Abstract
Background
Immunoglobulin E (IgE) is both a marker and mediator of allergic inflammation. Despite reported differences in serum total IgE levels by race-ethnicity, African American and Latino individuals have not been well represented in genetic studies of total IgE.
Objective
To identify the genetic predictors of serum total IgE levels.
Methods
We used genome wide association (GWA) data from 4,292 individuals (2,469 African Americans, 1,564 European Americans, and 259 Latinos) in the EVE Asthma Genetics Consortium. Tests for association were performed within each cohort by race-ethnic group (i.e., African American, Latino, and European American) and asthma status. The resulting p-values were meta-analyzed accounting for sample size and direction of effect. Top single nucleotide polymorphism (SNP) associations from the meta-analysis were reassessed in six additional cohorts comprising 5,767 individuals.
Results
We identified 10 unique regions where the combined association statistic was associated with total serum IgE levels (P-value <5.0×10−6) and the minor allele frequency was ≥5% in two or more population groups. Variant rs9469220, corresponding to HLA-DQB1, was the most significantly associated SNP with serum total IgE levels when assessed in both the replication cohorts and the discovery and replication sets combined (P-value = 0.007 and 2.45×10−7, respectively). In addition, findings from earlier GWA studies were also validated in the current meta-analysis.
Conclusion
This meta-analysis independently identified a variant near HLA-DQB1 as a predictor of total serum IgE in multiple race-ethnic groups. This study also extends and confirms the findings of earlier GWA analyses in African American and Latino individuals.
Keywords: meta-analysis, genome wide association study, total immunoglobulin E, race-ethnicity, continental population groups
INTRODUCTION
Immunoglobulin E (IgE) is an important mediator in allergic inflammation and is often elevated in individuals with atopic conditions, such as asthma.(1–3) Recently published genome wide association studies (GWAS) for asthma and serum total IgE levels suggest that there are some, but not many, overlapping top-hit single nucleotide polymorphisms (SNPs) in genes associated with these phenotypes.(4–6) The independence (i.e., lack of genetic pleiotropy) of these two phenotypes is also supported by minimal shared variance between the heritable components of serum total IgE and airway responsiveness.(7) In addition, although environmental exposures influence total IgE levels, studies in twins suggest that genetic inheritance accounts for the majority of variation in levels between individuals.(8;9)
Serum total IgE levels differ by population group in the U.S. with higher levels reported in both African American and Latino individuals when compared with non-Hispanic white individuals (henceforward referred to as European Americans).(10–13) As genetic ancestry has been shown to be associated with total IgE levels, it is possible that these population-level differences have some genetic underpinnings.(14) This is also supported by heterogeneity in linkage signals for total IgE among these three population groups.(15) However, despite these potential differences, African American and Latino individuals have not been well represented in genetic studies of total IgE to date.
The EVE consortium comprises nine research groups across the U.S. with genome wide genotype data on individuals with and without asthma. The assembled study participants are broadly representative of three major population groups (i.e., African American, European American, and Latino individuals) in North America and have data on asthma-associated phenotypes, including serum total IgE. We describe here the results of a meta-analysis of GWAS for serum total IgE, combining results from the various EVE investigators. Associations were first determined among subgroups of individuals defined by asthma status and self-identified race-ethnicity before combining so as to minimize the potential confounding effects of these characteristics.
METHODS
Study populations
The EVE consortium comprises nine investigative teams throughout the U.S., each with genome wide association data for asthma from case-control, trio, or extended family studies.(16) All of the cohorts represented in the meta-analysis (i.e., the discovery set) were part of the EVE consortium, and each of these individual study populations has been described in detail previously.(17–25) These study populations are broadly representative of the major North American population groups and were recruited through clinics and health systems in the U.S., Puerto Rico, Mexico, and Barbados. All studies included in the current analysis were approved by their respective institutional review boards.
Only the EVE samples in which the participants had serum total IgE measured at the time of screening or enrollment were included in the current meta-analysis. These included the following studies: the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE), the Genetic Research on Asthma in the African Diaspora (GRAAD), Genetics of Asthma in Latino Americans (GALA), the Childhood Asthma Management Program (CAMP), the Childhood Asthma Research and Education Network (CARE), the Chicago Asthma Genetics Study (CAG), the National Heart, Lung, and Blood Institute (NHLBI) Collaborative Studies of the Genetics of Asthma (CSGA), and the Severe Asthma Research Program (SARP). The latter three studies make up the NHLBI-supported SNP Typing for Association with Multiple Phenotypes from Existing Epidemiologic Data (STAMPEED) consortium (heretofore collectively referred to by that acronym).
SAPPHIRE is an ongoing prospective cohort study to determine the genetic determinants of inhaled corticosteroid response.(17;18) Participants were aged 12–56 years, had a physician diagnosis of asthma, demonstrated bronchodilator reversibility (>12% increase in forced expiratory volume at one second [FEV1]), and had no prior diagnosis (in the electronic medical record or by patient report) of chronic obstructive pulmonary disease (COPD) or congestive heart failure (CHF). All patients were recruited from a single, large health system covering southeast Michigan. A subset of these individuals was genotyped using the Affymetrix 6.0 GeneChip Array (Affymetrix, Inc., Santa Clara, California). Controls for SAPPHIRE were from the Wayne County Health Environment Allergy and Asthma Longitudinal Study, and included expectant women age ≥21 years from the Detroit metropolitan area and receiving care from the same health system. This group was genotyped using the Affymetrix 5.0 GeneChip Array or the 500K Mapping Array.
The GRAAD-AA study comprised African American individuals with asthma (self-reported) and non-asthmatic control patients from the Baltimore-Washington D.C. area.(19) The GRAAD-AC study consisted of extended families from Barbados with at least one asthma proband.(19) In this study, a diagnosis of asthma required a self-reported history, physician diagnosis, and a history of wheezing without an upper respiratory infection. Individuals were genotypied on the Illumina HumanHap650Y versions 1 and 3 BeadChips (Illumina, Inc., San Diego, California).
GALA is a trio study including asthma probands age 8–40 years with a physician diagnosis of mild to moderate-severe asthma and symptoms at least twice in the two years preceding enrollment.(20) Individuals were recruited from sites in San Francisco, New York City, Puerto Rico, and Mexico City. Individuals were genotyped on the Affymetrix 6.0 GeneChip Array.
The CAMP subjects were part of a multi-center clinical trial of children with mild to moderate asthma and airway responsiveness by methacholine challenge (≥20% drop in FEV1 with ≤12.5 mg/dl of methacholine).(23) While parents of these children were also recruited for this trio study, only the children with total IgE data were included in the current analysis. Individuals were genotyped on the Illumina HumanHap500 version 3 BeadChip.
CARE represents a series clinical trials, 5 of which contributed to the current meta-analysis (PEAK,(26) PACT,(27) CLIC,(28) AIMS,(29) and MARS(30)). All participants were children with physician diagnosed asthma. Parents of these subjects were also recruited to compose a trio study, but the only the children for whom we had total IgE data were included in this analysis. Individuals were genotyped using the Affymetrix 6.0 GeneChip array.
