Abstract
Steroid hormones are synthesized from cholesterol primarily in the adrenal gland and the gonads and play vital roles in normal physiology, the control of development, differentiation, metabolic homeostasis, and reproduction. The actions of these small lipophilic molecules are mediated by intracellular receptor proteins. It is just over 25 yr since the first cDNA for steroid receptors were cloned, a development that led to the birth of a superfamily of ligand-activated transcription factors: the nuclear receptors. The receptor proteins share structurally and functionally related ligand binding and DNA-binding domains but possess distinct N-terminal domains and hinge regions that are intrinsically disordered. Since the original cloning experiments, considerable progress has been made in our understanding of the structure, mechanisms of action, and biology of this important class of ligand-activated transcription factors. In recent years, there has been interest in the structural plasticity and function of the N-terminal domain of steroid hormone receptors and in the allosteric regulation of protein folding and function in response to hormone, DNA response element architecture, and coregulatory protein binding partners. The N-terminal domain can exist as an ensemble of conformers, having more or less structure, which prime this region of the receptor to rapidly respond to changes in the intracellular environment through hormone binding and posttranslation modifications. In this review, we address the question of receptor structure and function dynamics with particular emphasis on the structurally flexible N-terminal domain, intra- and interdomain communications, and the allosteric regulation of receptor action.
Introduction
-
Functional and Structural Organization of Steroid Hormone Receptors
Ligand binding domain (LBD) (AF2)
Hinge region
DNA binding domain (DBD)
N-terminal domain (NTD) (AF1)
Structural Analysis of Full-Length Nuclear Receptors
-
Steroid Hormone Receptors Function in a Ligand-, Cell-, and Promoter-Specific Manner
Selective utilization of AF1 and AF2: cell culture studies
Selective utilization of AF1 and AF2: in vivo studies
-
Intrinsically Disordered Structure and Steroid Hormone Receptor Action
What does it mean to be intrinsically disordered?
Coupled folding and binding of the steroid hormone receptor NTD
Intrinsic Disorder Can Optimize Allosteric Coupling
Structural Dynamics and the Design of Selective Small Molecule Modulators of Steroid Hormone Receptor Function
Conclusions and Future Challenges
I. Introduction
Steroid hormone receptors (SHRs) are ligand-regulated transcription factors that control a diverse array of physiological processes. The importance of SHRs in the regulation of cellular and developmental processes is well established (1–6), yet the mechanism by which transcription of target genes is controlled by steroid hormones and SHRs is not completely understood. In recent years, studies from many laboratories have revealed details of the mechanisms by which SHRs modulate transcription regulation and activate cell signaling pathways (for recent reviews, see Refs. 7–11). This has resulted in the identification of molecules with diverse activities that regulate receptor function and the elucidation of the role of SHRs in both normal physiological processes and pathological conditions, including cancer, cardiovascular disease, and several other disorders (12–19). However, despite remarkable progress, the mechanisms by which SHRs function in a tissue-specific manner remains unclear. To more fully understand how SHRs function as transcription factors, it is therefore necessary to know how these receptor proteins are activated by hormone and bind to specific DNA sequences in the regulatory regions of target genes to either up- or down-regulate the rate of transcriptional initiation by RNA Pol II. Identification of the specific factors and molecular mechanisms that contribute to the normal actions and aberrant pathological effects of SHRs is critical for the development of novel and potent therapeutic agents. It is worth noting that steroid hormones also mediate rapid actions through both classical SHR and membrane-associated receptors: these mechanisms have been discussed in a number of excellent reviews (20–23) and will not be specifically discussed here.
SHRs are part of a large superfamily of nuclear receptors that share structural and functional properties (24, 25). The cloning of the cDNA for different SHRs has greatly facilitated the molecular understanding of their actions as well as receptor protein structure and function (26–33). In animals, the main classes of steroid hormones are progestins (C22 steroids), corticosteroids (C21), androgens (C19), and estrogens (C18), whereas ecdysone (C27), the moulting hormone, is found in insects. Related molecules that also act as ligands for members of the nuclear receptor superfamily include bile acids and the sterol, vitamin D3. On the basis of amino acid sequence homology and DNA binding specificity, the classical SHRs are found in subfamily III, which includes estrogen receptors (ER) α and β (NR3A1 and A2 respectively); the estrogen-related receptors (ERR) α, β, and γ (NR3B1-3); the glucocorticoid receptor (GR; NR3C1); the mineralocorticoid receptor (MR; NR3C2); the progesterone receptor (PR; NR3C3); and the receptor for androgens (AR; NR3C4) (25). The receptors for vitamin D3 (VDR; NR1I1) and ecdysone (NR1H1) are found in subfamily I and act has heterodimers with retinoid X receptors (RXR) α, β, or γ (NR2B1-3) or the insect homolog ultraspiracle, respectively.
