Abstract
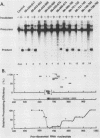
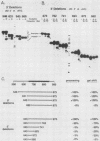
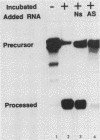
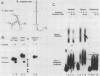
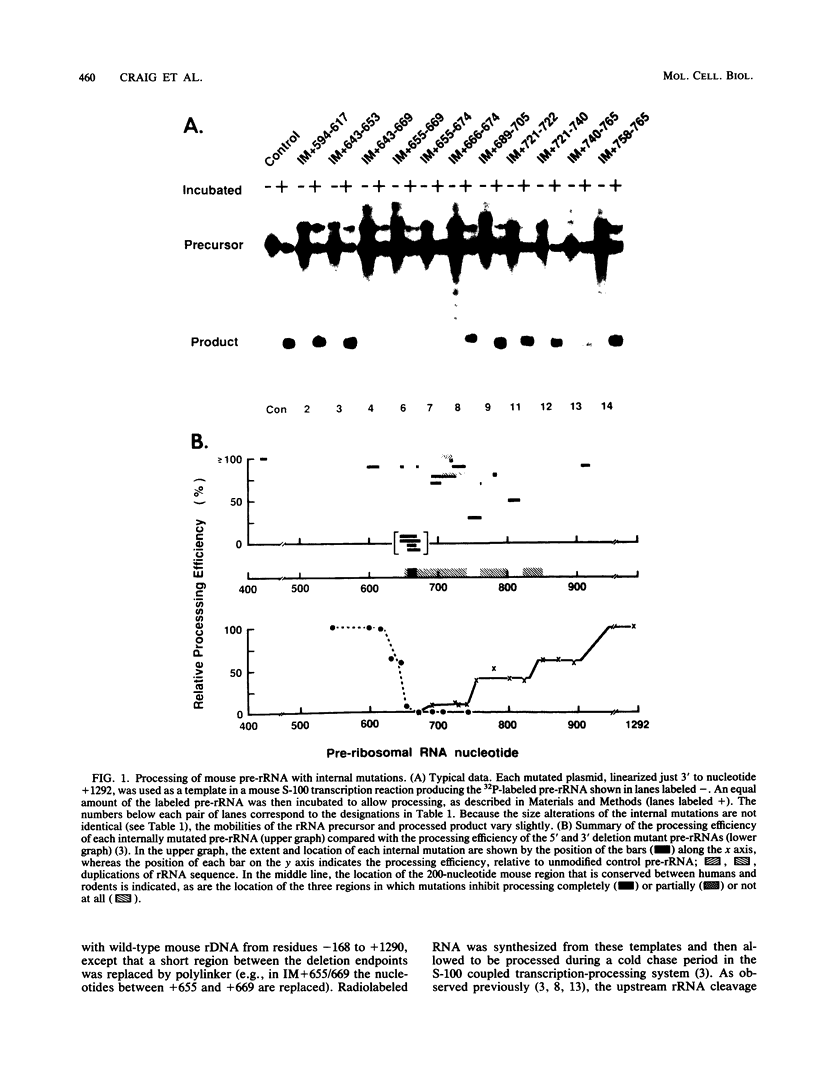
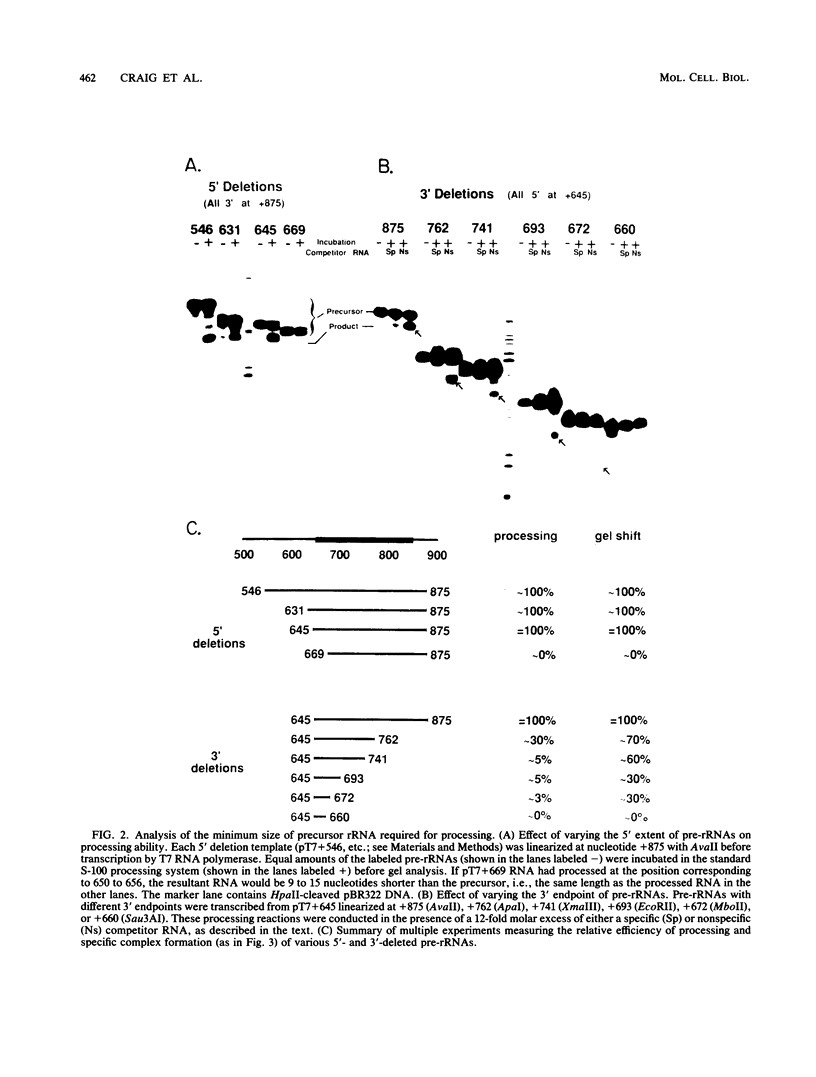
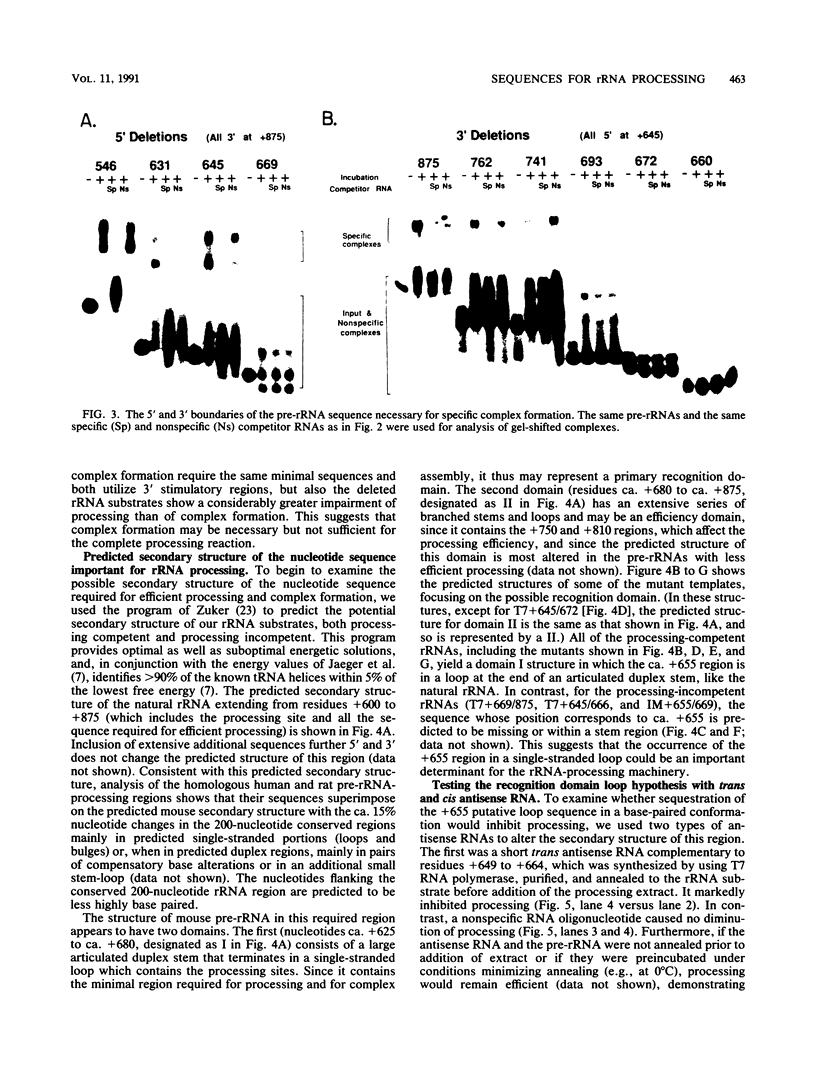
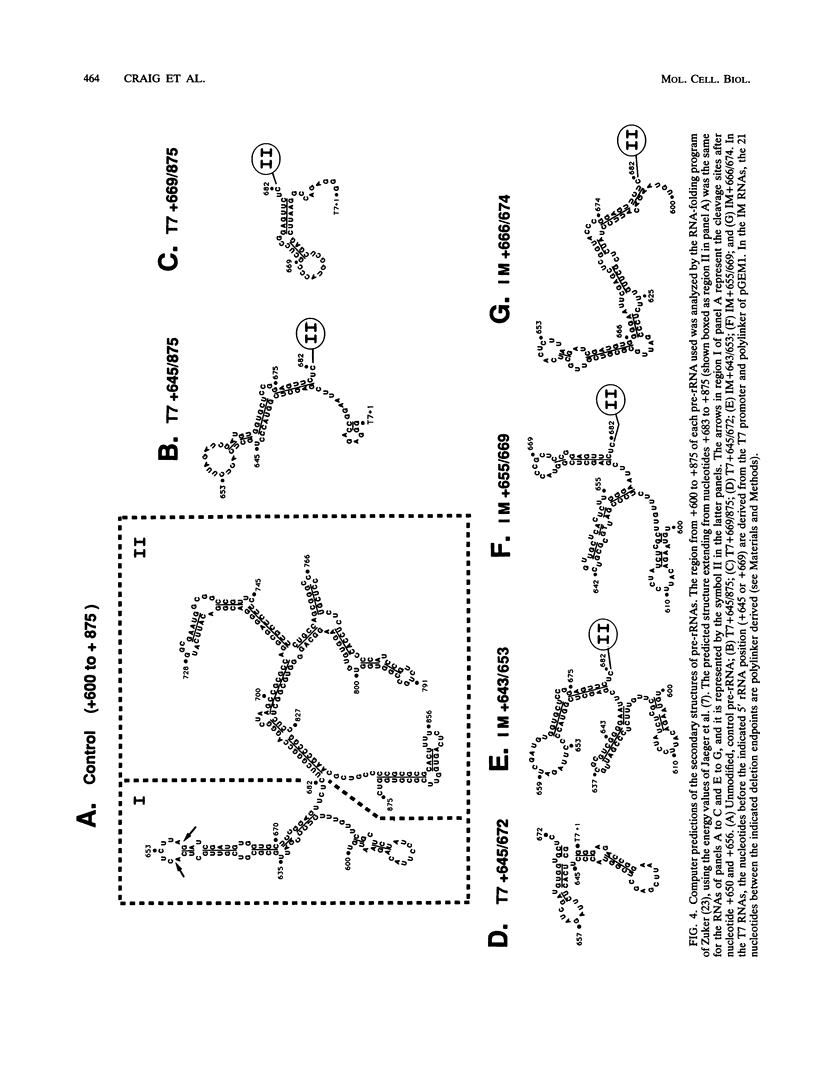
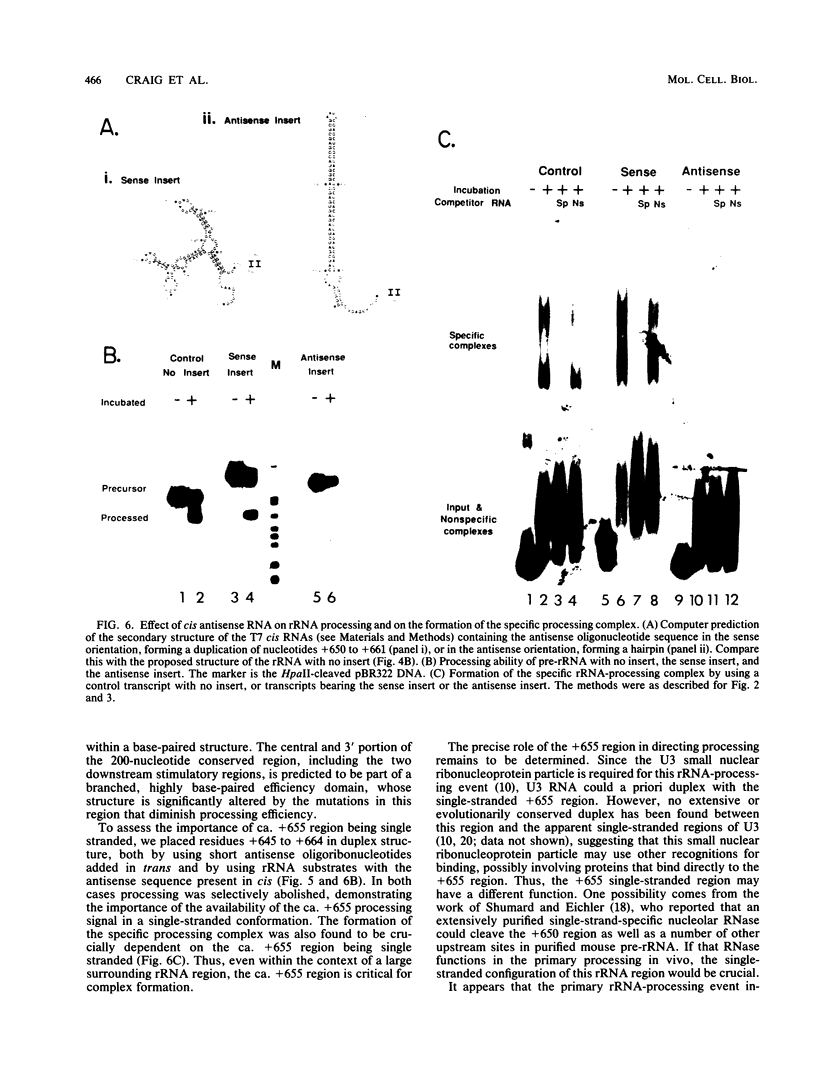
The first processing step in the maturation of mouse precursor rRNA involves cleavage at nucleotide ca. +650, at the 5' border of a 200-nucleotide region that is conserved across mammals and contains the sequences that direct the processing. To identify the relevant sequence elements, we used rRNAs with small internal mutations and short pre-rRNA substrates. Much of the region can be mutated without appreciable effect, but nucleotides +655 to +666 appear to be absolutely required and short segments surrounding +750 and +810 markedly stimulate processing. The minimal processing signal corresponds to rRNA nucleotides +645 to +672. Formation of a ribonucleoprotein complex of retarded electrophoretic mobility is evidently necessary but not sufficient for processing. Computer-assisted