CAG and CSGA samples comprise both children and adults with asthma (physician diagnosis, symptoms, and airway responsiveness by methacholine challenge or bronchodilator reversibility). Individuals in these studies were recruited from multiple locations, but only those recruited from clinics at the University of Chicago, University of Maryland – Baltimore, and Wake Forest University and who had genome wide genotype data were included in the current analysis. SARP included individuals with mild to severe asthma.(31) Non-asthmatic control subjects were recruited from the same clinic sites for these studies. The subjects were genotyped on the Illumina 1M version 1 BeadChip.
Meta-analysis for serum total IgE
Given the different platforms used to genotype the EVE cohorts, imputation was used to create a common and near complete set of SNP genotypes across all groups. Each research group used the program, MACH,(32) to impute genotypes in their respective study population(s) using the reference panels from HapMap phase 2, release 21. For European American individuals, haplotypes from the CEPH European Americans of Northern and Western European ancestry (CEU) were used as the reference. For African American and African Caribbean individuals, both the CEU and the Yoruba of West African (YRI) haplotypes served as reference. For Latino individuals, the haplotypes from all of the original HapMap population groups (i.e., CEU, YRI, Japanese from Tokyo [JPT], and the ethnic Han Chinese from Beijing [CHB]) were used as reference. Poorly imputed SNPs with MACH r-squared values <0.3 were removed from the analysis.
For analysis, study subjects were stratified by study cohort, race-ethnicity (i.e., African Caribbean and African American combined, European American, and Latino), and asthma status. The dependent variable, serum total IgE, was logarithmically (natural log) transformed to normalize its distribution and diminish the effect of outliers; five individuals with total IgE values <2 IU/ml were also excluded from the analysis. As has been done for other genetic analyses,(33) we used Studentized residuals to standardize the distributions of log-transformed IgE values across race-ethnic groups, asthma disease status, and study populations. For each within study population group, we generated Studentized residuals adjusted for age and sex, whereby the residuals represented the remaining variation in total IgE after adjustment. Because these residuals were normally distributed, these steps ensured a comparable scale for the regression coefficient summarizing the association between each SNP and total IgE, thus limiting bias in the meta-analysis arising from distributional differences in total IgE levels across groups. These residuals were then used as the primary phenotype for association with the allele dosage estimates for each genotype. We assumed an additive genetic model with the reference allele coded as zero. To account for underlying population structure, study-specific genome wide principal components generated by the program EIGENSOFT were included as covariates.(34) Given the extended pedigree structure of GRAAD-AC, a linear mixed effects model using a kinship matrix approach was employed for this cohort;(35) this method corrects for familial correlation (both known and unknown) by estimating the empirical kinship between all pairs of subjects.
We used the analytic software, METAL,(36) to perform the meta-analysis. This program used an inverse normal approach to combine stratum specific p-values, which accounted for both the direction of effect and the sample size of each stratum. The meta-analysis was performed for the overall sample and within the race-ethnic groups (i.e., African Caribbean and African American individuals combined, European Americans, and Latinos). The most significant SNP associations (P value <5.0 × 10−6) were promoted for replication if they were informative in at least two of the three race-ethnic groups (i.e., a minor allele frequency ≥5%). This threshold was similar to that suggested by others for genome-wide association studies,(37) but was less stringent than the Bonferroni corrected value (P value <5.0 × 10−8) which may overly conservative. We elected to use the former so as to provide a feasible number of SNPs for assessing replication. When multiple SNPs in a region met this criterion, we selected the polymorphism with the greatest overall statistical significance. We also assessed for heterogeneity of the top SNP associations by race-ethnicity and asthma disease status using the Cochran’s Q-test as implemented in METAL.(36;38)
Replication populations and analysis
The top 10 signals were replicated in six case-control and population-based samples from the U.S and Canada and a founder population in the U.S. The U.S. replication populations included a separate group of individuals from SAPPHIRE,(39) the Gene-Environments and Admixture in Latino Americans (GALA II) study, and the Study of African Americans, Asthma, Genes & Environments (SAGE [U.S.]).(40) The Canadian cohorts included the Canadian Asthma Primary Prevention Study (CAPPS)(41;42) and the Study of Asthma Genes and the Environment (SAGE [Canada]).(42;43) The U.S. founder population consisted of Hutterites who live in South Dakota.(44)
The SAPPHIRE study participants included in the replication were those without genome wide association data who were age 12–56 years, had a physician diagnosis of asthma, and no prior diagnosis of COPD or CHF. Replication controls were also age 12–56 years; had no prior diagnosis of asthma, COPD, or CHF; and were recruited within the same health care system.
The GALA II study is a large case-control study recruiting Latino patients with and without asthma (physician diagnosed) from clinical sites in San Francisco, New York City, Houston, Chicago, and Puerto Rico. Individuals were recruited at age 8–21 years. The SAGE (U.S.) study was overseen by the same research team, and recruited African American individuals with and without a physician diagnosis of asthma from the San Francisco Bay area. Case patients were age 8–40 years and had physician diagnosed asthma. Control patients in both GALA II and SAGE were enrolled from the same clinics concurrently and had no prior diagnosis of allergies, asthma, lung disease, symptoms consistent with asthma within the preceding 2 years, and <10 pack-year smoking history.
The Canadian SAGE study was a population-based birth cohort consisting of individuals born in Manitoba in 1995. From this group, a nested case-control study was selected, representing children with and without pediatrician diagnosed asthma. Serum total IgE was measured between the ages of 7–10 years. CAPPS, another birth cohort, included infants born between October 1994 and August 1996 who were deemed to be at high risk for developing allergies as a result of ≥1 1st degree relative with asthma or ≥2 first degree relatives with an IgE-mediated allergic disease.
The founder population comprised the Schmiedeleut Hutterites residing in primarily South Dakota. This extended pedigree has been used in prior investigations of allergy and asthma.(44–46)
Replication genotype data was generated from either allelic discrimination (TaqMan®, Applied Biosystems, Foster City, California) or commercially available high-throughput arrays. For the SAPPHIRE and SAGE cohorts all replication SNPs were genotyped by allelic discrimination. In GALA II genome wide array results were available from the Axiom LAT array (Affymetrix, Inc., Santa Clara, California) for rs9469220 and rs16977747, and the remaining SNP were genotyped using TaqMan. In the two Canadian cohorts, rs321588, rs7751374, rs10124954, and rs6499255 were genotyped on the Illumina Human610-Quad array; rs9469220, rs537526, and rs16977747 were imputed using MACH; rs2885872 was a genotyped proxy (CEU r2=1.0) used for rs2363709; and rs10043454 and rs4355582 were imputed proxies used for rs13361473 and rs10944017, respectively. Amongst the Hutterites, rs9469220, rs7751374, rs10124954, rs537526, and rs16977747 were genotyped on the Affymetrix 6.0 GeneChip, and rs613614, rs2101567, rs9314043, and rs11075734 were used as genotype proxies for rs321588, rs2363709, rs13361473, and rs6499255, respectively.