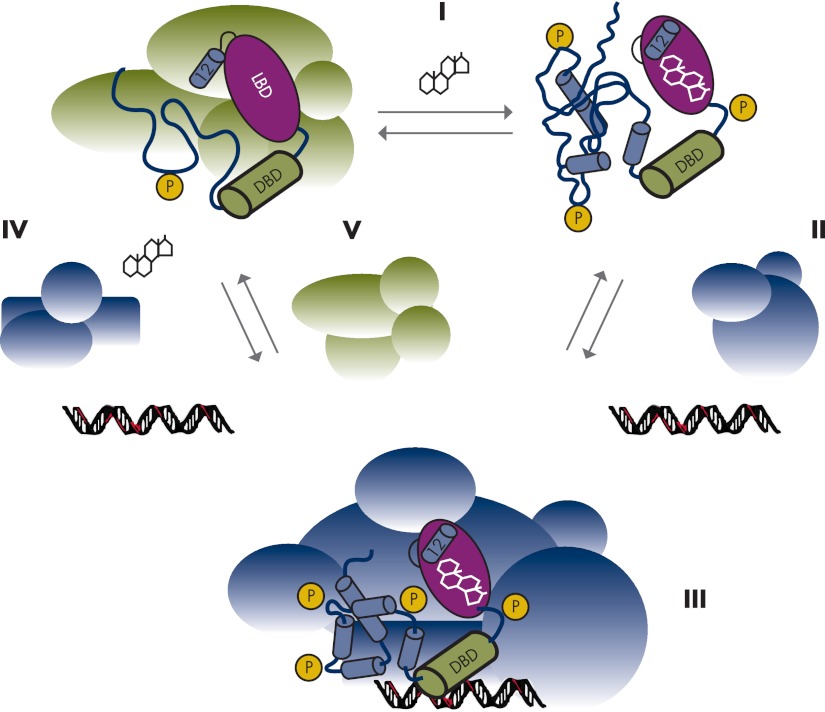
It is well established that transcriptional activation of genes involves the regulated assembly of multiprotein complexes on enhancers and promoters (34–36). However, despite an exponentially increasing number of reported coregulatory protein binding partners for SHRs and the mapping of receptor interacting domains within these proteins, the mechanisms by which SHRs function remains an area of important investigation. Furthermore, observations from both basic biochemical analysis and recent cell-based studies led to the inescapable conclusion that in cells, SHRs behave very dynamically, showing rapid and reversible interactions with partner proteins, together with chromatin and DNA (Fig. 1) (37–41). The coupling of the functional and structural dynamics of SHRs with differences in the local concentrations of potential coregulatory proteins is likely to result in receptors bringing differing sets of binding partners together in response to agonist or antagonist ligands, such that an agonist in one cell type can be an antagonist in another cell type (42–45). Binding of agonistic or antagonistic ligands leads to different allosteric changes in SHRs, making them competent to exert positive or negative effects on the expression of target genes by different mechanisms. The result is SHR-mediated modulation of specific responses in different tissues depending on the physiological and genetic context of the cell (46–48).
Figure 1.
General dynamics and mechanism of SHR action. SHR action is spatially, temporally, and structurally highly dynamic. In the absence of hormone, the receptor is complexed with co-chaperone molecules (green) in the cytoplasm. The NTD/AF1 exists in an ID conformation, compared with the well-ordered DBD and LBD. Factors affecting disorder-order transition of the SHR AF1/NTD are illustrated. The binding of hormone (I) causes rearrangement of regions in the LBD (helix 12, AF2), and this leads to translocation to the nucleus and binding to DNA response elements and coregulatory proteins (blue) (II). Under the influence of factors shown, NTD/AF1 undergoes disorder-order transition, resulting in the folding of NTD/AF1. In this conformation, AF1/NTD surfaces are well suited for the interaction with specific coactivators, binding of which further influences the conformation of NTD/AF1 and facilitates the assembly of the transcription initiation complex (blue) in a promoter-specific manner (III). The binding of DNA and/or coregulatory proteins may allosterically regulate ligand binding, which could lead to formation of SHR-transcription complexes (IV). The formation of transcriptionally competent complexes by the DNA-bound SHR (III) must be readily reversible, and the co-chaperone complex (green) may aid in dissociating the SHR-chromatin complex, recycling of the receptor protein to the cytoplasm, and stabilization of the ID NTD/AF1 (V). The SHR is also subject to posttranslational modifications, such as phosphorylation (P), which are likely to fine-tune the allosteric regulation of receptor structure and function.