analysis suggested a phylogenetic- and mutant-supported secondary structure in which the minimal processing signal forms a stem with the +655 region in the loop, and there is a separate branched duplex containing the downstream stimulatory sequences. Use of antisense RNA, in trans and in cis, to sequester the +655 region in a duplex supported the hypothesis that this critical region was needed in a single-stranded conformation for processing and for specific complex formation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowman L. H., Goldman W. E., Goldberg G. I., Hebert M. B., Schlessinger D. Location of the initial cleavage sites in mouse pre-rRNA. Mol Cell Biol. 1983 Aug;3(8):1501–1510. doi: 10.1128/mcb.3.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Muenchau P., Wolf R. E., Jr A method for cloning mixtures of long, synthetic oligodeoxynucleotides. Gene Anal Tech. 1987 Sep-Oct;4(5):105–110. doi: 10.1016/0735-0651(87)90003-3. [DOI] [PubMed] [Google Scholar]
- Craig N., Kass S., Sollner-Webb B. Nucleotide sequence determining the first cleavage site in the processing of mouse precursor rRNA. Proc Natl Acad Sci U S A. 1987 Feb;84(3):629–633. doi: 10.1073/pnas.84.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G. J., Maroney P. A., Branch A., Benenfield B. J., Robertson H. D., Nilsen T. W. Accurate processing of human pre-rRNA in vitro. Mol Cell Biol. 1989 Oct;9(10):4422–4431. doi: 10.1128/mcb.9.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., Turner D. H., Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Sollner-Webb B. The first pre-rRNA-processing event occurs in a large complex: analysis by gel retardation, sedimentation, and UV cross-linking. Mol Cell Biol. 1990 Sep;10(9):4920–4931. doi: 10.1128/mcb.10.9.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- King T. C., Sirdeskmukh R., Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986 Dec;50(4):428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K. The organization and transcription of eukaryotic ribosomal RNA genes. Prog Nucleic Acid Res Mol Biol. 1984;31:115–160. doi: 10.1016/s0079-6603(08)60376-1. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P. RNA processing comes of age. J Cell Biol. 1981 Dec;91(3 Pt 2):28s–38s. doi: 10.1083/jcb.91.3.28s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziuddin, Little R. D., Labella T., Schlessinger D. Transcription and processing of RNA from mouse ribosomal DNA transfected into hamster cells. Mol Cell Biol. 1989 Apr;9(4):1667–1671. doi: 10.1128/mcb.9.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumard C. M., Eichler D. C. Ribosomal RNA processing. Limited cleavages of mouse preribosomal RNA by a nucleolar endoribonuclease include the early +650 processing site. J Biol Chem. 1988 Dec 25;263(36):19346–19352. [PubMed] [Google Scholar]
- Shumard C. M., Torres C., Eichler D. C. In vitro processing at the 3'-terminal region of pre-18S rRNA by a nucleolar endoribonuclease. Mol Cell Biol. 1990 Aug;10(8):3868–3872. doi: 10.1128/mcb.10.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. The 5' end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989 Dec 5;210(3):497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- Vance V. B., Thompson E. A., Bowman L. H. Transfection of mouse ribosomal DNA into rat cells: faithful transcription and processing. Nucleic Acids Res. 1985 Oct 25;13(20):7499–7513. doi: 10.1093/nar/13.20.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]