared to SiN counterparts, Al2O3 surface is positively charged and exhibit superior mechanical attributes, better noise performance and higher lifetime[54–56]. The tunable thickness of the Al2O3 membranes and diameter of the nanopore makes it a competitive candidate for nanopore sensing.
(3) High dielectric-constant materials
Nanopores can be fabricated in titanium oxide (TiO2), and hafnium oxide (HfO2) membranes using ALD with angstrom level control[57]. These high dielectric-constant materials not only exhibit excellent electrical and mechanical properties, but also own a deposition temperature as low as 150°C. ALD of TiO2 onto graphene nanopores was shown to decrease the noise level of the graphene nanopore and improve its mechanical properties[58].
(4) Graphene sheets
Graphene is a 2D sheet of carbon atoms. Recently, 2–25 nm diameter pores have been fabricated in suspended graphene films, composed of one or two layers of carbon atoms and dsDNA was translocated through the pores (Fig. 3)[41,58–60]. Graphene nanopores are attractive for DNA sequencing due to its superior mechanical, electrical, and thermal properties, but more importantly they are of sub-nm thickness. As a result, the ionic current signal measured will be the convolution of only a few nucleotides in or around the pore, hence the higher spatial resolution versus other ‘thicker’ solid-state pores which accommodate significantly more DNA[61]. However, the access resistance (defined as the resistance through conducting solution from electrode to nanopore aperture) will contribute significantly to the measured conductance as the thickness of the pore approaches zero[62,63]. Furthermore, the graphene nanopore allows applying a lateral voltage across the nanopore membranes[60]. Lateral tunneling current measurements could theoretically be measured using nanofabricated graphene electrodes[64,65]. Since the tunneling current across the nanopore is related to the individual bases which go through the nanopore, the resolution of the nanopore DNA sequencing can be deeply enhanced.
Figure 3. DsDNA translocation through graphene nanopores.
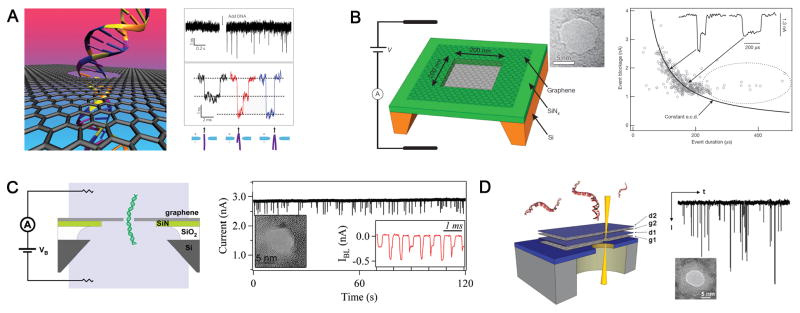
Illustration (left) and data (right) from (A) Dekker lab[41]; (B) Golovchenko lab[59]; (C) Drndic lab[58]; and (D) Bashir lab[60]. Figures reproduced with permissions from: (A) Refs.[41], © American Chemical Society, Refs.[153], © Nature Publishing Group; (B) Refs.[59], © Nature Publishing Group; (C) Refs.[58], © American Chemical Society; (D) Refs.[60], © American Chemical Society.
(C) Hybrid nanopores
The concept of hybrid pores was demonstrated by inserting α-hemolysin channel in SiN pores[66]. α-Hemolysin possesses an atomically precise structure and the potential for site-specific genetic engineering or chemical modifications. But α-hemolysin relies on fragile lipid bilayer membrane for mechanical support, which greatly limits their integration into wafer-scale devices. But this limitation is circumvented by placing a biological pore inside a mechanically robust solid-state nanopore thereby creating a hybrid pore with advantages of both worlds (Fig. 4A–C). The hybrid system was shown to be stable for several days under observation[66]. However, the biological pore deforms once in contact with the solid-state pore, loses the ability to discriminate single nucleotides and leakage currents are significant as ions can still flow through the regions between the solid-state and biological pore due to an imperfect seal. These factors limit this technology/approach.
Figure 4. DNA translocation through hybrid nanopores.
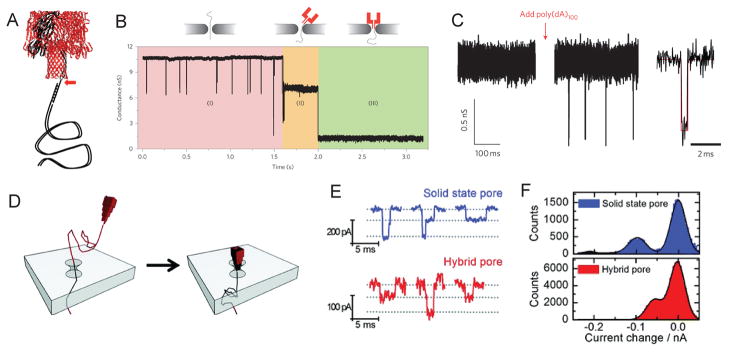
(A) α-hemolysin heteroheptamer with a 3 kbp dsDNA attached via a 12-nucleotide oligomer to one protein subunit. (B) Insertion of α-hemolysin protein pore into the solid state nanopore in three phases. (C) Current trace through a hybrid nanopore showing the baseline conductance directly after insertion (left) and upon addition of poly(dA)100 (middle). On the right is an expanded view of a typical event. (D) Schematic representation of the insertion of a DNA origami nanopore into a solid-state nanopore. (E) Typical events for the bare nanopore (blue) and the hybrid nanopore (red). (F) Current histograms indicating DNA translocations for the bare (blue) and hybrid nanopore (red). Figures reproduced with permissions from: (A–C) Ref. [66], © Nature Publishing Group; (D–F) Ref.[68], © American Chemical Society.
A interesting and novel approach is the insertion of 3D DNA origami structures into a conical SiN nanopore reproducibly and under controlled conditions (Fig. 4D–F) [67]. The method offers the possibility of programmable nanopores with tunable size and geometry for sensing applications. DNA origami based nanoplates has also been employed as gatekeepers of SiN membranes with additional chemical selectivity, albeit only proof of principle studies[68].
Comparison of biological and synthetic nanopores
While the basic principles of nanopores are the same, there are significant differences to note between biological and synthetic pores, as outlined below:
(1) Reproducibility
Biological channel proteins harvested in bacteria are extremely reproducible at the atomic level. The pore size cannot be reproduced as precisely by fabrication techniques in synthetic pores, although the precision with which nanopores in graphene can be formed is approaching atomic precision[69]. The size and shape of the synthetic pores can be tunable with sub-nanometer precision for desired applications, albeit variation can occur from batch to batch. Currently atomic layer deposition method of fabricating nanopores in ultra-thin membranes is very challenging due to ion current leakage[2].
(2) Channel size
Smaller channel such as MspA[22] may provide improved spatial resolution for DNA analysis due to its relatively short and narrow constriction, compared to α-hemolysin. However, larger channels, such as phi29 DNA packaging motor[33] could provide more room for modification and conjugation to display receptors for substrate binding.
(3) Stability of bedding or supporting substrate
A significant challenge for biological pores is the mechanically fragile nature of the lipid matrix due to weak intermolecular interactions, which can in part be circumvented by using polymer membranes[70], solid supports[71,72], or droplet-interface bilayer[73]. This is particularly relevant for integrating the biological pores in robust sensing solid state platforms for high throughput detection. Synthetic pore substrates on the other hand are far more durable.
(4) Pore Stability
Biological pores are often susceptible to extreme solution conditions, such as acidic/basic pH, low/high temperatures and low/high salt environments. Synthetic pores are highly stable chemically, thermally and mechanically over a wide range of experimental conditions. α-Hemolysin, MspA, and the connector protein channel of phi29 DNA packaging motor has been shown to be extremely robust and durable at room temperature. The protein samples from one production and purification batch can be stored in −80°C freezer for many years. Nevertheless, solid state nanopores should be stronger for manipulation.
(5) Cost
Biological pores can be harvested in cells in large quantities at low cost. Due to the overexpression and solubility, industrial large scale production have been possible. The cost of fabrication of synthetic pores has decreased a lot over the years; however the fabrication process is labor intensive. On the other hand, synthetic pores are reusable.
(6) Surface functionalization
Since the crystal structure of commonly used biological pores are known, site directed mutagenesis can be made to alter the size and charge properties of the channel. Furthermore, receptor modules can be conjugated to the pores at target locations within the channel lumen or on the surface with relative ease. Synthetic pores on the other hand generally lack chemical and location selectivity, although they can be functionalized to introduce chemical selectivity and location sensitivity[74–76].
(7) Integration into nanodevices
Biological nanopores are traditionally limited by the fragile nature of the supporting lipid substrate; however, polymer membranes, tethered substrates and even synthetic pores can allow them to be integrated into nanodevices. Solid-state nanopores can be readily integrated into nanofluidic or other nanodevices.
Current and Prospective Applications
Nanopore has the superiority to reach single molecule sensing due to its intrinsic properties, such as label-free, amplification-free, and potentials for high-throughput screening. As a result, they have become increasingly attractive for a wide range of applications, as discussed below.
(A) Sensing nucleic acid polymer structure and dynamics
The concept of nucleic acid transport in vitro via nanopores was first demonstrated more than 15 years ago using the α-hemolysin channel[77]. Single-stranded DNA was eletrophoretically driven through the pore and the translocation events were observed as current blockage events. Since then, using α-hemolysin system, the lengths of ssDNA and ssRNA have been identified[77]; homopolymers of poly-DNA and poly-RNA have been differentiated[78,79]. Over the last decade, the α-hemolysin channel has been extensively studied for probing the structure and dynamics of nucleic acids under a range of experimental conditions. Similar experiments have been performed uisng solid state nanopores to differentiate different lengths and even folded conformations of double-stranded DNA[80,81].
