Abstract
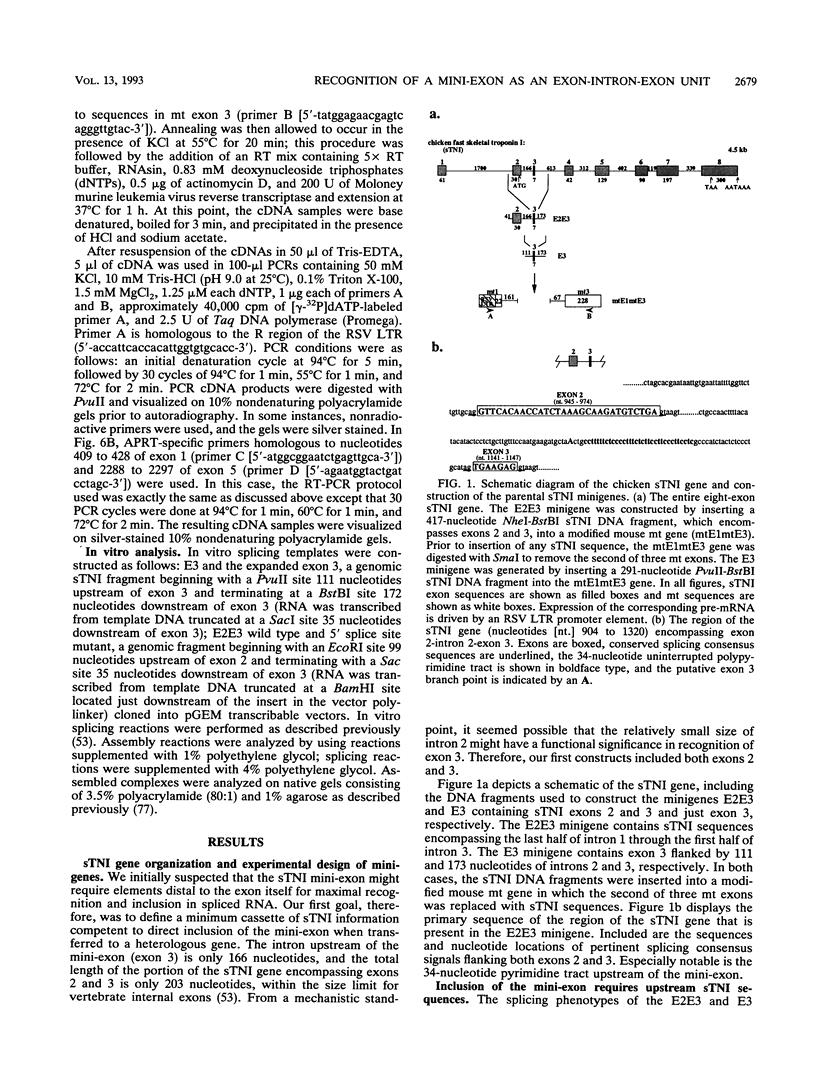
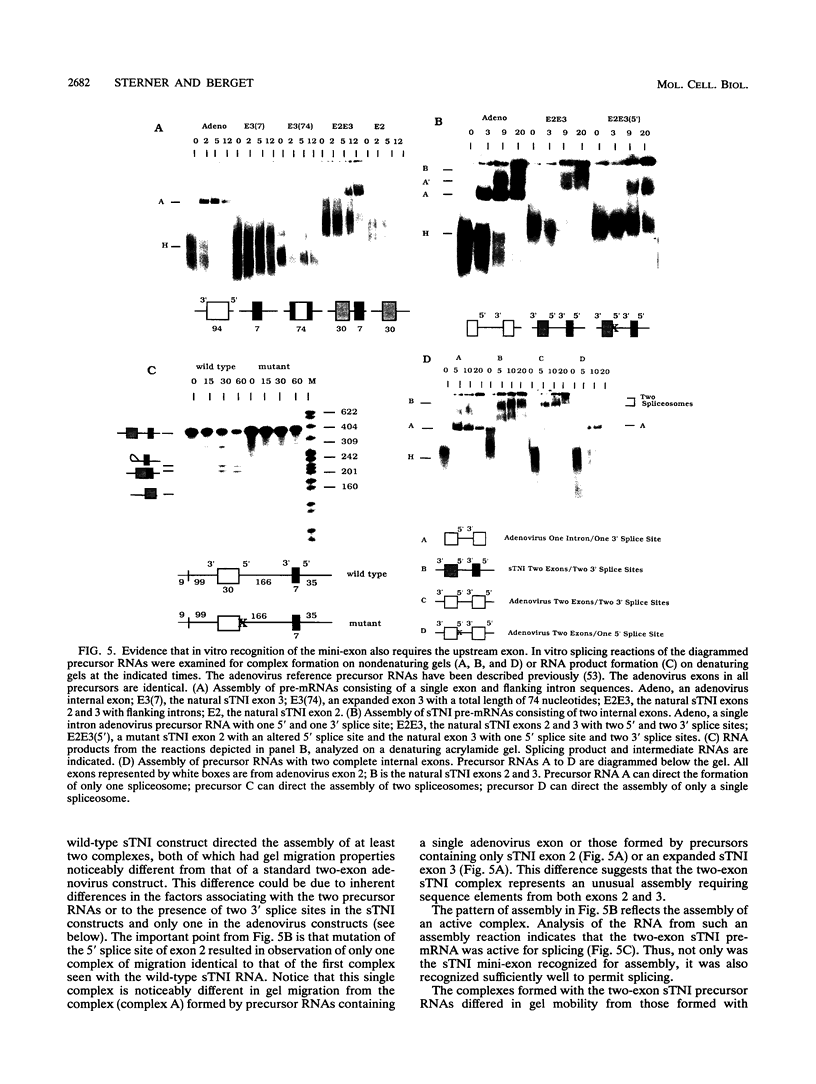
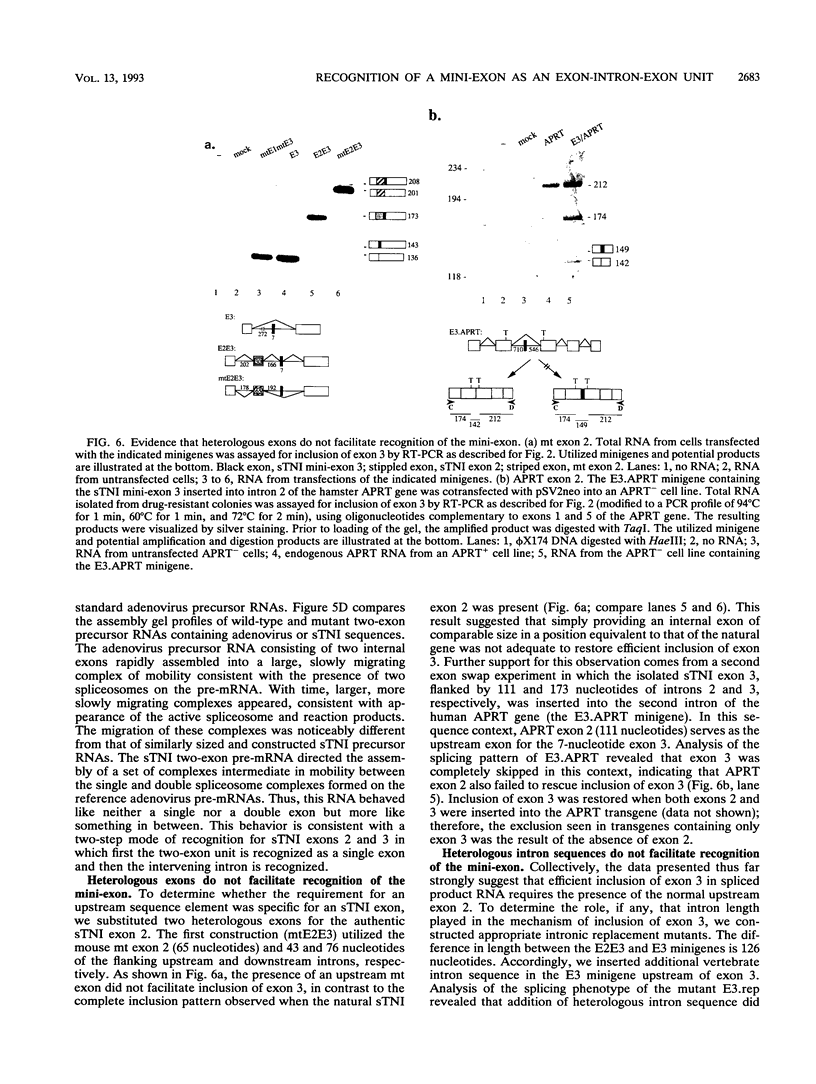
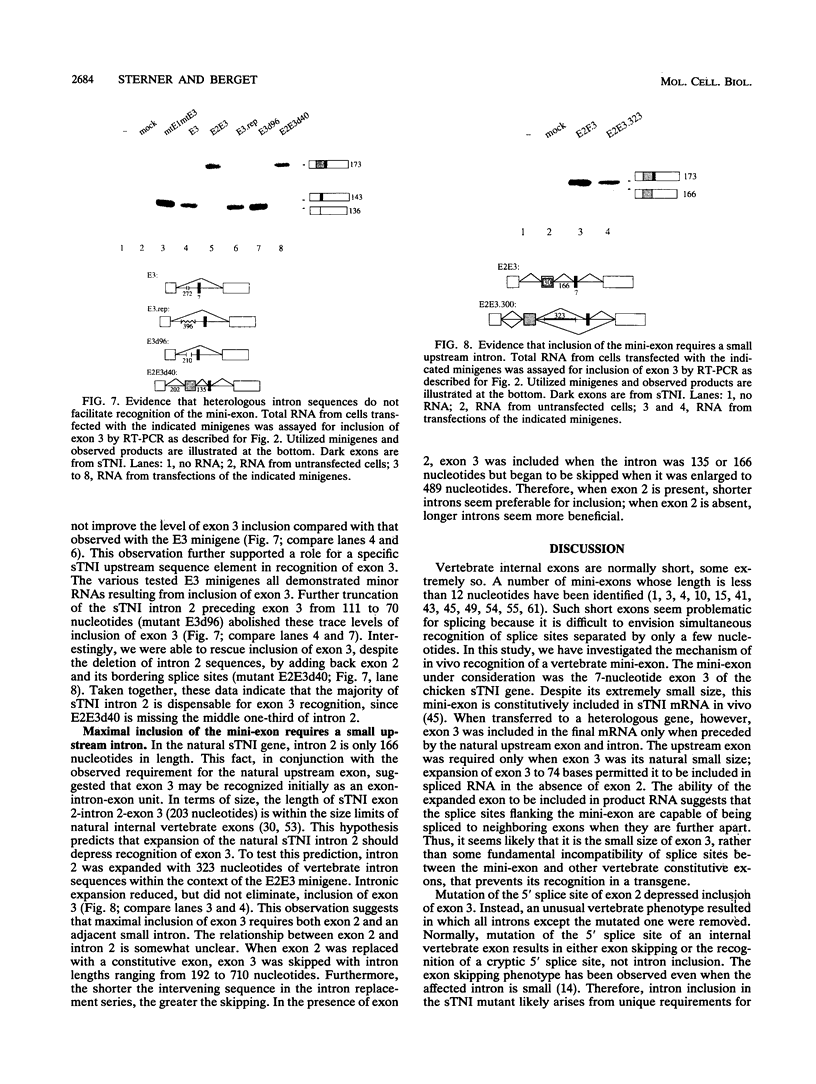
Very small vertebrate exons are problematic for RNA splicing because of the proximity of their 3' and 5' splice sites. In this study, we investigated the recognition of a constitutive 7-nucleotide mini-exon from the troponin I gene that resides quite close to the adjacent upstream exon. The mini-exon failed to be included in spliced RNA when placed in a heterologous gene unless accompanied by the upstream exon. The requirement for the upstream exon disappeared when the mini-exon was internally expanded, suggesting that the splice sites bordering the mini-exon are compatible with those of other constitutive vertebrate exons and that the small size of the exon impaired inclusion. Mutation of the 5' splice site of the natural upstream exon did not result in either exon skipping or activation of a cryptic 5' splice site, the normal vertebrate phenotypes for such mutants. Instead, a spliced RNA accumulated that still contained the upstream intron. In vitro, the mini-exon failed to assemble into spliceosome complexes unless either internally expanded or accompanied by the upstream exon. Thus, impaired usage of the mini-exon in vivo was accompanied by impaired recognition in vitro, and recognition of the mini-exon was facilitated by the presence of the upstream exon in vivo and in vitro. Cumulatively, the atypical in vivo and in vitro properties of the troponin exons suggest a mechanism for the recognition of this mini-exon in which initial recognition of an exon-intron-exon unit is followed by subsequent recognition of the intron.