We applied a similar analytic approach as used in the discovery cohorts to the assessment of the replication data. Groups were stratified according to cohort, race-ethnicity, and asthma status. Standardized residuals from the linear models associating natural log transformed serum total IgE levels with age and sex were used in second stage models in association with replication genotypes. The association with genotype was assessed assuming an additive genetic model. We again used METAL(36) to combined stratum specific p-values. Analyses were performed for the overall replication sample and within groups defined by race-ethnicity (i.e., for European American, African American, and Latino individuals).
RESULTS
The characteristics of the six study groups in the genome wide meta-analysis of serum total IgE levels are shown in Table 1 (individuals in GRAAD recruited from the U.S. and the Caribbean are shown separately). These cohorts comprised 4,292 study subjects which ranged in age from 1 to 84 years. The combined group included 2,425 (56.5%) females and 2,754 (64.1%) with a history of asthma. The race-ethnic breakdown of the total group included 2,469 (57.4%) African American and African Caribbean individuals, 259 (6.0%) Latino individuals, and 1,564 (36.4%) European American individuals. The distribution of serum total IgE levels by race-ethnicity, asthma status, and study is shown in the online supplement (eTable 1).
Table 1.
Baseline characteristics of individuals included in the meta-analysis of IgE stratified by study.
| Study Cohort | |||||||
|---|---|---|---|---|---|---|---|
| SAPPHIRE (n=276) |
GRAAD-AC (n=635) |
GRAAD-AA (n=806) |
GALA (n=215) |
CAMP (n=362) |
CARE (n=229) |
STAMPEED (n=1,769) |
|
| Age in years – mean ± SD (range) | 30.4 ± 10.5 (12–56) | 28.3 ± 16.4 (3–84) | 29.5 ± 18.3 (4–78) | 12.9 ± 5.5 (8–35) | 8.8 ± 2.1 (5–13) | 6.0 ± 4.1 (1–17) | 30.9 ± 14.7 (6–81) |
| Female – no. (%) | 216 (78.3) | 322 (50.7) | 405 (50.2) | 90 (41.9) | 134 (37.0) | 59 (25.8) | 1,199 (67.8) |
| Race-ethnicity – no. (%) | |||||||
| African American | 276 (100) | 0 (0) | 806 (100) | 0 (0) | 0 (0) | 0 (0) | 752 (42.5) |
| African Caribbean | 0 (0) | 635 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| European American | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 362 (100) | 185 (80.8) | 1,017 (57.5) |
| Latino | 0 (0) | 0 (0) | 0 (0) | 215 (100) | 0 (0) | 44 (19.2) | 0 (0) |
| History of Asthma – no. (%) | 149 (54.0) | 274 (43.2) | 458 (56.8) | 215 (100) | 362 (100) | 229 (100) | 1,067 (60.3) |
| Serum total IgE (IU/ml) – geometric mean ± geometric SD | 103.5 ± 4.1 | 188.7 ± 4.8 | 90.9 ± 5.0 | 247.2 ± 3.9 | 428.4 ± 4.6 | 34.1 ± 4.1 | 94.6 ± 4.8 |
SAPPHIRE denotes Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; GRAAD-AC, Genetic Research on Asthma in the African Diaspora-African Caribbean; GRAAD-AA, Genetic Research on Asthma in the African Diaspora-African American; GALA, Genetics of Asthma in Latino Americans; CAMP, Childhood Asthma Management Program; CARE, Childhood Asthma Research and Education Network; STAMPEED, SNP Typing for Association with Multiple Phenotypes from Existing Epidemiologic Data; SD, standard deviation; IgE, immunoglobulin E; and IU/ml, international units per milliliter.
STAMPEED comprises the Chicago Asthma Genetics Study (CAG), the National Heart, Lung, and Blood Institute Collaborative Studies of the Genetics of Asthma (CSGA), and the Severe Asthma Research Program (SARP).
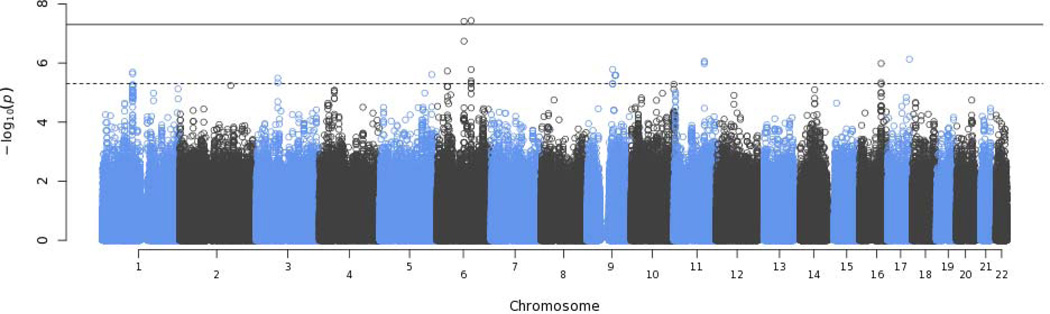
Association for total IgE was performed within groups stratified by race-ethnicity, asthma status, and study cohort. Within the cohorts with a single population group represented (i.e., SAPPHIRE, GRAAD-AC, GRAAD-AA, GALA, and CAMP), we stratified by asthma status alone. Association statistics were meta-analyzed accounting for the direction of the effect and sample size. The overall association results and the associations within each race-ethnic group are shown in Table 2 (parameter estimates for these associations are shown in eTable 2 of the online supplement). The Manhattan plot for the genome wide meta-analysis is shown in Figure 1 and the quantile-quantile plot is shown eFigure 1 of the online supplement. Ten distinct regions were identified where the combined P-value was <5.0 × 10−6 and the association was informative in ≥2 population groups. Most of the regions associated with total IgE levels were novel with the exception of HLA-DQB1.
Table 2.
Genome wide meta-analysis results of the top associations with serum total IgE levels stratified by race-ethnic group.*
| SNP | Chromosome | Position | Alleles† | Risk Allele‡ |
Nearest gene | P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| European American (n=1,564) |
African Caribbean and African American (n = 2,469) |
Latino (n = 259) |
All groups combined (n=4,292) |
||||||
| rs321588 | 1 | 96,688,818 | G/A | G | PTBP2 | 0.0019 | 0.0027 | 0.0137 | 2.05 × 10−6 |
| rs2363709 | 3 | 67,417,105 | G/A | A | SUCLG2 | 0.0224 | 1.92 ×10−5 | 0.8830 | 3.26 × 10−6 |
| rs13361473 | 5 | 164,170,729 | A/G | A | MAT2B | 0.0001 | 0.0024 | 0.7021 | 2.46 × 10−6 |
| rs9469220 | 6 | 32,658,310 | G/A | A | HLA-DQB1 | 0.0347 | 0.0017 | 6.43 × 10−6 | 1.87 × 10−6 |
| rs10944017 | 6 | 85,300,836 | C/T | C | TBX18 | --§ | 3.42 × 10−8 | 0.4222 | 3.88 × 10−8 |
| rs7751374 | 6 | 107,981,856 | G/A | G | SOBP | 0.0025 | 6.66 × 10−5 | 0.0072 | 3.68 × 10−8 |
| rs10124954 | 9 | 82,836,415 | T/C | T | TLE4 | 0.0132 | 3.22 × 10−5 | 0.3463 | 1.65 × 10−6 |
| rs537526 | 11 | 96,840,444 | G/A | A | CCDC82 | 0.0005 | 0.0004 | 0.5877 | 8.66 × 10−7 |
| rs6499255 | 16 | 69,830,328 | A/G | A | WWP2 | 0.0002 | 0.0035 | 0.1177 | 1.03 × 10−6 |
| rs16977747 | 17 | 71,699,790 | T/C | T | LINC00469 | --§ | 2.21 × 10−6 | 0.1467 | 7.42 × 10−7 |
Included the results from the following cohorts: the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE), Genetic Research on Asthma in the African Diaspora-African Caribbean (GRAAD-AC), the Genetic Research on Asthma in the African Diaspora-African American (GRAAD-AA), the Genetics of Asthma in Latino Americans (GALA), Childhood Asthma Management Program (CAMP), the Childhood Asthma Research and Education Network (CARE); the Chicago Asthma Genetics Study (CAG), the National Heart, Lung, and Blood Institute Collaborative Studies of the Genetics of Asthma (CSGA), and the Severe Asthma Research Program (SARP).