Four paramerters were primarily used either singly or in combination to charcaterize the DNA translocation events: current blockage amplitude; event duration or dwell time; exponential decay constant derived from the dwell time distribution; and distinctive electrical signature. Furthermore, the physics behind the DNA translocation process was investigated by force spectrosocpy using optical tweezers, which revealed a force of 0.23 pN mV−1 for dsDNA traversing through synthetic pores. Interestingly, the measured force was independent of the salt concentration used (0.2–1 M)[82].
(B) Towards DNA sequencing
The idea is rather simple: thread intact DNA strands through the nanopore and read off bases one at a time based on current amplitude to reveal the sequence of DNA. Nanopore-based DNA sequencing offers considerable advantage over traditional methods, since it is (i) label-free; (ii) does not require any amplifications; (iii) can provide high throughput reads; (iv) low cost; (v) requires very low sample volume; and, (vi) capable of achieving long DNA readouts. However, to resolve every single base in a long DNA strand has proven to be an insurmountable task. The limiting factor for the single pore DNA sequencing technology is that the ultra-fast DNA passage is beyond the temporal resolution of currently available optical and electrical technologies for detecting individual nucleotides with high sensitivity and confidence[1,2]. The key challenge therefore is to be able to slow the passage of DNA in a controlled fashion while maintaining high signal-to-noise ratio for accurately discriminating the bases. It is interesting to note that the speed of dsDNA translocation of the native phi29 DNA packaging nanomotor is within the detection limits (see perspectives).
Various passive methods either singly or in combination have been employed to slow the passage of DNA, such as (i) increasing the solution viscosity [83]; (ii) applying an asymetrical electro-osmotic flow gradient[84]; (iii) reducing applied voltage[85,86]; (iv) decreasing the temperature[79]; (v) tuning the charge distribution and size of the channel[20,37]; (vi) altering the pH of the conducting buffer[20]; (vii) using low melting point agarose matrix[87]; (viii) labeling of nucleotides[88,89]; (ix) introducing DNA hairpin bulges at the end of ssDNA[90]. However, in most cases, the signal-to-noise ratio was compromised with modest reduction in speed.
Recently, several active strategies have been employed to slow the passage of DNA in a controlled manner. These include the use of ssDNA terminated with a hairpin[19], electrically slowing down DNA using a gated nanopore[92,93]; using an enzyme motor/polymerase resulting in dsDNA synthesis at the pore[94]; and conjugation of a molecular adaptor, β-cyclodextrin (β-CD)[10,91] into the modified α-hemolysin channel via disulfide linkage to introduce specificity and constrict the channel and thereby identify all four mononucleotides[95,96].
The real potential for the development of next-generation DNA sequencing has so far been demonstrated by α-hemolysin[97], and MspA biological nanopore systems[23]. Controlled base-by-base ratcheting of ssDNA at slow speed was achieved by incorporating DNA polymerase to both channels. Oxford Nanopore is developing GridION and MinION systems on a chip to commercialize strand sequencing[98]. These systems are discussed in depth below.
(1) α-hemolysin channel
The nanopore DNA sequencing could be dated back to 1996, when Branton and colleagues first demonstrated the translocation of polynucleotide molecules through an α-hemolysin nanopore[77]. Early studies had shown that DNA/RNA homo- and copolymers could be readily differentiated by α-hemolysin, judging from their characteristic plot features, such as dwell time and current blockage amplitude[79]. Ideally, the ion current modulation should be limited to one nucleotide at a time to achieve single base resolution. However, the 5-nm long β-barrel of α-hemolysin can accommodate ca. 15 bases, all of which would contribute to the current blockades, precluding the possibility of base recognition[99]. Through immobilizing ssDNA in the nanochannel via biotin-streptavidin conjugation, three recognition sites in the β-barrel were disclosed: near the constriction, in the middle of the barrel, and near the trans-entrance[99]. Moreover, their recognition abilities could be finely tuned by rationally substituting amino acid residues in the recognition sites to modulate the electrostatic energy barriers for ion movement and enhance interactions of side chains with DNA[100]. Besides the four standard nucleobases, two modified bases of epigenetic significance, methylcytosines and hydroxymethylcytosines, were also readily identified in the nanopore[101]. Although this immobilization strategy renders α-hemolysin nanopores superior single-base identification in a given DNA strand, the discrimination is strictly limited to the position where the DNA base meets recognition site inside α-hemolysin.
DNA strands traverse α-hemolysin nanopore at a rate of several microseconds per base[79]. Several attempts have been made to slow down the translocation of DNA through α-hemolysin, including decreasing temperature[79,102], increasing viscosity[83], adding organic salts[103], equipping internal molecular brakes[88], and binding to an enzyme[94,104–106]. Among them, using of a DNA polymerase is of special interest which could halt the rapid slip of ssDNA and ratchet the template DNA through the nanochannel during synthesis, in which case the DNA translocation is governed by replication rate[106]. Initial success was achieved by using a bacteriophage T7 DNA polymerase, where successive current steps assigned to sequential nucleotide incorporation were observed when the DNA-polymerase complex was held on top of the nanopore. However, T7 polymerase suffers from low stability with DNA template under electrostatic force, resulting in a short detection window[107]. Later on, it was discovered that phi29 DNA polymerase could overcome this drawback, since it remains bound to DNA template while captured in the nanopore and sustains normal elongation ability[97]. The DNA replication rate is determined to be tens of milliseconds, which is long enough for accurate current probing, yet conditions need to be optimized to reduce insertion and deletion errors. Furthermore, this effective DNA control strategy could be readily integrated into other sequencing platforms[108].
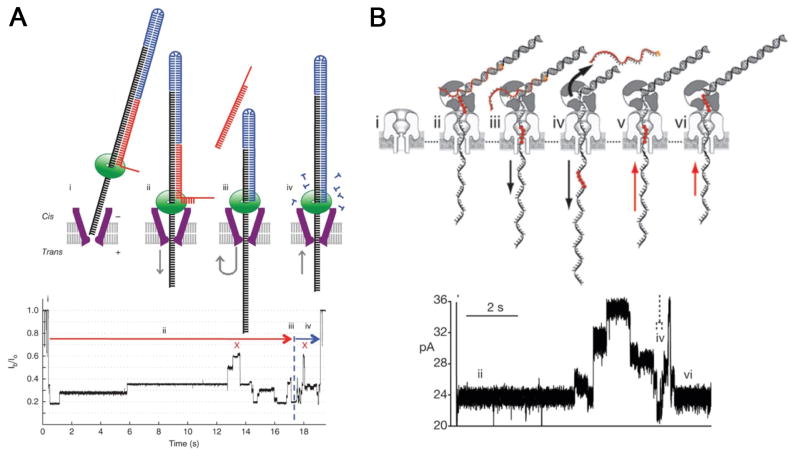
Other than the strand nanopore sequencing discussed above, another strategy involves the combination of an exonuclease which digests the DNA strand and a nanopore which discriminates different nucleotides, as proposed in the α-hemolysin system (Fig. 5A). The potential for accurate base discrimination was demonstrated by supplying mononucleotides into α-hemolysin equipped with a covalently attached amino-β-cyclodextrin (Fig. 5)[91,96]. However, no published data is available yet on direct exonuclease-nanopore sequencing and it still remains an open question as to how to appropriately fix the exonuclease enzyme at the pore entrance and guarantee every cleaved nucleotide being fed into the nanopore for sequencing in the correct order.
Figure 5. Proposed exonuclease-sequencing with a chimera of α-hemolysin and exonucleoase.
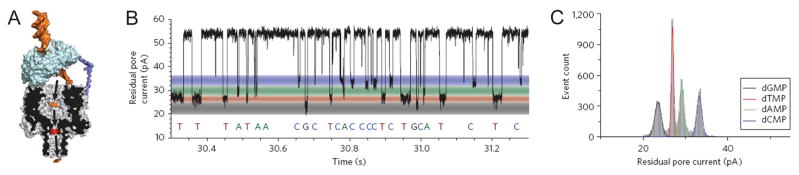
(A) An exonuclease (pale blue) attached to the top of an α-hemolysin pore through a genetically encoded (deep blue), or chemical, linker sequentially cleaves dNMPs (gold) off the end of a DNA strand. A dNMP’s identity (A, T, G or C) is determined by the level of the current blockade it causes when driven into an aminocyclodextrin adaptor (red) lodged within the pore. (B) Single-channel recording from the WT-(M113R/N139Q)6(M113R/N139Q/L135C)1-am6amDP1bCD pore upon addition of mono-nucleotides showing dGMP, dTMP, dAMP and dCMP discrimination, with colored bands added to represent the residual current distribution for each nucleotide. (C) Corresponding residual current histogram of nucleotide binding events, including Gaussian fits. Figures reproduced with permissions from (A) Ref. [1], © Nature Publishing Group; (B-C) Refs. [96], © Nature Publishing Group.
Oxford Nanopore Technologies
Early this year, the Oxford Nanopore Technologies, a company founded by Hagan Bayley, announced their intention to release a nanopore based sequencing platform for high throughput and ultra-long strand reads[98]. Their proposed technology revolves around strand sequencing, and preliminary data was presented at the AGBT (Advances in Genomic Biology and Technology) meeting in 2012. Exonuclease sequencing was initially proposed, but no data has been published yet.
(2) MspA pore
Gundlach’s group focused on sequencing with another biological pore, Mycobacterium smegmatis porin A (MspA). MspA is an outer membrane porin, containing a single constriction about 1.2 nm wide and 0.6 nm long[18], which could afford improved spatial resolution, compared to α-hemolysin. Prior to deciphering DNA, wild type MspA were engineered to remove the negative charges at the constriction and allow DNA translocation[20]. Without a barrel geometry to interact with DNA molecule, the average translocation rate of each nucleotide in MspA is less than one microsecond, even faster than the rate observed in α-hemolysin. Therefore, direct strand sequencing of DNA was unrealistic. Similar to the study in α-hemolysin, the immobilized ssDNA was held inside the MspA nanopore to study the nucleotide discrimination ability, either by a DNA duplex or a biotin-streptavidin complex, and a single recognition site covering four nucleotides was determined[19,20]. Single-nucleotide discrimination was achieved in MspA and the residual current differences between different bases observed in MspA were much larger than that in α-hemolysin, nearly a ten-fold enhancement. A real-time sequencing study with MspA has been reported recently, in which the robust phi29 DNA polymerase was employed to ratchet template DNA across MspA constriction base by base (Fig. 6A)[23]. A good correlation was observed between current levels and the sequence of a known DNA, and a single nucleotide substitution in a repeated motif could dramatically alter the current pattern, indicating a powerful base recognition ability of MspA. It should be noted that extraction of encoded information from a current pattern for an unknown sequence is by no means easy due to missing current steps as well as double counting. Nevertheless, the results demonstrated the great potential of MspA in nanopore sequencing.