Full text
PDF

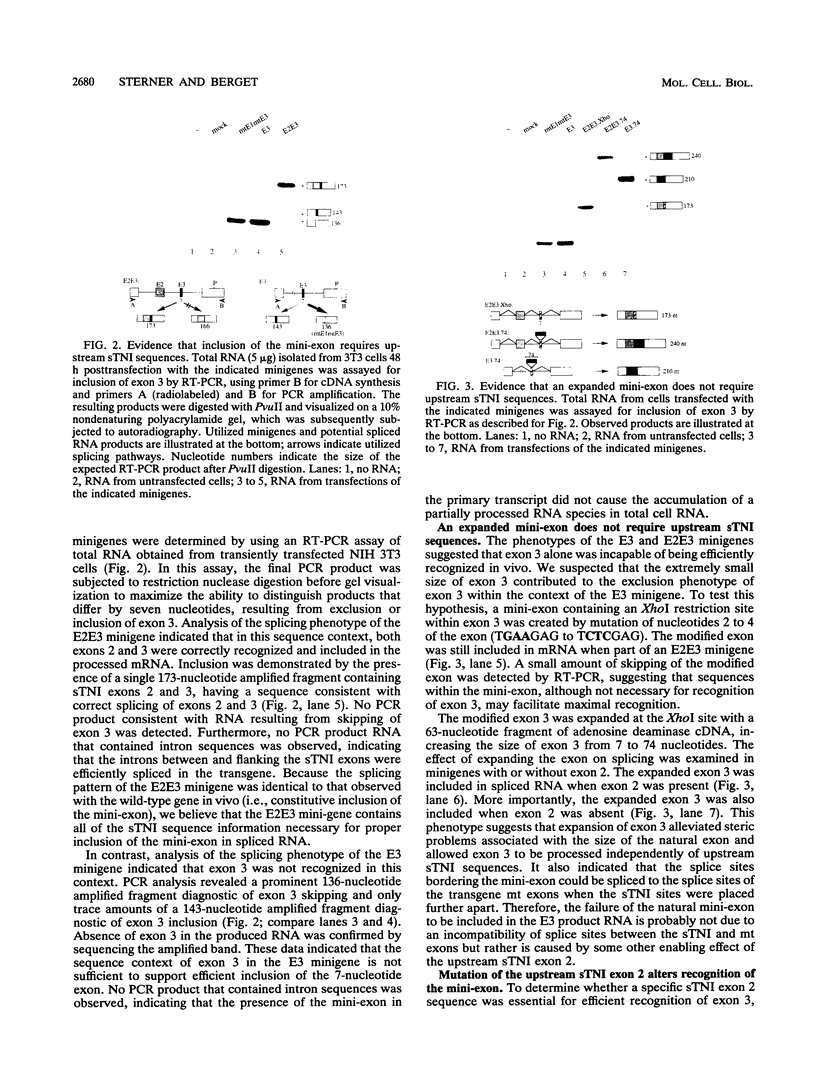




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aho S., Tate V., Boedtker H. Location of the 11 bp exon in the chicken pro alpha 2(I) collagen gene. Nucleic Acids Res. 1984 Aug 10;12(15):6117–6125. doi: 10.1093/nar/12.15.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi A., Guglielmi P., Siebenlist U., Ravetch J. V., Jensen J. P., Korsmeyer S. J. A DNA insertion/deletion necessitates an aberrant RNA splice accounting for a mu heavy chain disease protein. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2689–2693. doi: 10.1073/pnas.83.8.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Kittler E. L., Emerson C. P., Jr Structure, evolution, and regulation of a fast skeletal muscle troponin I gene. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8080–8084. doi: 10.1073/pnas.82.23.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthels D., Santoni M. J., Wille W., Ruppert C., Chaix J. C., Hirsch M. R., Fontecilla-Camps J. C., Goridis C. Isolation and nucleotide sequence of mouse NCAM cDNA that codes for a Mr 79,000 polypeptide without a membrane-spanning region. EMBO J. 1987 Apr;6(4):907–914. doi: 10.1002/j.1460-2075.1987.tb04837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992 May 29;69(5):795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Black D. L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991 Mar;5(3):389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- Bonadio J., Ramirez F., Barr M. An intron mutation in the human alpha 1(I) collagen gene alters the efficiency of pre-mRNA splicing and is associated with osteogenesis imperfecta type II. J Biol Chem. 1990 Feb 5;265(4):2262–2268. [PubMed] [Google Scholar]
- Brandt C. R., Morrison S. L., Birshtein B. K., Milcarek C. Loss of a consensus splice signal in a mutant immunoglobulin gene eliminates the CH1 domain exon from the mRNA. Mol Cell Biol. 1984 Jul;4(7):1270–1277. doi: 10.1128/mcb.4.7.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Complete nucleotide sequence of the fast skeletal troponin T gene. Alternatively spliced exons exhibit unusual interspecies divergence. J Mol Biol. 1986 Apr 5;188(3):313–324. doi: 10.1016/0022-2836(86)90157-9. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991 Jul 5;220(1):49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- Casanova J. L., Pannetier C., Jaulin C., Kourilsky P. Optimal conditions for directly sequencing double-stranded PCR products with sequenase. Nucleic Acids Res. 1990 Jul 11;18(13):4028–4028. doi: 10.1093/nar/18.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Liao Y. C., Smith D. H., Barrera-Saldaña H. A., Gelinas R. E., Seeburg P. H. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics. 1989 May;4(4):479–497. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985 Sep 15;260(20):11140–11148. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992 May;12(5):2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991 Dec;11(12):6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer G. A., O'Brien J. P., Hurwitz J. Alterations in the polypyrimidine sequence affect the in vitro splicing reactions catalyzed by HeLa cell-free preparations. J Biol Chem. 1989 Sep 5;264(25):14631–14637. [PubMed] [Google Scholar]