The first allele corresponds to the “reference/other” allele in dbSNP build 125.
The risk allele denotes the allele associated with high serum total IgE levels for all groups combined.
These SNPs were uninformative based on our within group threshold requiring a minor allele frequency ≥5%.
Figure 1.
Manhattan plot of the genome wide association results for serum total IgE from the meta-analysis. The significance level used to promote single nucleotide polymorphisms for replication (P < 5.0 × 10−6) is denoted by the dashed line. The solid line shows the genome wide significance level of P < 5.0 × 10−8.
Six study populations consisting of 5,767 individuals were available to replicate the findings of the discovery set (Table 3). The replication set comprised 2,961 (51.3%) African American individuals, 1,477 (25.6%) Latino individuals, and 1,329 (23.0%) European American individuals; 3,930 (68.1%) had a history of asthma. The association statistics for replication set were combined in a manner analogous to the discovery set. As shown in Table 4, of the ten SNPs analyzed for replication, only rs9469220 near HLA-DQB1 reached marginal significance (P-value = 0.0074 [replication set]; P-value = 2.45 × 10−7 [combined discovery and replication set]) (parameter estimates for these associations are shown in eTable 3 of the online supplement). There was also some evidence for possible heterogeneity at rs16977747 on chromosome 17 by population group and asthma status (i.e., strongest association for both Latino individuals and those with asthma – eTables 4 and 5 in the online supplement).
Table 3.
Characteristics of individuals included in the meta-analysis replication stratified by study.
| Study Cohort | ||||||
|---|---|---|---|---|---|---|
| SAPPHIRE* (United States) (n = 1,761) |
SAGE (United States) (n = 1,200) |
GALA II (United States) (n = 1,477) |
SAGE (Canada) (n = 423) |
CAPPS (Canada) (n = 226) |
Hutterites (United States) (n = 680) |
|
| Age in years – mean ± SD (range) | 34.1 ± 14.1 (12–57) | 15.3 ± 5.7 (7–41) | 12.8 ± 3.3 (8–21) | 9.0 ± 0.5 (7–10) | 7.2 ± 0.2 (6–8) | 29.8 ± 16.7 (6–78) |
| Female – no. (%) | 1,117 (63.4) | 574 (47.8) | 727 (49.2) | 187 (44.2) | 101 (44.7) | 352 (51.8) |
| Race-ethnicity – no (%) | ||||||
| African American | 1,761 (100) | 1,200 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| African Caribbean | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| European American | 0 (0) | 0 (0) | 0 (0) | 423 (100) | 226 (100) | 680 (100) |
| Latino | 0 (0) | 0 (0) | 1,477 (100) | 0 (0) | 0 (0) | 0 (0) |
| History of Asthma | 1,427 (64.5) | 916 (76.3) | 856(58.0) | 157 (37.1) | 47 (20.8) | 153 (22.5) |
| Serum total IgE (IU/ml) – geometric mean ± geometric SD | 120.3 ± 4.5 | 119.1 ± 4.8 | 162.4 ± 5.0 | 8.3 ± 15.4 | 14.8 ± 14.96 | 23.8 ± 4.6 |
SAPPHIRE denotes Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SAGE (United States), Study of African Americans, Asthma, Genes & Environments; GALA II, Gene-Environments and Admixture in Latino Americans; SAGE (Canada), Study of Asthma Genes and the Environment; CAPPS, Canadian Asthma Primary Prevention Study; SD, standard deviation; IgE, immunoglobulin E; and IU/ml, international units per milliliter.
SAPPHIRE individuals in the replication set excluded those who were part of the discovery set.
Table 4.
Replication results for genome wide meta-analysis of serum total IgE levels stratified by race-ethnic group.*
| SNP | Chromosome | Nearest gene |
P-value | ||||
|---|---|---|---|---|---|---|---|
| European American (n=1,329) |
African Caribbean and African American (n=2,961) |
Latino (n=1,477) |
All groups – replication set (n=5,767) |
All groups – discovery and replication set combined† (n=10,059) |
|||
| rs321588 | 1 | PTBP2 | 0.4129 | 0.8124 | 0.3737 | 0.9062 | 0.0011 |
| rs2363709 | 3 | SUCLG2 | 0.5518 | 0.8460 | 0.0093 | 0.1896 | 4.55 × 10−6 |
| rs13361473 | 5 | MAT2B | 0.6795 | 0.2455 | 0.6848 | 0.4083 | 0.0002 |
| rs9469220 | 6 | HLA-DQB1 | 0.0592 | 0.1084 | 0.2255 | 0.0074 | 2.45 × 10−7 |
| rs10944017 | 6 | TBX18 | --‡ | 0.7110 | 0.9087 | 0.8132 | 0.0003 |
| rs7751374 | 6 | SOBP | 0.6858 | 0.5180 | 0.0550 | 0.7589 | 0.0006 |
| rs10124954 | 9 | TLE4 | 0.1098 | 0.5325 | 0.5034 | 0.5516 | 0.0044 |
| rs537526 | 11 | CCDC82 | 0.3519 | 0.6422 | 0.4353 | 0.7894 | 0.0022 |
| rs6499255 | 16 | WWP2 | 0.9524 | 0.1110 | 0.0308 | 0.0230 | 1.16 × 10−6 |
| rs16977747 | 17 | LINC00469 | --‡ | 0.0865 | 0.0373 | 0.8508 | 0.0031 |
The replication set included the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) (i.e., those not in the discovery set), the Study of African Americans, Asthma, Genes & Environments (SAGE - United States), the Gene-Environments and Admixture in Latino Americans cohort (GALA 2), the Study of Asthma Genes and the Environment (SAGE - Canada); the Canadian Asthma Primary Prevention Study (CAPPS), and the Hutterites.
The discovery set included the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE), Genetic Research on Asthma in the African Diaspora-African Caribbean (GRAAD-AC), the Genetic Research on Asthma in the African Diaspora-African American (GRAAD-AA), the Genetics of Asthma in Latino Americans (GALA), Childhood Asthma Management Program (CAMP), the Childhood Asthma Research and Education Network (CARE); the Chicago Asthma Genetics Study (CAG), the National Heart, Lung, and Blood Institute Collaborative Studies of the Genetics of Asthma (CSGA), and the Severe Asthma Research Program (SARP).