Figure 6.
Strategies for reading DNA at single-nucleotide resolution using phi29 DNA polymerase to ratchet template DNA across (A) MspA nanopore and (B) α-hemolysin pore. Figures reproduced with permissions from: Ref. [23], © Nature Publishing Group; (B) Ref. [97], © Nature Publishing Group.
(3) Solid-state nanopore
As for solid-state nanopore-based technique, the magnitude of the ion current is mainly dependent on two factors: the effective pore diameter and the charge distribution inside the nanopore. Consequently, by suitably tuning the size, shape and thickness of the nanopore, the translocation events of DNA can be promptly and exactly detected through the decrease of the ion current. This is primarily due to the effective pore diameter being partially blocked by the translocating DNA molecules. Although direct sequencing has not yet been achieved due to the major challenges in the sequencing field, much effort has been directed toward developing highly effective nanopore-based platform for slowing down the DNA translocation speed and increasing the sensitivity of all four DNA nucleotides.
SiN has been a traditional material of choice owing to its low mechanical stress and high chemical resistance. Meller’s group reported a proof-of-principle studies of an ‘opti-pore’ technique capable of multi-color readout of DNA sequence[109]. Each of the four bases in the target DNA is represented by a binary code (2 bits/base), which is recognized by molecular beacons harboring two different fluorophores. The converted DNA and hybridized beacons is then translocated through 3–5 nm SiN nanopores, where the beacons are sequentially unzipped and the fluorescence is recorded using a custom TIRF (Total Internal Reflection) setup at the location of the nanopore. The method offers the possibility of implementing a four-color readout for indentifying the four bases, although technical challenges remain. Furthermore, multiple pores can potentially be probed at the same time for high-throughput readouts.
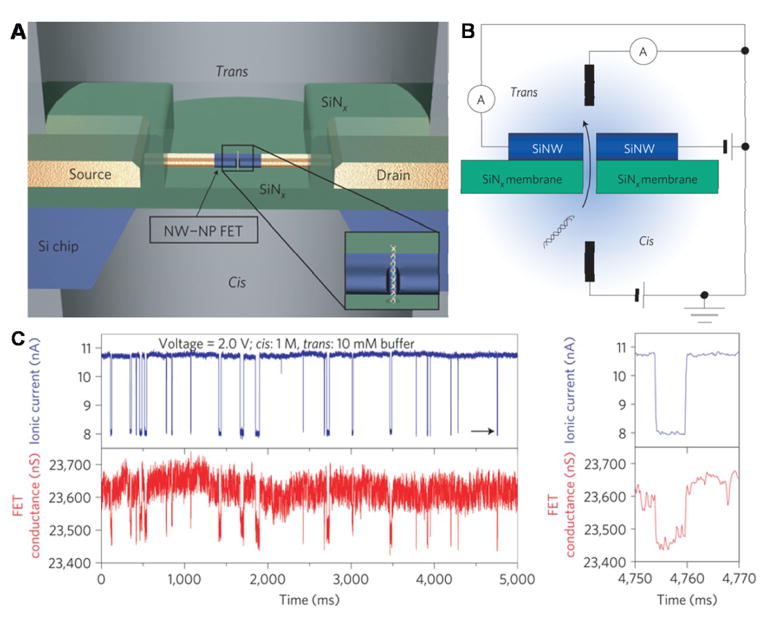
Xie et al. described a nanowire–nanopore field-effect transistors (FET) sensor comprising a silicon nanowire field-effect transistor on a SiN nanopore (Fig. 7)[110]. Notably, continuous DNA translocation events were observed simultaneously in both FET channel and ionic current channel without signal interference. Furthermore, data obtained from those two testing models showed nearly perfect consistency. Combining with excellent properties of FET, such as high intrinsic speed and high sensitivity, the sensor could enable large-scale integration and multiplexing with a high intrinsic bandwidth. Significantly, this nanowire–nanopore FET sensor also represents a promising designation for developing DNA sequencing devices with the capability of fast and sensitive sequencing by integrating and taking advantage of other useful techniques.
Figure 7. Nanowire–nanopore field-effect transistors (FET) sensor.
(A) Schematic of the sensor setup. Insert: magnified view at the nanopore. (B) Schematic illustration of the sensing circuit. (C) The real-time change of ionic current and FET conductance signals at 2.4 V voltage. Figures reproduced with permissions from Ref. [110], © Nature Publishing Group.
Golovchenko[59], Dekker[41] and Drndic[58] labs reported their works about DNA detection based on the graphene nanopore platform almost at the same time in 2010. They fabricated the graphene nanopore by using mechanical exfoliation from graphite method and chemical vapor deposition (CVD) method separately. Obvious ionic current blockages were observed when the double strand DNA translated through the graphene nanopore[41,58–60] (Fig. 3). The ionic current noise level of graphene nanopore was measured to be several orders of magnitude larger than those for silicon nitride nanopores. Drndic’s group found that by means of depositing several nanometers of titanium dioxide over the graphene nanopore, the noise could be significantly reduced. At the same time, the devices’ mechanical was greatly improved.
An alternative approach relies on identifying individual nucleotides as DNA passes through a nanogap between a pair of nanoelectrodes. The principle is based on electron transport through single nucleotides via tunneling current. Taniguchi and Kawai’s groups demonstrated that the transverse electrical conductivity was statistically different for each base, due to differences in highest occupied molecular orbital (HOMO) and lowest occupied molecular orbital (LOMO) of individual bases[108]. More recently, the same group reported a transverse electric field dragging approach to slow DNA translocation in a SiO2 nanopore[93]. Lindsay’s group reported that gold scanning tunneling microscope probe and substrate functionalized with a benzamide recognition reagent could identify single bases in an intact short DNA oligomer (Fig. 8A). Characteristic burst of tunneling current blockade events were observed and quantified based on frequency and amplitudes [111]. Further refinements associated with electron tunneling are needed to thread the DNA through the nanogap at a controlled fashion. The weak tunneling current signatures are often buried in the transmitted ionic currents as well. The transverse electric field also substantially changes the DNA translocation kinetics due to the contribution of an additional lateral electrostatic force, but this may be useful for probing dynamics of biomolecules. More recently, tunneling detectors have been integrated with nanopores, albeit only proof of principle studies (Fig. 8B)[112].
Figure 8. Electron tunneling approach for single base detection.
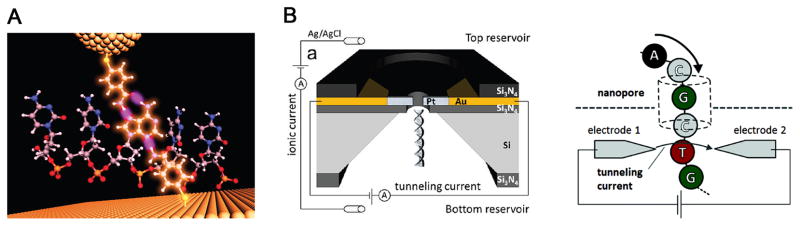
(A) Electron tunneling approach using a benzamide recognition group to identify single bases within a short DNA oligomer d(CCACC). A characteristic signal given by the shortest tunneling path is highlighted for the connection to the single A in d(CCACC). (B) Schematic of concurrent tunneling detection and ionic current detection of DNA molecules in a SiN nanopore platform. Figures reproduced with permissions from: (A) Ref [111], © Nature Publishing Group; (B) Ref. [112], © American Chemical Society.
(5) Translocation of dsDNA through membrane embedded phi29 motor channel
DsDNA translocation through the phi29 nanochannel was studied by electrophysiological measurements[33–37]. In presence of linear dsDNA of varying lengths (12–5000 bp), numerous transient current blockade events, each representing the translocation of a single dsDNA molecule were observed (Fig. 2A). The channel blockage percentage by one dsDNA was centered ~32% (Fig. 2B), which is consistent with the dimensions of the channel (3.6 nm in diameter at its narrowest end) and the dsDNA that is ~2 nm in diameter. Each transient current blockade event was observed to be nearly identical as observed by a sharp Gaussian distribution (Fig. 2B). When circular plasmid DNA (Cx43) was added, no such current blockade events were observed. However, upon digestion of circular plasmid DNA with DNaseI, a burst of DNA translocation events were observed, demonstrating that the phi29 channel is only capable of translocating linear dsDNA.
Challenges
(1) Speed of translocation
A bottleneck in all nanopore sequencing techniques is the ultra-fast translocation of DNA, which is well beyond the electronic detection limits of current technologies. Speeds of >10 nt/μs in solid state pores and >1 nt/μs in α-hemolysin and MspA pores have been reported[2]. Compared to freely translocating DNA, ratcheting ssDNA using DNA polymerase reduced the translocation speed by around four orders of magnitude.
(2) Spatial resolution
Typically the nanochannels investigated are several nanometers long; as a result 10–20 nucleotides reside within the channel lumen at any given point, which makes it difficult to distinguish the signal arising from a single nucleotide. Even with MspA pores, where the DNA is restricted to only a few bases at the constriction point, the current signatures were even more complicated with multiple current levels generated by the neighboring nucleotides.
(3) Temporal resolution
DNA motion is stochastic and there are fluctuations in translocation kinetics. In addition, non-specific interactions with the channel wall gives rise to a broad distribution of dwell times and different electrical signatures for identical length and sequences of DNA. These variations in kinetics generate uncertainties in the number of bases traversing the pore. Single-stranded DNA also displays different conformational dynamics which adds further complexities.