- Fu X. D., Katz R. A., Skalka A. M., Maniatis T. The role of branchpoint and 3'-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991 Feb;5(2):211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. Inhibition of splicing but not cleavage at the 5' splice site by truncating human beta-globin pre-mRNA. Proc Natl Acad Sci U S A. 1986 Feb;83(4):927–931. doi: 10.1073/pnas.83.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs R. A., Nguyen P. N., Edwards A., Civitello A. B., Caskey C. T. Multiplex DNA deletion detection and exon sequencing of the hypoxanthine phosphoribosyltransferase gene in Lesch-Nyhan families. Genomics. 1990 Jun;7(2):235–244. doi: 10.1016/0888-7543(90)90545-6. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Nasim F. U., Kuo H. C., Burch R. Combinatorial splicing of exon pairs by two-site binding of U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1991 Dec;11(12):5919–5928. doi: 10.1128/mcb.11.12.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Kunisada T., Ogawa M., Yamaguchi K., Nishikawa S. Exon skipping by mutation of an authentic splice site of c-kit gene in W/W mouse. Nucleic Acids Res. 1991 Mar 25;19(6):1267–1271. doi: 10.1093/nar/19.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka Y., Palella T. D., O'Toole T. E., Tarlé S. A., Kelley W. N. Human adenine phosphoribosyltransferase. Identification of allelic mutations at the nucleotide level as a cause of complete deficiency of the enzyme. J Clin Invest. 1987 Nov;80(5):1409–1415. doi: 10.1172/JCI113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges P. E., Rosenberg L. E. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4142–4146. doi: 10.1073/pnas.86.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg M. J., Gennissen A. M., Vos H. L., Hilkens J. A single nucleotide polymorphism in an exon dictates allele dependent differential splicing of episialin mRNA. Nucleic Acids Res. 1991 Jan 25;19(2):297–301. doi: 10.1093/nar/19.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvit J., DiLella A. G., Brayton K., Ledley F. D., Robson K. J., Woo S. L. GT to AT transition at a splice donor site causes skipping of the preceding exon in phenylketonuria. Nucleic Acids Res. 1987 Jul 24;15(14):5613–5628. doi: 10.1093/nar/15.14.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Masumura T., Nishio H., Nakajima T., Kitoh Y., Takumi T., Koga J., Nakamura H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Invest. 1991 Jun;87(6):2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineo I., Clarke P. R., Sabina R. L., Holmes E. W. A novel pathway for alternative splicing: identification of an RNA intermediate that generates an alternative 5' splice donor site not present in the primary transcript of AMPD1. Mol Cell Biol. 1990 Oct;10(10):5271–5278. doi: 10.1128/mcb.10.10.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Fukamizu A., Hirose S., Hayashi T., Hori H., Ohkubo H., Nakanishi S., Murakami K. Structure of the human renin gene. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5999–6003. doi: 10.1073/pnas.81.19.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Nikovits W., Jr, Kuncio G., Ordahl C. P. The chicken fast skeletal troponin I gene: exon organization and sequence. Nucleic Acids Res. 1986 Apr 25;14(8):3377–3390. doi: 10.1093/nar/14.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Suzuki K. Multiple abnormal beta-hexosaminidase alpha chain mRNAs in a compound-heterozygous Ashkenazi Jewish patient with Tay-Sachs disease. J Biol Chem. 1988 Dec 5;263(34):18563–18567. [PubMed] [Google Scholar]
- Parker B. A., Stark G. R. Regulation of simian virus 40 transcription: sensitive analysis of the RNA species present early in infections by virus or viral DNA. J Virol. 1979 Aug;31(2):360–369. doi: 10.1128/jvi.31.2.360-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991 Jan 11;64(1):181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Prediger E. A., Hoffman S., Edelman G. M., Cunningham B. A. Four exons encode a 93-base-pair insert in three neural cell adhesion molecule mRNAs specific for chicken heart and skeletal muscle. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9616–9620. doi: 10.1073/pnas.85.24.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina R. L., Morisaki T., Clarke P., Eddy R., Shows T. B., Morton C. C., Holmes E. W. Characterization of the human and rat myoadenylate deaminase genes. J Biol Chem. 1990 Jun 5;265(16):9423–9433. [PubMed] [Google Scholar]