These SNPs were uninformative based on our within group threshold requiring a minor allele frequency ≥5%.
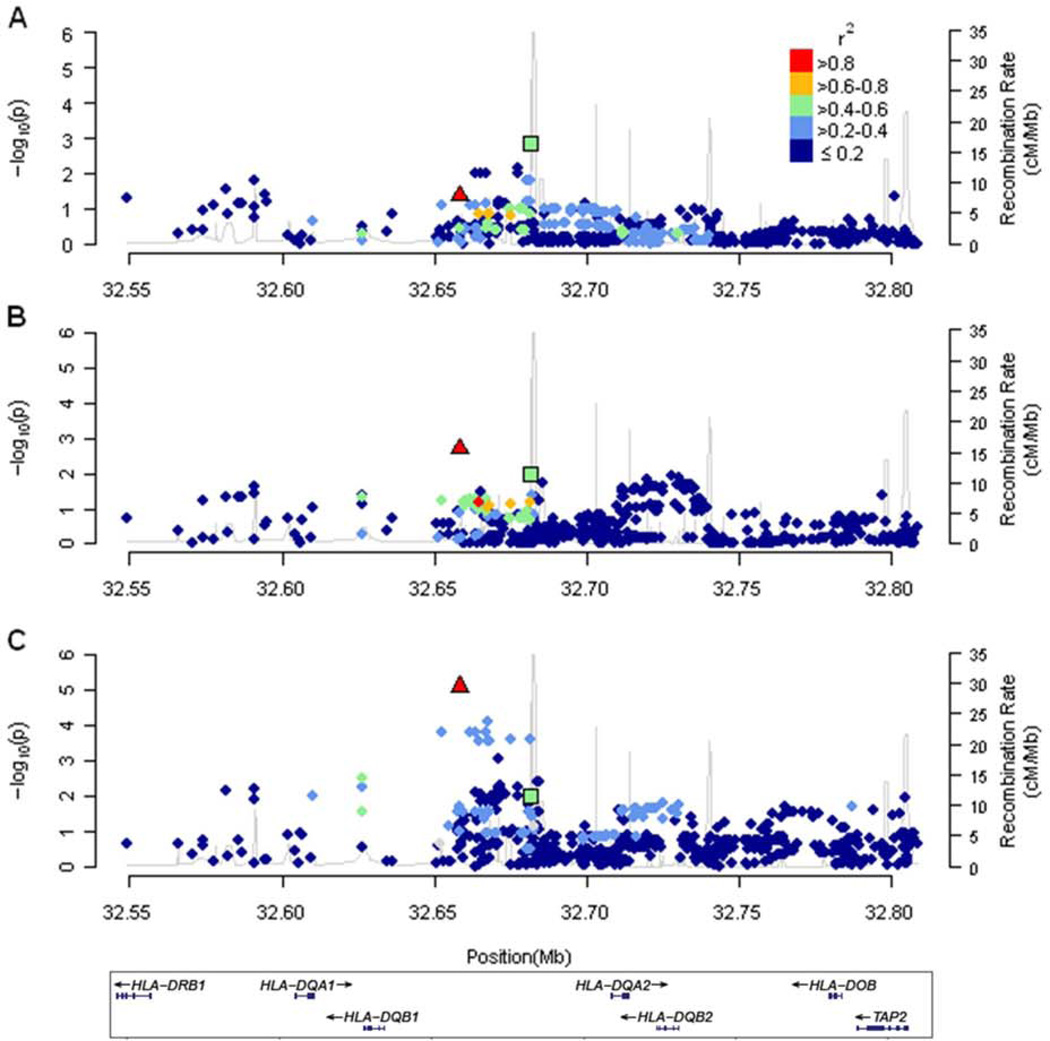
We also assessed variants in genes described in earlier genome wide association studies of serum total IgE levels (Table 5). We found that many of the genes (and variants) identified in these earlier studies replicated in the current meta-analysis. Genes with evidence of replication in all of our study groups combined included DARC, FCER1A, RAD50, IL13, HLA-DQA2, and STAT6. Not surprisingly, some of the variants showed differences in statistical significance by population group, whereas others (e.g., rs2858331 in HLA-DQA2) showed a consistent pattern in all population groups. Interestingly, the variant, rs2858331, which has been ascribed to HLA-DQA2,(47) is located ~20 kilobases (kb) from our top hit, rs9469220. Figure 2 shows the linkage disequilibrium (LD) between rs9469220 and rs2858331 and the other SNPs in the HLA-DR/DQ region. eTable 6 in the supplement shows the SNP association statistics for polymorphisms where we had data and which were located within 20 kb of the transcription start and stop site for genes shown in Table 5.
Table 5.
Replication results from earlier genome wide association studies for serum total IgE levels in the current meta-analytic group stratified by race-ethnicity.*
| Gene | SNP | Chromosome | Position | Alleles† | Risk Allele‡ |
P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| European American (n=1,564) |
African Caribbean and African American (n = 2,469) |
Latino (n = 259) |
All groups combined (n=4,292) |
||||||
| DARC | rs13962¶ | 1 | 159175527 | G/A | G | 0.0019 | -- | 0.0590 | 0.0003 |
| FCER1A | rs2427837§** | 1 | 159258545 | G/A | -- | -- | -- | -- | -- |
| rs2494264¶ | 1 | 159266838 | T/A | T | 0.0115 | 0.3614 | 0.1527 | 0.2345 | |
| rs2251746§¶ | 1 | 159272060 | T/C | T | 0.0039 | 0.6669 | 0.0123 | 0.0072 | |
| rs2252226‖ | 1 | 159276153 | C/T | C | 0.0060 | 0.3452 | 0.1306 | 0.0060 | |
| rs4656784¶** | 1 | 159326880 | A/G | -- | -- | -- | -- | -- | |
| RAD50 | rs2706347§ | 5 | 131905117 | G/T | T | 0.0121 | 0.0301 | 0.0792 | 0.0064 |
| rs3798135§ | 5 | 131965109 | C/T | T | 0.0127 | 0.0002 | 0.1158 | 8.38 × 10−5 | |
| rs2040704§ | 5 | 131973177 | A/G | G | 0.0084 | 0.0039 | 0.1076 | 0.0007 | |
| rs7737470§ | 5 | 131974063 | T/A | A | 0.0094 | 0.1311 | 0.0929 | 0.0214 | |
| IL13 | rs20541‖¶ | 5 | 131995964 | A/G | A | 0.0003 | 0.8945 | 0.5571 | 0.0246 |
| rs2243297¶** | 5 | 131999171 | T/A | -- | -- | -- | -- | -- | |
| HLA-G | rs2523809¶ | 6 | 29849619 | G/T | T | 0.5319 | 0.9972 | 0.2351 | 0.4701 |
| HLA-A | rs2517754¶** | 6 | 29896680 | T/C | -- | -- | -- | -- | -- |
| rs2571391¶ | 6 | 29923838 | A/C | A | 0.4381 | 0.5206 | 0.5674 | 0.2730 | |
| HLA-DRB1 | rs9271300‖** | 6 | 32581582 | C/G | -- | -- | -- | -- | -- |
| HLA-DQA2 | rs2858331¶ | 6 | 32681277 | A/G | G | 0.0014 | 0.0102 | 0.0099 | 6.30 × 10−6 |
| STAT6 | rs167769‖¶ | 12 | 57503775 | C/T | T | 0.0574 | 8.81 × 10−5 | 0.6933 | 2.48 × 10−5 |
| rs12368672§ | 12 | 57512470 | C/G | G | 0.0357 | 0.0255 | 0.7028 | 0.0022 | |
| IL4R/IL21R | rs1859308‖ | 16 | 27397998 | A/G | A | 0.6262 | 0.1098 | 0.4543 | 0.2186 |
Includes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE), the Genetic Research on Asthma in the African Diaspora-African Caribbean (GRAAD-AC) study, the Genetic Research on Asthma in the African Diaspora-African American (GRAAD-AA) study, the Genetics of Asthma in Latino Americans (GALA) study; the Childhood Asthma Management Program (CAMP), the Childhood Asthma Research and Education Network (CARE); the Chicago Asthma Genetics (CAG) study, the National Heart, Lung, and Blood Institute Collaborative Studies of the Genetics of Asthma (CSGA), and the Severe Asthma Research Program (SARP). Associations were first assessed in groups stratified by race-ethnicity, asthma status, and study before meta-analytically combining to generate the overall and population group results.