(4) Accuracy rates
The length of DNA that can be sequenced and the accuracy rate of reading the bases are of paramount importance for the successful application of the nanopores. This is a major hurdle not yet addressed in any systems to date. The α-hemolysin pore reported an error of 10–25%, while the MspA pore relies on a computational reference map to assign individual nucleotides. Furthermore, the processive fidelity of the DNA polymersase or exonucleases at longer DNA read lengths remain to be determined. With the Oxford Nanopore Technologies, an error rate of 4% was reported.
(C) A novel system for studying the mechanism of viral DNA packaging
The lipid-embedded phi29 connector represents a new system for understanding the mechanism of viral DNA packaging. The connector channel exercises a one-way traffic property for dsDNA translocation from N-terminal entrance (narrower-end) to C-terminal exit (wider-end), as demonstrated by voltage ramping, electrode polarity switching, and sedimentation force assessment (Fig. 2D–F)[34]. Interestingly, this is the same direction of DNA packaging in live virus. Furthermore, packaged viral dsDNA remains within the procapsid even under strong centrifugation forces[113]. Naturally, this phenomenon brings up a stimulating question of how DNA is expelled from the virus during the infection process. Indeed the connector exercises three step discrete conformational changes in regulating the direction of DNA translocation [37].
Structural studies of the phi29 connector revealed that there are 48 lysine residues in the inner channel (four 12-lysine rings from the twelve gp10 subunits). It was speculated that these positively charged rings play an important role in DNA translocation through the channel in that they may interact with the negatively charged phosphate backbone of dsDNA during DNA translocation [25,114]. Contrary to expectations, mutation of basic lysines to neutral alanine residues and changing of pH to extremely acidic or basic environments did not significantly affect the motor in DNA packaging. The results indicated that the four lysine rings within the phi29 connector channel are not involved in the active translocation of dsDNA [36]. The one-way traffic property in combination with studies on pRNA and the ATPase gp16 of phi29 DNA packaging motor elucidated the “Push through One-way valve” mechanism of viral DNA packaging[34,36,115,116]. In this model, the connector remains static; DNA translocation is induced by a DNA packaging enzyme or terminase, which pushes a certain length of DNA into the procapsid, and then shifts to bind to a far distal region of the DNA and inserts an additional section. The one-way traffic makes the DNA enter the procapsid and does not allow it to come out, similar to the pumping of blood into the heart and the use of valves to control the flow of blood.
(D) Real-time sensing of chemicals via molecular adapters and non-covalent/covalent interactions
Engineered trans-membrane channels have the potential for stochastic detection, an approach relying on the real-time observation of individual binding events between single substrate molecules and a receptor[117–119]. Protein pores can be selectively functionalized with various probes that can bind individual target molecules with high selectivity and sensitivity[120–122]. The characteristic binding and distinctive current signatures can reveal the identity and concentration of the target analyte[117,122]. In addition, the dynamic interactions between the analyte and the binding sites can be studied in real-time at high resolution using single channel conduction assays. Nanopores can overcome many of the well known limitations with regards to sensitivity and accuracy arising from background noise and nonspecific reactions. This is particularly relevant in cases where the target substrate is at a very low concentration or when impurities are present at high concentrations.
The application of nanopores as robust sensing devices for detecting biomolecules or chemicals was demonstrated recently using reengineered phi29 motor channel. Ethane (57 Da), thymine (167 Da), and benzene (105 Da) with reactive thioester moieties and maleimide were clearly discriminated upon interaction with the available set of cysteine residues introduced into the channel lumen via mutagenesis[38]. These studies demonstrate the feasibility of constructing multiple probes by engineering within a single pore for concurrent detection of multiple targets on a robust platform.
Incorporation of cyclodextrin molecular adapters into the α-hemolysin channel allowed the sensing of organic molecules and enantiomers of drug molecules in solution[10,123], and attachment of cyclic peptides enabled the binding of polyanions[124]. α-Hemolysin can be engineered to contain binding sites or reaction sites for specific analyte sensing. Braha and collegues engineered α-hemolysin to have four adjacent histidines for capturing divalent metal ions[8,125]. Current recordings showed that this single sensor element was able to simultaneously determine up to five different divalent metal ions, each of which producing characteristic signatures. Assays of binary and tertiary mixtures revealed that each type of metal ion interacts with α-hemolysin pores independently, and therefore their respective concentration could be determined. Meanwhile, Cheley et al. designed a mutant α-hemolysin with a ring of seven positively charged arginines near the constriction, which could specifically recognize phosphate ions[126]. With the incorporation of another seven arginines into the ring, inositol 1,4,5-trisphosphate, the Ca2+-mobilizing second messenger, could be detected at nanomolar concentration and free of interference from its analogs cAMP and ATP. Such noncovalent sensor elements were also prepared for trinitrotoluene sensing, and sample purity determination[12,127].
Other than non-covalent binding, covalent reactions have also been investigated for nanopore sensing in several cases, usually on cysteine residues. Shin et al. observed that 4-sulfophenylarsane oxide could reversibly form covalent bond with the thiol group on the cysteine residue in the β-barrel, and thus produce reversible current drops[128]. A linear relationship was derived between event frequency and analyte concentration, providing the basis for quantitative analysis. Wu et al. conducted detection of nitrogen mustards via the reactions between those toxic chemicals and single or multiple cysteine residues inside the α-hemolysin mutant[11]. Up to five reaction steps were observed from the sequential reactions of nitrogen mustards. Other examples of single molecular detection of chemicals include porphyrin macromolecules[129], immunoglobulin E, bioterrorist agent ricin[130], DNA polymerase[131], and so on[91,132].
Challenges
The limitation factor of various outlined techniques is the need for labels or tags for selective capture, such as biotin, streptavidine, histidine, and NTA-Ni2+, which is not present in vast majority of proteins or antigens. Low capture rate of analytes by the probes due to steric hindrance effects is another potential drawback. Nevertheless, the nanopore technique offers the capture and fingerprinting of analytes in real time at single molecule level, which cannot be achieved by traditional ensemble methods.
(E) Medical diagnostic applications
The principle of nanopore diagnostic approaches was demonstrated in solid-state pores for profiling microRNA expressions. The detection of specific miRNA sequences enriched from cellular tissues was comparable to conventional microarray technologies. The studies can be extended in the future for screening cancer biomarkers for early diagnosis and for montioring the progression of the diseases. Many of the medical diagnostic applications have been reviewed in detail in previous nanopore reviews[1,2].
RNA interference is becoming the focus of attention as it is highly related with RNA-mediated diseases. Drndic and co-workers reported a nanopore-based method for the analysis of RNA/Antibiotic complexes on the single molecule level[42]. Aptamers have been recently introduced into nanopore sensing field by the Gu group[133]. The thrombin-binding aptamer forms quadruplex structure in the presence of various monovalent and divalent metal ions, which can be trapped by the α-hemolysin cavity, offering single-molecule level folding/unfolding kinetic data[133,134]. Based on a similar rationale, the same group designed a programmable aptamer-like oligonucleotide probe for specific detection of a circulating microRNA, miR-155 (Fig. 9A)[135]. The hybridization of probe DNAs with miR-155 produced characteristic multilevel current signals upon translocating through α-hemolysin nanopores. This sensing platform enables monitoring microRNAs down to subpicomolar levels. Around the same time, Wen et al. reported a Hg2+-sensitive nanopore sensor based on high affinity DNA probes[9]. A properly designed random-coiled single-stranded DNA (ssDNA) was transformed into a stable hairpin conformation in the presence of Hg2+, which significantly altered its behavior during the translocation through α-hemolysin nanopore. Unlike hairpins, the three-way junction structures formed by cocaine and its aptamer could not be unzipped in α-hemolysin and caused a permanent blockage, based on which a high-speed cocaine sensor was created[136].
Figure 9. Medical diagnostics and sensing applications.
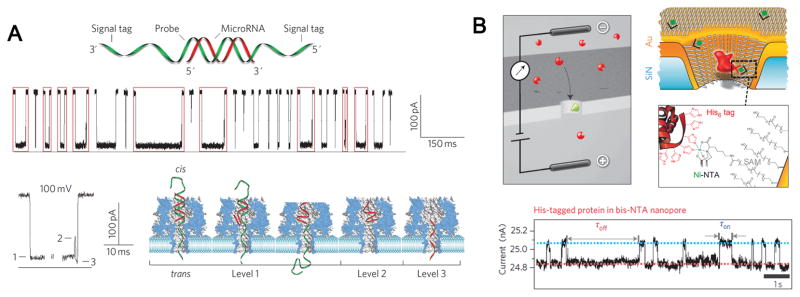
(A) Data showing α-hemolysin pore based detection of miRNA (miR-155) using programmable oligonucleotide probe (P155). The hybridization of probe with miRNA produced characteristic multi-level current signals upon translocating through α-hemolysin nanopores. (B) Gold-coated SiN nanopore functionalized with a mixed SAM of SC15EG3 matrix thiols and NTA receptor thiols for stochastic sensing of proteins. Charged analyte molecules (red) are electrokinetically driven through the pore and are detected by transient blockades in the ion current. When the pore is equipped with a specific receptor site (green), the blockade time reflects the binding time of the analyte (ligand) to the receptor. Figures reproduced with permissions from: (A) Ref [135], © Nature Publishing Group; (B) Ref. [76], © Nature Publishing Group.
Nanopore based detection of abnormal DNA methylation (epigenetic analysis) has been demonstrated using synthetic nanopores. DNA methylation is a valuable biomarker for assessing the metastatic potentials for many tumors. Methylation of MS3 and BRCA1, two DNA fragments, can cause disordered gene expression, leading to mutations and tumorigenesis. Mirsaidov et al. measured the translocation of the two DNA fragments across a SiN nanopore in their unmethylated, hemimethylated, and fully methylated state respectively. The results revealed the voltage threshold of the DNA across the membrane is highly related to its methylation level[137]. The nanopore based detection has the potential to be developed for applications in epigenetic diagnosis, since it simplified the analysis of methylation on a large scale compared to existing methods, such as MeDIP (methylated DNA immunoprecipitation) or HELP (HpaII tiny fragment enrichment by ligation-mediated PCR).