- Santoni M. J., Barthels D., Vopper G., Boned A., Goridis C., Wille W. Differential exon usage involving an unusual splicing mechanism generates at least eight types of NCAM cDNA in mouse brain. EMBO J. 1989 Feb;8(2):385–392. doi: 10.1002/j.1460-2075.1989.tb03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980 Aug 21;286(5775):779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Sikder S. K., Kabat E. A., Morrison S. L. Alternative splicing patterns in an aberrantly rearranged immunoglobulin kappa-light-chain gene. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4045–4049. doi: 10.1073/pnas.82.12.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate V. E., Finer M. H., Boedtker H., Doty P. Chick pro alpha 2 (I) collagen gene: exon location and coding potential for the prepropeptide. Nucleic Acids Res. 1983 Jan 11;11(1):91–104. doi: 10.1093/nar/11.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Streuli M., Saito H. Integrity of the exon 6 sequence is essential for tissue-specific alternative splicing of human leukocyte common antigen pre-mRNA. Mol Cell Biol. 1989 Oct;9(10):4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull-Ross A. D., Else A. J., Eperon I. C. The dependence of splicing efficiency on the length of 3' exon. Nucleic Acids Res. 1988 Jan 25;16(2):395–411. doi: 10.1093/nar/16.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu N., Kobayashi H., Miyatake T., Tsuji S. A novel exon mutation in the human beta-hexosaminidase beta subunit gene affects 3' splice site selection. J Biol Chem. 1992 Feb 5;267(4):2406–2413. [PubMed] [Google Scholar]
- Weil D., Bernard M., Combates N., Wirtz M. K., Hollister D. W., Steinmann B., Ramirez F. Identification of a mutation that causes exon skipping during collagen pre-mRNA splicing in an Ehlers-Danlos syndrome variant. J Biol Chem. 1988 Jun 25;263(18):8561–8564. [PubMed] [Google Scholar]
- Weil D., D'Alessio M., Ramirez F., Steinmann B., Wirtz M. K., Glanville R. W., Hollister D. W. Temperature-dependent expression of a collagen splicing defect in the fibroblasts of a patient with Ehlers-Danlos syndrome type VII. J Biol Chem. 1989 Oct 5;264(28):16804–16809. [PubMed] [Google Scholar]
- Weil D., D'Alessio M., Ramirez F., de Wet W., Cole W. G., Chan D., Bateman J. F. A base substitution in the exon of a collagen gene causes alternative splicing and generates a structurally abnormal polypeptide in a patient with Ehlers-Danlos syndrome type VII. EMBO J. 1989 Jun;8(6):1705–1710. doi: 10.1002/j.1460-2075.1989.tb03562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson J. M., Grand R. J. Comparison of amino acid sequence of troponin I from different striated muscles. Nature. 1978 Jan 5;271(5640):31–35. doi: 10.1038/271031a0. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zhang L. H., Vrieling H., van Zeeland A. A., Jenssen D. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J Mol Biol. 1992 Feb 5;223(3):627–635. doi: 10.1016/0022-2836(92)90979-t. [DOI] [PubMed] [Google Scholar]
- Zhuang Y. A., Goldstein A. M., Weiner A. M. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2752–2756. doi: 10.1073/pnas.86.8.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Zapp M. L., Berget S. M. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol. 1988 Feb;8(2):814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]