The first allele corresponds to the “reference/other” allele in dbSNP build 125.
The risk allele denotes the allele associated with high serum total IgE levels for all groups combined.
Identified by Weidinger et al.(4)
Identified by Moffatt et al.(6)
Identified by Granada et al.(47)
Not imputed
Figure 2.
Locus zoom plots of rs9469220 (triangle) located between HLA-DQB1 and HLA-DQA2 on chromosome 6 and its association with serum total IgE levels. The plots show the association P-values (derived from the genome wide meta-analysis) and the linkage disequilibrium (with rs9469220) in color for the surrounding single nucleotide polymorphisms in European American individuals (A), African American individuals (B), and Latino individuals (C). The polymorphism near HLA-DQA2, rs2858331, identified by Granada et al.(47) is also shown (square).
DISCUSSION
We are aware of two genome wide association studies(4;47) and one genome wide meta-analysis examining genetic variants associated with serum total IgE levels.(6) However, to our knowledge this is the first of such studies to include substantial numbers of participants who are not primarily European in ancestry (i.e., African American and Latino individuals).
Our findings confirm the role of variants in the class II major histocompatibility complex (MHC) region on serum total IgE levels. In particular, we found that a polymorphism, rs9469220, located between HLA-DQB1 and HLA-DQA2 was associated with total IgE levels. The diversity of our study population further supported the importance of this locus on IgE levels, as the relationship was consistent across multiple population groups and was robust to differences in asthma-disease status.
Moffatt and colleagues as part of the GABRIEL (A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community) Consortium also recently described markers around HLA-DRB1 and HLA-DQB1 to be associated with total IgE levels, although the latter was more strongly associated with asthma.(6) A recent GWAS of participants enrolled in the Framingham Study also found that a polymorphism just upstream of HLA-DQA2 (rs2858331) was significantly associated with total IgE levels.(47) Others testing more limited numbers of SNPs have found relationships between polymorphisms in the HLA-DR/DQ region and IgE levels.(48;48;49) Therefore, our results support the findings of these earlier studies and extend them to encompass most major North American population groups.
Total IgE production is promoted through elaboration of type 2 helper T cell (Th2) cytokines, such as interleukin (IL)-4 and IL-5.(50) Type 1 helper T cell (Th1) and Th2 activation may in turn depend on the number of MHC class II molecules involved in presentation and the presence or absence of co-stimulation through CD28.(51) Curiously, querying gene expression results from lymphoblastoid cell lines derived from HapMap individuals,(52) we found that our most significant polymorphism in the HLA-DR/DQ region, rs9469220, is related to the expression of a number of MHC class II molecules. This same SNP has also been shown to be associated with Crohn’s disease,(53) which is considered at least in part to be a Th1 condition.(54)
We found evidence for replication in genes previously identified in other genome wide analyses and candidate gene studies as being associated with total IgE levels. As shown in Table 5, these genes included DARC,(47;55) FCER1A,(4;6;47) RAD50,(4;49) IL13,(4;6;47;49;56) HLA-DQA2,(47) STAT6,(4;6;47;57) and IL4R/IL21R.(6) Most importantly, we now demonstrate that these relationships are present in multiple population groups.
We also identified at least 2 other loci (rs2363709 on Chromosome 3 and rs16977747 on Chromosome 17) that were associated with serum total IgE levels with possible heterogeneous associations across population groups and disease state. Further studies will be needed to assess whether these represent true ancestry-specific or asthma-specific (or non-asthma specific) determinants of IgE levels.
This study was not without limitations. First, to remove the potential confounding influence of race-ethnicity, asthma-status, and study cohort, we first stratified our analytic groups by these categories and then recombined the resultant association statistics via meta-analysis. Therefore, despite our large overall study size, the power within each population-specific stratum was limited. Second, although we accounted for the important confounders such as sex and age,(58) many other factors which can influence IgE levels were not available for inclusion in our models, such as the season in which an IgE sample was drawn,(59;60) use of systemic corticosteroids, and exposure to tobacco smoke.(11;61) While these missing factors may have confounded our analysis, it is heartening to see that we could replicate many of the previously described associations. In addition, we cannot say with certainty that the variants we identified by association are causally related to IgE levels; rather, these polymorphism may be in LD with the true causal variant. Because patterns of LD may vary by race-ethnicity, it is also possible that the true causal variant still differs by population group.(62) Determining these causal variants will likely require additional, finer scale genotyping and follow-up functional studies.
In summary, our findings provide further evidence for a role of the HLA-DR/DQ region in serum total IgE levels. Importantly, this study suggests that this region has similar determinative import in European American, African American, and Latino individuals. Our study also replicates many of the previously identified genetic associations with total IgE in these three population groups. Preliminary evidence for genetic predictors which vary by race-ethnicity and asthma status will need to be replicated in the respective population groups. Nevertheless, this study underscores the need to study diverse populations so as to identify both shared and group-specific genetic predictors.
Supplementary Material
ACKNOWLEDGEMENTS
We would especially like to thank all the study participants and recruiters whose time and effort made this work possible.