Single nucleotide polymorphisms (SNPs) are the cause of the phenotypic differences among individuals. There are many reports on the relationship between SNP and tumor development or progression. Thus, the detection of SNPs is of great significance to realize the early diagnosis of cancer. Nanopore can be used as a powerful tool to distinguish the SNPs due to its ability of DNA sensing. Timp’s group found that the voltage threshold for the enzyme-bound dsDNA permeating through ~5 nm SiN nanopore is sequence dependent. Based on this discovery, a single mutation in the recognition site was distinguished by comparing the voltage threshold. This practicable detection method provides a powerful tool to realize the point-of-care diagnostics[48].
For many diseases, the pathogen needs to be identified on the genomic level prior to medical therapy. The work reported by Meller and co-workers makes a contribution to the development of genomic level HIV subtype classification. They employed two γPNAs as probes. By altering the distance between two γPNA/DNA sites, the resulting time (δt) was observed to change. Thus the localization of the γPNA/DNA sites along the DNA molecule and effectively barcoding the target can be realized consequently. Four different γPNA oligomers was designed to successfully distinguish HIV-1/B from HIV-1/C through the barcoding method they built[138]. This method can be further developed to a feasible, label-free, diagnostic platform for other pathogen classification.
(F) Elucidate single molecule dynamics in RNA/protein, protein-protein or protein-ligand interactions
Gold plated SiN nanopores were uniformly coated with ethylene glycols, with a small fraction containing NTA functional groups (Fig. 9B)[76]. His-tagged Protein A, an immune pathogen were selectively captured onto nitrilotriacetic acid (NTA) receptor groups and then screened for the binding affinities of a variety of antibodies. Similar concept of capture and fingerprinting was demonstrated in biological pores, such as phi29 nanochannels. In presence of a His-tag at the C-terminal of the connector, the binding of proteins, such as Anti-His-tag antibody or Ni-NTA nanogold induced a conformational change of the connector, characterized by a 33% reduction in conductance per analyte binding[37].
Modulating the movement of a polymer chain inside and/or outside the lumen of α-hemolysin nanopore upon target molecule binding offers another approach for stochastic sensing. This concept was first demonstrated by Movileanu and collegues who functionalized α-hemolysin with a polyethylene glycol (PEG) chain bearing a biotin group at its free end[15]. The strong interactions between wild type streptavidin and biotin resulted in total diminishment of the large-amplitude spikes caused by the movement of free PEG chain (streptavidin, cis) or permanent partial channel block (streptavidin, trans). Xie et al. tethered an inhibitor peptide near the entrance of β-barrel, and successfully detected the binding of the catalytic subunit of cAMP-dependent protein kinase, characterized by long lifetime blockades[139]. Later on, a genetically coded kinase sensor was reported, in which the inhibitor peptide codons were integrated into the α-hemolysin subunit genome and expressed along with the protein. This provides a facile tool to construct α-hemolysin/peptide ligand hybrids and facilitates the study of ligand-protein interaction[16]. More recently, thrombin-binding aptamer was anchored near the mouth of α-hemolysin through hybridization to a covalently attached DNA adapter. Binding of the target thrombin to the aptamer was then demonstrated. Different analytes can therefore be detected simply by hybridizing corresponding aptamer ligands[140].
Some proteins, especially enzymes, can form complexes with their DNA/RNA substrates and participate in the subsequent DNA/RNA replication, repair, and transcription. Based on this knowledge, several protein sensors were reported. RNA-binding ARPase P4, a viral packaging motor, was able to recognize and bind to oligoribonucleotides. The P4-RNA complex was driven through α-hemolysin pore under voltage and induced current blockades with durations of hundreds of milliseconds, which was clearly different from bare RNA translocation[141]. Meanwhile, Hornblower et al. studied the ssDNA-exonuclease I interactions using nanopore force spectroscopy (NFS)[104]. Upon capturing a molecule, NFS automatically ramps the applied potential and an abrupt rise of current indicates rupture of bonds or dissociation of a complex. Energy barriers can be obtained by studying the relationship between most probable rupture voltages and loading rate. Cockroft and collegues performed a delicate experiment to monitor DNA polymerase activity[131]. They built a biotinylated ssDNA-PEG hybrid structure which was capped by streptavidin outside the β-barrel. The sequential nucleotide incorporation altered the ratio of DNA to PEG occupying the lumen, thus resulting in different current responses. A nine-base elongation process was resolved, where the cumulative current was only 1.3 pA.
(G) Nanopore for biomimetic applications
The ion channel embedded within the cell membranes is the communication medium with the extracellular world. These channels modulate the physical process by responding to the environmental stimuli and trigger conformational change by opening and closing the channel. Inspired by this principle, biomimetic nanopores have been widely developed as stimulate responsive switches[142], biological sensors[143], nanotubes[144], nanofilters[145] and energy harvest devices[146]. For a detailed review of biomimetic pores interested readers are referred to an excellent review[147].
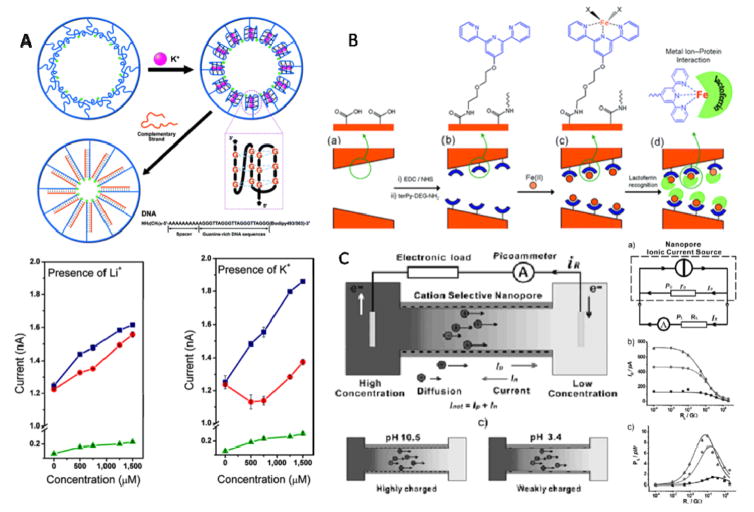
To make the solid-state nanopore realize its biomimetic potential and respond to the environmental stimuli, two major methods have been applied: (i) altering the interfacial structures or (ii) modifying the inner space of the nanopore with ‘smart’ materials. By adopting the former method, many biomimetic nanopores that can respond to the environmental stimulation have been developed, for example, pH, ions, light, temperature and so on. Jiang’s group reported a biomimetic nanochannel that can respond to potassium ions and the responsive signal is concentration dependent in a nonlinear fashion (Fig. 10A)[148]. In vivo, the G-rich telomere overhangs undergo conformational change when attached to the chromosome. Therefore, the grafting of the G-rich DNA onto the inner surface of the nanopore can closely imitate this condition. Accordingly, the G-quadruplex DNA was immobilized onto the inner surface of the PET nanopore. A nonlinear current–concentration curve was obtained indicating that the DNA strand undergoes conformational change from a loosely packed single-stranded structure to a densely packed rigid quadruplex structure. Such a system may spark the work towards simulation of the ion transport in living organism and this model can be generalized to more complex biomimetic nanochannel device in the future.
Figure 10. Examples of biomimetic application of solid-state nanopores.
(A) Top: Schematic representation of G4 DNA undergoing conformational change in the presence of K+, and after adding the complementary DNA, a closely packed double-strand DNA was formed, all processes are shown within the nanopore; Bottom: Current-concentration (I-C) characters of the single-conical PET nanopore before and after the G4 DNA molecule modified onto the nanopore inner space. (B) Schematic representation of nanopore-based lactoferrin sensor via the biorecognition of metal-chelating ligand. (C) Left: concentration gradient caused ion diffusion across the cation-selective nanopore; Right: supplying the electrical power produced by the nanopore system to an electric resistance RL, three lines represent different concentration gradient respectively. Figures reproduced with permissions from: (A) Ref. [148], © American Chemical Society; (B) Ref. [149], © American Chemical Society; (C) Ref. [150] © WILEY-VCH Verlag GmbH & Co.KGaA.
With grafting biorecognizable ligand onto the inner surface, nanopore-based sensing devices have been constructed for the biological detection ranging from protein, nucleic acids to drug compounds. A lactoferrin biosensor was developed using track-etched polyethylene terephthalate membranes (Fig. 10B)[149]. The nanopore was used to construct a nanobiosensor by covalently immobilizing amine-terminated terpyridine ligand-iron via carbodiimide coupling chemistry onto the nanopore inner surface for the protein recognition through the lactoferrin-iron specific interactions. This method can also be applied to other protein analysis system whose polypeptide backbone has the metal ion receptor sites. The label-free and amplitude-free nanopore-based detection of these molecules can be applied to the early diagnose of related diseases.
Miniaturization of energy supply devices is necessary for developing micro-electrical devices to meet growing industrial needs for portable personal electronic equipments. Inspired by the nature, an ionic gradient can be appropriately converted into sustainable electricity with the nanopore-based platform, as demonstrated by a bioelectricity system built using the conical polyimide nanopore membranes (Fig. 10C)[150]. Adopting a nanopore whose diameter is thinner than the electrical double layer inside the nanopore, it becomes cation-selective owing to the overlapping electrical double layer. Using different KCl electrolyte in the cis- and trans-chambers respectively, the spontaneous ion diffusion across the nanopore with preference for the cations can generate electrical power whose output can reach as high as 26 pW. What’s more, by tailoring the shape, surface chemistry, material and the symmetry of the nanopore, this device can be even more effective and produce higher throughput electric power which makes it an ideal candidate for clean-energy cell.
(H) Investigate translocations of other biopolymers, such as peptides and proteins
Translocation of polypeptides has added molecular complexities compared to polynucleic acids. The current blockage signature is dependent on the intrinsic properties of the polypeptide, such as length, cross-sectional area, charge, hydrophobic groups, secondary structure and the specific sequence of amino acids. β-Hairpin peptides displaying various stabilities were electrically driven through the α-hemolysin pore and their characteristic signature profiles were analyzed. Highly unfolded polypeptides traversed the pore in a single file fashion, while partially/fully folded ones displayed 2–3 fold longer dwell time[151]. The kinetics of polypeptide translocation were further examined by engineering electrostatic traps at the entry and exit of the α-hemolysin β-barrel[152]. However, energetic and entropic contributions need to be carefully examined to work out the complex interplay between polypeptide-pore interactions and pore mediated folding and unfolding at single molecule resolution. These studies are essential to lay the groundwork for future nanopore based proteomics.
Perspectives
Several first, second and third generation DNA sequencing approaches exist in the market, but they often require substantial biochemical labeling, extensive sample preparations, low throughput, requires massive data processing, costly, and not practical for long reads lengths. Nanopore technology has the potential to overcome many of the aforementioned challenges and significant strides has been towards the goal, even though challenges remain. The most promising data is from MspA and α-hemolysin channels utilizing DNA polymerase to rachet DNA base by base[23,97]. The DNA translocation speed was substantially slower and more importantly was within the limit of current detection technologies. However, the approach relies heavily on the processing ability of the enzyme. Controlling the kinetics of enzymatic activity is a major hurdle to overcome in order to achieve long read lengths in a reproducible manner. In addition, typically ~10–15 nt occupy the channel at any given time and give rise to complicated current signatures with a wide range of different current levels. Exonuclease based nanopore sequencing offers an attractive alternative. However, one still has to ensure high processing ability of the enzyme, and more importantly all cleaved nucleotides must enter the pore in the correct order.
An intriguing alternative is graphene, which is incredibly thin with only a few carbon atoms. The thickness is comparable to the spacing between nucleotides in single-stranded DNA. This notable characteristic makes graphene a promising candidate for nucleic acid detection and sequencing by using the nanopore platform. It’s electrical conductivity offers new possibilities, such as the application of tunneling current to probe the bases of DNA molecules. However, tunneling current approaches to detect individual bases has certain limitations, such as the electronic contrast between the bases; orientation of the bases relative to the detector; and need for relatively longer residence time to overcome electrical noise and stochastic molecular motions.
Interestingly, the speed of dsDNA translocation of the native phi29 DNA packaging motor is well within the current detection limit[153]. Using one ATP, the motor can package two base pairs of DNA [36,116,154]. The initial packaging rate was found to be 6 ms per base pair, which is 1000 fold slower than the speed of DNA driven by current through the single pore, and 10–20 fold slower than that of the motor of lambda and T4 phages [153]. Construction of a biomimetic artificial DNA packaging motor of bacteriophage phi29 offers the possibility of developing an active ATP-driven DNA-sequencing apparatus to advance the field of nucleic acid delivery by combining the best features of the native viral motor and the solid state silicon pore, while avoiding their accompanying limitations.
Other bottlenecks in the development of a practical nanopore technology is the instrumentation technology, which is limited by sampling frequency; current detection sensitivity; the depth of the focus, if optical detection is considered; and the channel size of the chamber, if a tunneling current is applied. Parallel readout of multiple nanopores is yet to be achieved for throughput readouts. Nevertheless, nanopore technology has made tremendous strides and we can fully expect wide ranging impact in the upcoming years, not only for DNA sequencing and sensing/diagnostic applications, but also in proteomics.
HIGHLIGHTS.
Nanopore based single molecule analysis is currently an area of great interest in many disciplines.
We review the concept of nanopores as well as types and attributes of various biological and synthetic nanopores.
We discuss the current and potential applications of nanopores in nanomedicine, biotechnology, and nanotechnology.
Acknowledgments
We thank Tao Zeng, Jing Yan, Juan Liu, for assistance in preparation of this review. Research in P.G. lab was supported by NIH R01 EB003730, R01 EB012135, U01 CA151648, R01 GM059944, and NIH Nanomedicine Development Center: Phi29 DNA Packaging Motor for Nanomedicine, through the NIH Roadmap for Medical Research (PN2 EY 018230) directed by P.G. Research in J.L. lab was supported by National Basic Research Program of China (No. 2011CB935704). Research in H-C W’s lab was supported by the “100 Talents” program of the Chinese Academy of Sciences, the National Natural Science Foundation of China (No. 21175135) and 973 programs (2009CB930200, 2010CB933600). P.G. is a co-founder of Kylin Therapeutics, Inc, and Biomotor and Nucleic Acids Nanotech Development, Ltd.
Biographies

Farzin Haque - Ph.D.
Dr. Haque is a Research Assistant Professor in the University of Kentucky College of Pharmacy, Department of Pharmaceutical Sciences. He received his B.A. degree in Biochemistry and Mathematics (2004) from Lawrence University and a Ph.D. degree in Chemistry (2008) from Purdue University. He held a postdoctoral appointment (2009–2011) at the University of Cincinnati, with Professor Peixuan Guo. Dr. Haque’s scholarly interest broadly focuses on Nanoscience and Nanotechnology in Biology and Medicine. These include, nanopore-based technology for single molecule detection and sensing of chemicals and biopolymers; and RNA Nanotechnology - construction of RNA nanoparticles for therapeutic and diagnostic applications.

Jinghong Li - Ph.D.
Dr. Li is Cheung Kong Professor in the Department of Chemistry at Tsinghua University, China. He received his Ph.D. from Changchun Institute of Applied Chemistry (CIAC), Chinese Academy of Sciences (CAS) in 1996. He held a postdoc or research scientist position at University of Illinois at Urbana-Champaign, University of California at Santa Barbara, Clemson University, and Evonyx Inc., USA (1997–2001). He has received several awards including National Science Fund for Distinguished Young Scholars, National Excellent Doctoral Dissertation of China, Distinguished Young Scholars for Chinese Academy of Sciences, the Young Electrochemistry Prize of Chinese Chemical Society, and the Li Foundation Prize, USA. His research interests include electroanalytical chemistry, bioelectrochemistry and sensors, physical electrochemistry and interfacial electrochemistry, electrochemical materials science and nanoscopic electrochemistry, fundamental aspects of energy conversion and storage, advanced battery materials, and photoelectrochemistry. He has published over 230 papers in international, peer-reviewed journals with >10 invited review articles. http://www.researcherid.com/rid/D-4283-2012.

Hai-Chen Wu - Ph.D.
Dr. Wu obtained his Ph.D. in organic chemistry from University of Cambridge (Cambridge, UK) in 2005. He then moved to Oxford and carried out his postdoctoral research under the supervision of Prof. Hagan Bayley in Department of Chemistry, University of Oxford until the end of 2008. Since early 2009, he has been a principal investigator at the Key Laboratory for Biomedical Effects of Nanomaterials & Nanosafety, Institute of High Energy Physics, Chinese Academy of Sciences (Beijing, China). Dr Wu’s main research interest focuses on modification of nonmaterials and applying them in biosensing studies.

Xing-Jie Liang - Ph.D.
Dr. Liang obtained his Ph.D at National Key Laboratory of Biomacromolecules, Institute of Biophysics at CAS. He finished his postdoc at Center for Cancer Research, NCI, NIH, and worked as a Research Fellow at Surgical Neurology Branch, NINDS. Dr. Liang worked on Molecular imaging at School of Medicine, Howard University before joining as deputy director of CAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, National Center for Nanoscience and Technology of China. Dr. Liang is current editorial board member of Acta Biophysica Sinica, and Current Nanoscience. Developing drug delivery strategies for prevention/treatment of AIDS and cancers are ongoing research projects in Dr. Liang’s lab based on understanding of basic physio-chemical and biological processes of nanomedicine.

Peixuan Guo, Ph.D.