This work was supported by grants from the National Heart, Lung, and Blood Institute (HL101651 to Drs. Ober and Nicolae; HL087665 to Dr. Nicolae; HL085197, HL087665, HL072414, and HL49596 to Dr. Ober; HL064307 and HL064313 to Dr. Martinez; HL075419, HL65899, HL083069, HL066289, HL087680, HL101543, and HL101651 to Dr. Weiss; HL079055 to Dr. Williams; HL087699, HL049612, HL075417, HL04266, and HL072433 to Dr. Barnes; HL061768, HL076647, to Dr. Gilliland; HL087680 to Dr. Gauderman; HL078885 and HL088133 to Dr. Burchard; HL087665 to Dr. Meyers; and HL069167 to Dr. Bleecker), the National Institutes of Allergy and Infectious Disease (AI070503 to Dr. Ober; AI079139 and AI061774 to Dr. Williams; AI050024, AI044840, and AI041040 to Dr. Barnes; and AI077439 to Dr. Burchard), the National Institute of Diabetes and Digestive and Kidney Diseases to Dr. Williams (DK064695); the National Institutes of Environmental Health Sciences (ES09606, ES018176, and ES015903 to Dr. Barnes; ES007048, ES009581, R826708, RD831861, and ES011627 to Dr. Gilliland; ES015794 to Dr. Burchard; and the Division of Intramural Research, Z01 ES049019, to Dr. London); the National Center for Research Resources (RR03048 to Dr. Barnes), the Environmental Protection Agency (83213901 and R-826724 to Dr. Barnes), the American Asthma Foundation and the Fund for Henry Ford Hospital (to Dr. Williams), Mary Beryl Patch Turnbull Scholar Program (to Dr. Barnes), the National Council of Science and Technology (Mexico) (26206-M to Dr. Romieu), the Centers for Disease Control, U.S. (to Dr. Romieu); and the Flight Attendant Medical Research Institute (FAMRI), RWJF Amos Medical Faculty Development Award, the American Asthma Foundation, and the Sandler Foundation (to Dr. Burchard). The Canadian studies (SAGE and CAPPS) were supported by AllerGen NCE Inc. (the Allergy, Genes and Environment Network which is a member of the Networks of Centres of Excellence) and by a grant from the Canadian Institutes for Health Research. Dr. Daley is supported by a Tier II Canadian Research Chair and a scholar award from the Michael Smith Foundation for Health Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- HLA
Human leukocyte antigen
- IgE
Immunoglobulin E
- GWAS
Genome wide association study
- LD
Linkage disequilibrium
- MHC
Major histocompatibility complex
- SNP
Single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Clinical Implications: This study suggests that many of the genetic risk factors for serum total IgE are shared among North American race-ethnic groups.
REFERENCES
- 1.Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin-test reactivity to allergens. N Engl J Med. 1989;320(5):271–277. doi: 10.1056/NEJM198902023200502. [DOI] [PubMed] [Google Scholar]
- 2.Sears MR, Burrows B, Flannery EM, Herbison GP, Hewitt CJ, Holdaway MD. Relation between airway responsiveness and serum IgE in children with asthma and in apparently normal children. N Engl J Med. 1991;325(15):1067–1071. doi: 10.1056/NEJM199110103251504. [DOI] [PubMed] [Google Scholar]
- 3.Sunyer J, Anto JM, Castellsague J, Soriano JB, Roca J. Total serum IgE is associated with asthma independently of specific IgE levels. The Spanish Group of the European Study of Asthma. Eur Respir J. 1996;9(9):1880–1884. doi: 10.1183/09031936.96.09091880. [DOI] [PubMed] [Google Scholar]
- 4.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8) doi: 10.1371/journal.pgen.1000166. e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125(2):328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer LJ, Burton PR, Faux JA, James AL, Musk AW, Cookson WO. Independent inheritance of serum immunoglobulin E concentrations and airway responsiveness. Am J Respir Crit Care Med. 2000;161(6):1836–1843. doi: 10.1164/ajrccm.161.6.9805104. [DOI] [PubMed] [Google Scholar]
- 8.Hopp RJ, Bewtra AK, Watt GD, Nair NM, Townley RG. Genetic analysis of allergic disease in twins. J Allergy Clin Immunol. 1984;73(2):265–270. doi: 10.1016/s0091-6749(84)80018-4. [DOI] [PubMed] [Google Scholar]
- 9.Hanson B, McGue M, Roitman-Johnson B, Segal NL, Bouchard TJ, Jr, Blumenthal MN. Atopic disease and immunoglobulin E in twins reared apart and together. Am J Hum Genet. 1991;48(5):873–879. [PMC free article] [PubMed] [Google Scholar]
- 10.Yang JJ, Burchard EG, Choudhry S, Johnson CC, Ownby DR, Favro D, et al. Differences in allergic sensitization by self-reported race and genetic ancestry. J Allergy Clin Immunol. 2008;122(4):820–827. doi: 10.1016/j.jaci.2008.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gergen PJ, Arbes SJ, Jr, Calatroni A, Mitchell HE, Zeldin DC. Total IgE levels and asthma prevalence in the US population: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2009;124(3):447–453. doi: 10.1016/j.jaci.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litonjua AA, Celedon JC, Hausmann J, Nikolov M, Sredl D, Ryan L, et al. Variation in total and specific IgE: effects of ethnicity and socioeconomic status. J Allergy Clin Immunol. 2005;115(4):751–757. doi: 10.1016/j.jaci.2004.12.1138. [DOI] [PubMed] [Google Scholar]
- 13.Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, et al. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108(3):357–362. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 14.Vergara C, Caraballo L, Mercado D, Jimenez S, Rojas W, Rafaels N, et al. African ancestry is associated with risk of asthma and high total serum IgE in a population from the Caribbean Coast of Colombia. Hum Genet. 2009;125(5–6):565–579. doi: 10.1007/s00439-009-0649-2. [DOI] [PubMed] [Google Scholar]
- 15.Mathias RA, Freidhoff LR, Blumenthal MN, Meyers DA, Lester L, King R, et al. Genome-wide linkage analyses of total serum IgE using variance components analysis in asthmatic families. Genet Epidemiol. 2001;20(3):340–355. doi: 10.1002/gepi.5. [DOI] [PubMed] [Google Scholar]
- 16.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43(9):887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y, Hu D, Peterson EL, Eng C, Levin AM, Wells K, et al. Dual-specificity phosphatase 1 as a pharmacogenetic modifier of inhaled steroid response among asthmatic patients. J Allergy Clin Immunol. 2010;126(3):618–625. doi: 10.1016/j.jaci.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar R, Seibold MA, Aldrich MC, Williams LK, Reiner AP, Colangelo L, et al. Genetic ancestry in lung-function predictions. N Engl J Med. 2010;363(4):321–330. doi: 10.1056/NEJMoa0907897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125(2):336–346. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169(3):386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 21.Galanter JM, Torgerson D, Gignoux CR, Sen S, Roth LA, Via M, et al. Cosmopolitan and ethnic-specific replication of genetic risk factors for asthma in 2 Latino populations. J Allergy Clin Immunol. 2011;128(1):37–43. doi: 10.1016/j.jaci.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet. 2009;84(5):581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, et al. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Ampleford EJ, Howard TD, Moore WC, Li H, Busse WW, et al. The C11orf30-LRRC32 region is associated with total serum IgE levels in asthmatic patients. J Allergy Clin Immunol. 2012;129(2):575–578. doi: 10.1016/j.jaci.2011.09.040. 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guilbert TW, Morgan WJ, Krawiec M, Lemanske RF, Jr, Sorkness C, Szefler SJ, et al. The Prevention of Early Asthma in Kids study: design, rationale and methods for the Childhood Asthma Research and Education network. Control Clin Trials. 2004;25(3):286–310. doi: 10.1016/j.cct.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Sorkness CA, Lemanske RF, Jr, Mauger DT, Boehmer SJ, Chinchilli VM, Martinez FD, et al. Long-term comparison of 3 controller regimens for mild-moderate persistent childhood asthma: the Pediatric Asthma Controller Trial. J Allergy Clin Immunol. 2007;119(1):64–72. doi: 10.1016/j.jaci.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 28.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115(2):233–242. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Bacharier LB, Phillips BR, Bloomberg GR, Zeiger RS, Paul IM, Krawiec M, et al. Severe intermittent wheezing in preschool children: a distinct phenotype. J Allergy Clin Immunol. 2007;119(3):604–610. doi: 10.1016/j.jaci.2006.12.607. [DOI] [PubMed] [Google Scholar]