Dr. Guo is William Farish Endowed Chair in Nanobiotechnology, and director of the Nanobiotechnology center at the university of Kentucky, and director of NIH/NCI Cancer Nanotechnology Platform Partnership Program: “RNA Nanotechnology for Cancer Therapy”. He obtained his Ph.D from University of Minnesota, and postdoctoral training at NIH, joined Purdue University in 1990, was tenured in 1993, became a full Professor in 1997, and was honored as a Purdue Faculty Scholar in 1998. He constructed phi29 DNA-packaging motor, discovered phi29 motor pRNA, pioneered RNA nanotechnology, incorporated phi29 motor channel into lipid membranes for single-molecule sensing with potential for high-throughput dsDNA sequencing. He is a member of two US prominent national nanotech initiatives sponsored by NIH, NSF, NIST, and National Council of Nanotechnology, director of one NIH Nanomedicine Development Center from 2006–2011. His work was featured hundreds of times over radio, TV such as ABC, NBC, newsletters NIH, NSF, MSNBC, NCI, and ScienceNow. http://www.eng.uc.edu/nanomedicine/peixuanguo.html
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. Nat Biotechnol. 2008;26:1146. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkatesan BM, Bashir R. Nature Nanotechnology. 2011;6:615. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 3.Healy K. Nanomedicine. 2007;2:459. doi: 10.2217/17435889.2.4.459. [DOI] [PubMed] [Google Scholar]
- 4.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Current Opinion in Biotechnology. 2010;21:439. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iqbal SM, Bashir R. Nanopores: Sensing and Fundamental Biological Interactions. Springer; London: 2011. [Google Scholar]
- 6.Coulter WH. 1953 [Google Scholar]
- 7.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 8.Braha O, Gu LQ, Zhou L, Lu X, Cheley S, Bayley H. Nat Biotechnol. 2000;18:1005. doi: 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- 9.Wen S, Zeng T, Liu L, Zhao K, Zhao Y, Liu X, Wu HC. J Am Chem Soc. 2011;133:18312. doi: 10.1021/ja206983z. [DOI] [PubMed] [Google Scholar]
- 10.Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]
- 11.Wu HC, Bayley H. J Am Chem Soc. 2008;130:6813. doi: 10.1021/ja8004607. [DOI] [PubMed] [Google Scholar]
- 12.Guan X, Gu LQ, Cheley S, Braha O, Bayley H. Chembiochem. 2005;6:1875. doi: 10.1002/cbic.200500064. [DOI] [PubMed] [Google Scholar]
- 13.Howorka S, Cheley S, Bayley H. Nat Biotechnol. 2001;19:636. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 14.Japrung D, Henricus M, Li Q, Maglia G, Bayley H. Biophys J. 2010;98:1856. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Movileanu L, Howorka S, Braha O, Bayley H. Nature Biotechnology. 2000;18:1091. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- 16.Cheley S, Xie H, Bayley H. Chembiochem. 2006;7:1923. doi: 10.1002/cbic.200600274. [DOI] [PubMed] [Google Scholar]
- 17.Kang XF, Gu LQ, Cheley S, Bayley H. Angew Chem Int Ed Engl. 2005;44:1495. doi: 10.1002/anie.200461885. [DOI] [PubMed] [Google Scholar]
- 18.Faller M, Niederweis M, Schulz GE. Science. 2004;303:1189. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 19.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Proc Natl Acad Sci U S A. 2010;107:16060. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Proc Natl Acad Sci U S A. 2008;105:20647. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz C, Engelhardt H, Niederweis M. J Biol Chem. 2003;278:8678. doi: 10.1074/jbc.M212280200. [DOI] [PubMed] [Google Scholar]
- 22.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Proceedings of the National Academy of Sciences. 2008;105:20647. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. Nat Biotechnol. 2012;30:349. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossman MG. Acta Cryst D. 2001;57:1260. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- 25.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. J Mol Biol. 2002;315:663. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Blocker F, Guo P. J Nanosci Nanotechnol. 2005;5:856. doi: 10.1166/jnn.2005.143. [DOI] [PubMed] [Google Scholar]
- 27.Carazo J, Santisteban A, Carrascosa J. J Mol Biol. 1985;183:79. doi: 10.1016/0022-2836(85)90282-7. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez J, Santisteban A, Carazo JM, Carrascosa JL. Science. 1986;232:1113. doi: 10.1126/science.3754654. [DOI] [PubMed] [Google Scholar]
- 29.Carazo JM, Donate LE, Herranz L, Secilla JP, Carrascosa JL. J Mol Biol. 1986;192:853. doi: 10.1016/0022-2836(86)90033-1. [DOI] [PubMed] [Google Scholar]
- 30.Valle M, Kremer L, Martinez A, Roncal F, Valpuesta JM, Albar JP, Carrascosa JL. J Mol Biol. 1999;288:899. doi: 10.1006/jmbi.1999.2731. [DOI] [PubMed] [Google Scholar]
- 31.Lee CS, Guo P. J Virol. 1995;69:5024. doi: 10.1128/jvi.69.8.5024-5032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu C, Prevelige P. Virology. 2009;394:149. doi: 10.1016/j.virol.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wendell D, Jing P, Geng J, Subramaniam V, Lee TJ, Montemagno C, Guo P. Nature Nanotechnology. 2009;4:765. doi: 10.1038/nnano.2009.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing P, Haque F, Shu D, Montemagno C, Guo P. Nano Lett. 2010;10(9):3620. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jing P, Haque F, Vonderheide A, Montemagno C, Guo P. Molecular BioSystems. 2010;6:1844. doi: 10.1039/c003010d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang H, Jing P, Haque F, Guo P. Biophysical Journal. 2012;102:127. doi: 10.1016/j.bpj.2011.11.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geng J, Fang H, Haque F, Zhang L, Guo P. Biomaterials. 2011;32:8234. doi: 10.1016/j.biomaterials.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haque F, Lunn J, Fang H, Smithrud D, Guo P. ACS Nano. 2012;6:3251. doi: 10.1021/nn3001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den HM, Hall AR, Wu MY, Zandbergen HW, Dekker C, Dekker NH. nanotechnology. 2010;21:115304. doi: 10.1088/0957-4484/21/11/115304. [DOI] [PubMed] [Google Scholar]
- 40.Lu N, Wang JG, Floresca HC, Kim MJ. Carbon. 2012;50:2961. [Google Scholar]
- 41.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LM, Dekker C. Nano Lett. 2010;10:3163. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 42.Wanunu M, Bhattacharya S, Xie Y, Tor Y, Aksimentiev A, Drndic M. ACS Nano. 2011;5:9345. doi: 10.1021/nn203764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo W, Xia HW, Cao LX, Xia F, Wang ST, Zhang GZ, Song YL, Wang YG, Jiang L, Zhu DB. Advanced Functional Materials. 2010;20:3561. [Google Scholar]
- 44.Yuan JH, He FY, Sun DC, Xia XH. Chemistry of Materials. 2004;16:1841. [Google Scholar]
- 45.Siwy Z, Gu Y, Spohr HA, Baur D, Wolf-Reber A, Spohr R, Apel P, Korchev YE. Europhysics Letters. 2002;60:349. [Google Scholar]
- 46.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Nature. 2001;412:166. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 47.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nat Mater. 2003;2:537. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Nano Letters. 2007;7:1680. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, Grinkova YV, Sligar S, Schulten K, Timp G. Biophysical Journal. 2004;87:2905. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White HS, Bund A. Langmuir. 2008;24:2212. doi: 10.1021/la702955k. [DOI] [PubMed] [Google Scholar]
- 51.Gyurcsanyi RE. Trac-Trends in Analytical Chemistry. 2008;27:627. [Google Scholar]
- 52.Kim MJ, Wanunu M, Bell DC, Meller A. Adv Mater. 2006;18:3149. [Google Scholar]
- 53.Dekker C. Nature Nanotechnology. 2007;2:209. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 54.Chen P, Mitsui T, Farmer DB, Golovchenko J, Gordon RG, Branton D. Nano Letters. 2004;4:1333. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkatesan BM, Dorvel B, Yemenicioglu S, Watkins N, Petrov I, Bashir R. Adv Mater. 2009;21:2771. doi: 10.1002/adma.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Venkatesan BM, Shah AB, Zuo JM, Bashir R. Advanced Functional Materials. 2010;20:1266. doi: 10.1002/adfm.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knez M, Niesch K, Niinisto L. Advanced Materials. 2007;19:3425. [Google Scholar]
- 58.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson AT, Drndic M. Nano Lett. 2010;10:2915. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 59.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Nature. 2010;467:190. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae MH, Aluru NR, Pop E, Bashir R. ACS Nano. 2012;6:441. doi: 10.1021/nn203769e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells DB, Belkin M, Comer J, Aksimentiev A. Nano Lett. 2012;12:4117. doi: 10.1021/nl301655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall JE. J Gen Physiol. 1975;66:531. doi: 10.1085/jgp.66.4.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vodyanoy I, Bezrukov SM. Biophys J. 1992;62:10. doi: 10.1016/S0006-3495(92)81762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Postma HW. Nano Lett. 2010;10:420. doi: 10.1021/nl9029237. [DOI] [PubMed] [Google Scholar]
- 65.Prasongkit J, Grigoriev A, Pathak B, Ahuja R, Scheicher RH. Nano Lett. 2011;11:1941. doi: 10.1021/nl200147x. [DOI] [PubMed] [Google Scholar]
- 66.Hall AR, Scott A, Rotem D, Mehta KK, Bayley H, Dekker C. Nature Nanotechnology. 2010;5:874. doi: 10.1038/nnano.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei R, Martin TG, Rant U, Dietz H. Angew Chem Int Ed Engl. 2012;51:4864. doi: 10.1002/anie.201200688. [DOI] [PubMed] [Google Scholar]
- 68.Bell NA, Engst CR, Ablay M, Divitini G, Ducati C, Liedl T, Keyser UF. Nano Lett. 2012;12:512. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- 69.Song B, Schneider GF, Xu Q, Pandraud G, Dekker C, Zandbergen H. Nano Lett. 2011;11:2247. doi: 10.1021/nl200369r. [DOI] [PubMed] [Google Scholar]
- 70.Shenoy DK, Barger WR, Singh A, Panchal RG, Misakian M, Stanford VM, Kasianowicz JJ. Nano Lett. 2005;5:1181. doi: 10.1021/nl050481q. [DOI] [PubMed] [Google Scholar]
- 71.Cornell BA, Braach-Maksvytis VL, King LG, Osman PD, Raguse B, Wieczorek L, Pace RJ. Nature. 1997;387:580. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 72.Schiller SM, Naumann R, Lovejoy K, Kunz H, Knoll W. Angew Chem Int Ed Engl. 2003;42:208. doi: 10.1002/anie.200390080. [DOI] [PubMed] [Google Scholar]
- 73.Holden MA, Needham D, Bayley H. J Am Chem Soc. 2007;129:8650. doi: 10.1021/ja072292a. [DOI] [PubMed] [Google Scholar]