- 30.Strunk RC, Bacharier LB, Phillips BR, Szefler SJ, Zeiger RS, Chinchilli VM, et al. Azithromycin or montelukast as inhaled corticosteroid-sparing agents in moderate-to-severe childhood asthma study. J Allergy Clin Immunol. 2008;122(6):1138–1144. doi: 10.1016/j.jaci.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. American Thoracic Society. Am J Respir Crit Care Med. 2000;162(6):2341–2351. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolerman ES, Manning AK, McAteer JB, Dupuis J, Fox CS, Cupples LA, et al. Haplotype structure of the ENPP1 Gene and Nominal Association of the K121Q missense single nucleotide polymorphism with glycemic traits in the Framingham Heart Study. Diabetes. 2008;57(7):1971–1977. doi: 10.2337/db08-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.Lourenco VM, Pires AM, Kirst M. Robust linear regression methods in association studies. Bioinformatics. 2011;27(6):815–821. doi: 10.1093/bioinformatics/btr006. [DOI] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 38.Cochran WG. The combination of esimates from different experiments. Biometrics. 1954;10(1):101–129. [Google Scholar]
- 39.Williams LK, Peterson EL, Wells K, Ahmedani BK, Kumar R, Burchard EG, et al. Quantifying the proportion of severe asthma exacerbations attributable to inhaled corticosteroid nonadherence. J Allergy Clin Immunol. 2011;128(6):1185–1191. doi: 10.1016/j.jaci.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, et al. Beta 2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet. 2006;119(5):547–557. doi: 10.1007/s00439-006-0169-2. [DOI] [PubMed] [Google Scholar]
- 41.Becker A, Watson W, Ferguson A, mich-Ward H, Chan-Yeung M. The Canadian asthma primary prevention study: outcomes at 2 years of age. J Allergy Clin Immunol. 2004;113(4):650–656. doi: 10.1016/j.jaci.2004.01.754. [DOI] [PubMed] [Google Scholar]
- 42.Begin P, Tremblay K, Daley D, Lemire M, Claveau S, Salesse C, et al. Association of urokinase-type plasminogen activator with asthma and atopy. Am J Respir Crit Care Med. 2007;175(11):1109–1116. doi: 10.1164/rccm.200607-1012OC. [DOI] [PubMed] [Google Scholar]
- 43.Kozyrskyj AL, HayGlass KT, Sandford AJ, Pare PD, Chan-Yeung M, Becker AB. A novel study design to investigate the early-life origins of asthma in children (SAGE study) Allergy. 2009;64(8):1185–1193. doi: 10.1111/j.1398-9995.2009.02033.x. [DOI] [PubMed] [Google Scholar]
- 44.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67(5):1154–1162. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan A, Newman DL, Shon AM, Schneider DH, Kuldanek S, Ober C. Variation in the type I interferon gene cluster on 9p21 influences susceptibility to asthma and atopy. Genes Immun. 2006;7(2):169–178. doi: 10.1038/sj.gene.6364287. [DOI] [PubMed] [Google Scholar]
- 46.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358(16):1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granada M, Wilk JB, Tuzova M, Strachan DP, Weidinger S, Albrecht E, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J Allergy Clin Immunol. 2012;129(3):840–845. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woszczek G, Kowalski ML, Borowiec M. Association of asthma and total IgE levels with human leucocyte antigen-DR in patients with grass allergy. Eur Respir J. 2002;20(1):79–85. doi: 10.1183/09031936.02.01002001. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA, et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol. 2010;125(2):328–335. doi: 10.1016/j.jaci.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothers J, Halonen M, Stern DA, Lohman IC, Mobley S, Spangenberg A, et al. Adaptive cytokine production in early life differentially predicts total IgE levels and asthma through age 5 years. J Allergy Clin Immunol. 2011;128(2):397–402. doi: 10.1016/j.jaci.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holzer U, Kwok WW, Nepom GT, Buckner JH. D ifferential antigen sensitivity and costimulatory requirements in human Th1 and Th2 antigen-specific CD4+ cells with similar TCR avidity. J Immunol. 2003;170(3):1218–1223. doi: 10.4049/jimmunol.170.3.1218. [DOI] [PubMed] [Google Scholar]
- 52.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, et al. SCAN: SNP and copy number annotation. Bioinformatics. 2010;26(2):259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brand S. Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut. 2009;58(8):1152–1167. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 55.Vergara C, Tsai YJ, Grant AV, Rafaels N, Gao L, Hand T, et al. Gene encoding Duffy antigen/receptor for chemokines is associated with asthma and IgE in three populations. Am J Respir Crit Care Med. 2008;178(10):1017–1022. doi: 10.1164/rccm.200801-182OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105(3):506–513. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 57.Schedel M, Carr D, Klopp N, Woitsch B, Illig T, Stachel D, et al. A signal transducer and activator of transcription 6 haplotype influences the regulation of serum IgE levels. J Allergy Clin Immunol. 2004;114(5):1100–1105. doi: 10.1016/j.jaci.2004.07.048. [DOI] [PubMed] [Google Scholar]
- 58.Jarvis D, Luczynska C, Chinn S, Potts J, Sunyer J, Janson C, et al. Change in prevalence of IgE sensitization and mean total IgE with age and cohort. J Allergy Clin Immunol. 2005;116(3):675–682. doi: 10.1016/j.jaci.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Yunginger JW, Gleich GJ. Seasonal changes in serum and nassal IgE concentrations. J Allergy Clin Immunol. 1973;51(3):174–186. doi: 10.1016/0091-6749(73)90022-5. [DOI] [PubMed] [Google Scholar]
- 60.Salkie ML, Weimer N. The influence of season and of sex on the serum level of total IgE and on the distribution of allergen-specific IgE. Clin Biochem. 1984;17(6):362–366. doi: 10.1016/s0009-9120(84)90734-3. [DOI] [PubMed] [Google Scholar]
- 61.Oryszczyn MP, nnesi-Maesano I, Charpin D, Paty E, Maccario J, Kauffmann F. Relationships of active and passive smoking to total IgE in adults of the Epidemiological Study of the Genetics and Environment of Asthma, Bronchial Hyperresponsiveness, and Atopy (EGEA) Am J Respir Crit Care Med. 2000;161(4 Pt 1):1241–1246. doi: 10.1164/ajrccm.161.4.9905027. [DOI] [PubMed] [Google Scholar]
- 62.Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, et al. Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet. 2001;69(5):1080–1094. doi: 10.1086/323922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.