- 74.Kowalczyk SW, Kapinos L, Blosser TR, Magalhaes T, van NP, Lim RY, Dekker C. Nat Nanotechnol. 2011;6:433. doi: 10.1038/nnano.2011.88. [DOI] [PubMed] [Google Scholar]
- 75.Iqbal SM, Akin D, Bashir R. Nature Nanotechnology. 2007;2:243. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 76.Wei R, Gatterdam V, Wieneke R, Tampe R, Rant U. Nat Nanotechnol. 2012;7:257. doi: 10.1038/nnano.2012.24. [DOI] [PubMed] [Google Scholar]
- 77.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci U S A. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci U S A. 2000;97:1079. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 81.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. Nature Mater. 2003;2:611. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 82.Keyser UF, Koeleman BN, Van Dorp S, Krapf D, Smeets RMM, Lemay SG, Dekker NH, Dekker C. Nature Physics. 2006;2:473. [Google Scholar]
- 83.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Nano Lett. 2005;5:1734. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Nature Nanotechnology. 2010;5:160. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henrickson SE, Misakian M, Robertson B, Kasianowicz JJ. Phys Rev Lett. 2000;85:3057. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 86.Meller A, Branton D. Electrophoresis. 2002;23:2583. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 87.Heron AJ, Thompson JR, Cronin B, Bayley H, Wallace MI. J Am Chem Soc. 2009;131:1652. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 88.Rincon-Restrepo M, Mikhailova E, Bayley H, Maglia G. Nano Lett. 2011;11:746. doi: 10.1021/nl1038874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchell N, Howorka S. Angew Chem Int Ed Engl. 2008;47:5565. doi: 10.1002/anie.200800183. [DOI] [PubMed] [Google Scholar]
- 90.Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nat Biotech. 2001;19:248. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 91.Astier Y, Braha O, Bayley H. J Am Chem Soc. 2006;128:1705. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 92.Paik KH, Liu Y, Tabard-Cossa V, Waugh MJ, Huber DE, Provine J, Howe RT, Dutton RW, Davis RW. ACS Nano. 2012;6:6767. doi: 10.1021/nn3014917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsutsui M, He Y, Furuhashi M, Rahong S, Taniguchi M, Kawai T. Sci Rep. 2012;2:394. doi: 10.1038/srep00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benner S, Chen RJA, Wilson NA, bu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Nature Nanotechnology. 2007;2:718. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu HC, Astier Y, Maglia G, Mikhailova E, Bayley H. J Am Chem Soc. 2007;129:16142. doi: 10.1021/ja0761840. [DOI] [PubMed] [Google Scholar]
- 96.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Nat Nanotechnol. 2009;4:265. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 97.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Nat Biotechnol. 2012;30:344. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eisenstein M. Nat Biotechnol. 2012;30:295. doi: 10.1038/nbt0412-295. [DOI] [PubMed] [Google Scholar]
- 99.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Proc Natl Acad Sci U S A. 2009;106:7702. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stoddart D, Heron AJ, Klingelhoefer J, Mikhailova E, Maglia G, Bayley H. Nano Lett. 2010;10:3633. doi: 10.1021/nl101955a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Chem Commun (Camb) 2010;46:8195. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Proc Natl Acad Sci U S A. 2005;102:12377. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.de Zoysa RS, Jayawardhana DA, Zhao Q, Wang D, Armstrong DW, Guan X. J Phys Chem B. 2009;113:13332. doi: 10.1021/jp9040293. [DOI] [PubMed] [Google Scholar]
- 104.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Nat Methods. 2007;4:315. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 105.Hurt N, Wang H, Akeson M, Lieberman KR. J Am Chem Soc. 2009;131:3772. doi: 10.1021/ja809663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Nat Nanotechnol. 2010;5:798. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. J Am Chem Soc. 2010;132:17961. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Nat Nanotechnol. 2010;5:286. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 109.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Nano Lett. 2010;10:2237. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie P, Xiong Q, Fang Y, Qing Q, Lieber CM. Nat Nanotechnol. 2012;7:119. doi: 10.1038/nnano.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang S, He J, Chang S, Zhang P, Liang F, Li S, Tuchband M, Fuhrmann A, Ros R, Lindsay S. Nat Nanotechnol. 2010;5:868. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov AP, Instuli E, McGilvery CM, Baldwin G, McComb DW, Albrecht T, Edel JB. Nano Lett. 2011;11:279. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shu D, Guo P. Virology. 2003;309(1):108. doi: 10.1016/s0042-6822(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 114.Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. J Mol Biol. 2005;347:895. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 115.Zhang H, Schwartz C, De Donatis GM, Guo P. Adv Virus Res. 2012;83:415. doi: 10.1016/B978-0-12-394438-2.00009-8. [DOI] [PubMed] [Google Scholar]
- 116.Schwartz C, Fang H, Huang L, Guo P. Nucleic Acids Res. 2012;40:2577. doi: 10.1093/nar/gkr841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bayley H, Cremer PS. Nature. 2001;413:226. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 118.Raj A, van OA. Annu Rev Biophys. 2009;38:255. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perkins TJ, Swain PS. Mol Syst Biol. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thomson K, Amin I, Morales E, Winters-Hilt S. BMC Bioinformatics. 2007;8(Suppl 7):S11. doi: 10.1186/1471-2105-8-S7-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Winters-Hilt S. BMC Bioinformatics. 2006;7(Suppl 2):S21. doi: 10.1186/1471-2105-7-S2-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bayley H, Martin CR. Chem Rev. 2000;100:2575. doi: 10.1021/cr980099g. [DOI] [PubMed] [Google Scholar]
- 123.Kang XF, Cheley S, Guan X, Bayley H. J Am Chem Soc. 2006;128:10684. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 124.Sanchez-Quesada J, Ghadiri MR, Bayley H, Braha O. J Am Chem Soc. 2000;122:11757. [Google Scholar]
- 125.Braha O, Walker B, Cheley S, Kasianowicz JJ, Song L, Gouaux JE, Bayley H. Chem Biol. 1997;4:497. doi: 10.1016/s1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- 126.Cheley S, Gu LQ, Bayley H. Chem & Biol. 2002;9:829. doi: 10.1016/s1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- 127.Krishantha DM, Breitbach ZS, Padivitage NL, Armstrong DW, Guan X. Nanoscale. 2011;3:4593. doi: 10.1039/c1nr10974j. [DOI] [PubMed] [Google Scholar]
- 128.Shin SH, Luchian T, Cheley S, Braha O, Bayley H. Angew Chem Int Ed Engl. 2002;41:3707. doi: 10.1002/1521-3773(20021004)41:19<3707::AID-ANIE3707>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 129.Heins EA, Siwy ZS, Baker LA, Martin CR. Nano Lett. 2005;5:1824. doi: 10.1021/nl050925i. [DOI] [PubMed] [Google Scholar]
- 130.Ding S, Gao C, Gu LQ. Anal Chem. 2009:6649. doi: 10.1021/ac9006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cockroft SL, Chu J, Amorin M, Ghadiri MR. J Am Chem Soc. 2008;130:818. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baaken G, Ankri N, Schuler AK, Ruhe J, Behrends JC. ACS Nano. 2011;5:8080. doi: 10.1021/nn202670z. [DOI] [PubMed] [Google Scholar]
- 133.Shim JW, Gu LQ. J Phys Chem B. 2008;112:8354. doi: 10.1021/jp0775911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shim JW, Tan Q, Gu LQ. Nucleic Acids Res. 2009;37:972. doi: 10.1093/nar/gkn968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nat Nanotechnol. 2011;6:668. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kawano R, Osaki T, Sasaki H, Takinoue M, Yoshizawa S, Takeuchi S. J Am Chem Soc. 2011;133:8474. doi: 10.1021/ja2026085. [DOI] [PubMed] [Google Scholar]
- 137.Mirsaidov U, Timp W, Zou X, Dimitrov V, Schulten K, Feinberg AP, Timp G. Biophysical Journal. 2009;96:L32–L34. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nano Lett. 2010;10:738. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xie H, Braha O, Gu LQ, Cheley S, Bayley H. Chem Biol. 2005;12:109. doi: 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 140.Rotem D, Jayasinghe L, Salichou M, Bayley H. J Am Chem Soc. 2012;134:2781. doi: 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Astier Y, Kainov DE, Bayley H, Tuma R, Howorka S. Chemphyschem. 2007;8:2189. doi: 10.1002/cphc.200700179. [DOI] [PubMed] [Google Scholar]
- 142.Xia F, Guo W, Mao Y, Hou X, Xue J, Xia H, Wang L, Song Y, Ji H, Ouyang Q, Wang Y, Jiang L. J Am Chem Soc. 2008;130:8345. doi: 10.1021/ja800266p. [DOI] [PubMed] [Google Scholar]
- 143.Han C, Hou X, Zhang H, Guo W, Li H, Jiang L. J Am Chem Soc. 2011;133:7644. doi: 10.1021/ja2004939. [DOI] [PubMed] [Google Scholar]
- 144.Kohli P, Harrell CC, Cao Z, Gasparac R, Tan W, Martin CR. Science. 2004;305:984. doi: 10.1126/science.1100024. [DOI] [PubMed] [Google Scholar]
- 145.Goyal S, Kim YT, Li Y, Iqbal SM. Biomed Microdevices. 2010;12:317. doi: 10.1007/s10544-009-9387-4. [DOI] [PubMed] [Google Scholar]
- 146.Wu JZ, Emergo RLS, Wang X, Xu G, Haugan TJ, Barnes PN. Applied Physics Letters. 2008;93 [Google Scholar]
- 147.Kowalczyk SW, Blosser TR, Dekker C. Trends Biotechnol. 2011;29:607. doi: 10.1016/j.tibtech.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 148.Hou X, Guo W, Xia F, Nie FQ, Dong H, Tian Y, Wen L, Wang L, Cao L, Yang Y, Xue J, Song Y, Wang Y, Liu D, Jiang L. J Am Chem Soc. 2009;131:7800. doi: 10.1021/ja901574c. [DOI] [PubMed] [Google Scholar]
- 149.Ali M, Nasir S, Nguyen QH, Sahoo JK, Tahir MN, Tremel W, Ensinger W. J Am Chem Soc. 2011;133:17307. doi: 10.1021/ja205042t. [DOI] [PubMed] [Google Scholar]
- 150.Guo W, Cao LX, Xia JC, Nie FQ, Ma W, Xue JM, Song YL, Zhu DB, Wang YG, Jiang L. Advanced Functional Materials. 2010;20:1339. [Google Scholar]
- 151.Goodrich CP, Kirmizialtin S, Huyghues-Despointes BM, Zhu A, Scholtz JM, Makarov DE, Movileanu L. J Phys Chem B. 2007;111:3332. doi: 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- 152.Wolfe AJ, Mohammad MM, Cheley S, Bayley H, Movileanu L. J Am Chem Soc. 2007;129:14034. doi: 10.1021/ja0749340. [DOI] [PubMed] [Google Scholar]
- 153.Siwy ZS, Davenport M. Nat Nanotechnol. 2010;5:697. doi: 10.1038/nnano.2010.198. [DOI] [PubMed] [Google Scholar]
- 154.Guo P, Peterson C, Anderson D. J Mol Biol. 1987;197:229. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]