Although some of these complex behaviors have been explained by identifying cell-specific binding partners for SHRs, it is as yet unclear whether other dynamic considerations must be taken into account while defining the underlying mechanisms, especially given the ubiquitous nature of a large number of known coregulatory proteins. We propose that the often disparate effects of selective ligand modulators of SHR function, the many and varied nonconsensus DNA response elements for SHRs, and the effects of proteins that bind reversibly to various sites of SHRs can be explained by the intra- and interdomain coupling and structural dynamics within the receptor protein. Although conformational stability of most proteins determines their biological function, it has been proposed that the structural plasticity of the SHR proteins can explain much about their observed behaviors (49–51). The good and bad effects of steroids on physiology, pathophysiology, and in therapeutic use are well documented. We contend that understanding the differential effects of ligands, response element sequences, and various protein binding partners on the dynamic structure of SHRs will provide essential knowledge for those seeking to design specific steroidal (and nonsteroidal) selective SHR modulators (SRMs), which minimize adverse side effects.
A functionally competent structure can be achieved by the SHR protein through different regions of the receptor rapidly and reversibly adopting various conformations, controlled by ligand and DNA binding. This in turn may help to create protein surfaces that are readily available for selective binding to coregulatory proteins, resulting in SHR-mediated transcriptional regulation of target genes (Fig. 1). How and under what conditions such surfaces are created to include or exclude coregulatory proteins in a promoter- and cell- specific manner is an open question. The answer may in part lie with the dynamic structure of SHRs combined with allosteric regulation mediated through inter- and/or intradomain cross talk, which can be influenced by hormone binding, DNA architecture, and the binding of coregulatory proteins. In this review article, we discuss the properties of the SHRs that allow this discriminatory action, which is critical to SHR transduction of different signals. It is timely in light of recent advances in our understanding of SHR protein structure and the growing interest in targeting regions of the receptor distinct from the ligand binding pocket, with small molecule modulators that may ultimately lead to a new portfolio of tailored therapeutic treatments for hormone-dependent diseases.
II. Functional and Structural Organization of Steroid Hormone Receptors
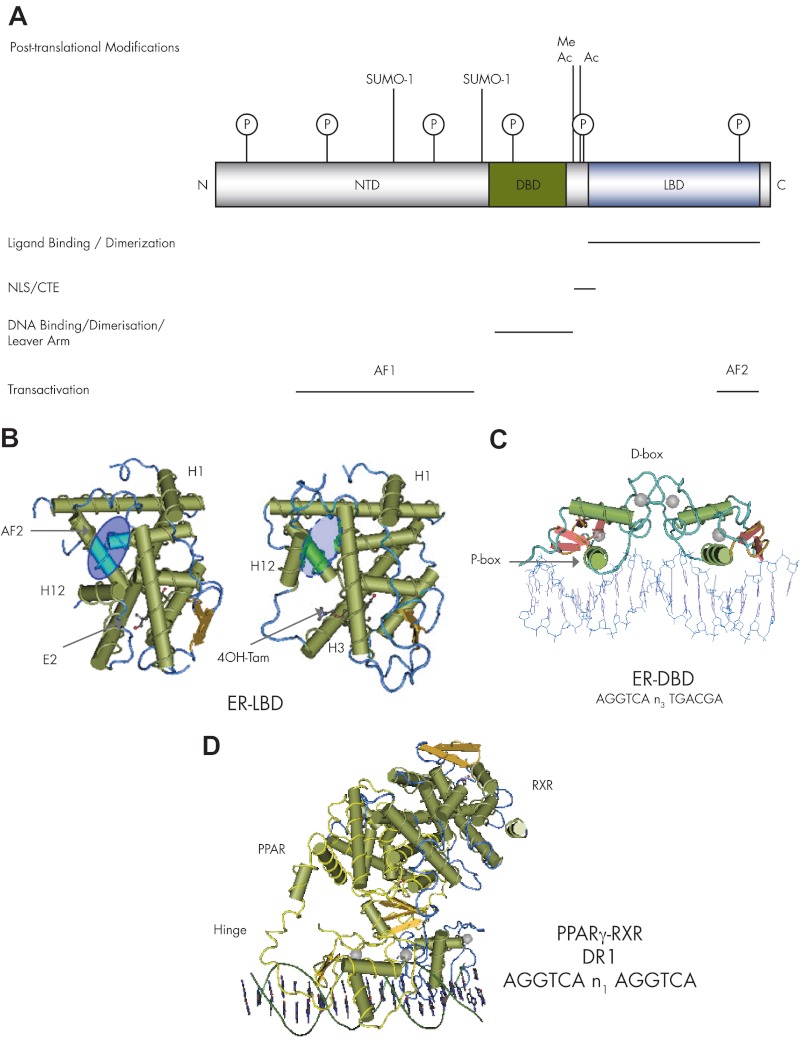
From early biochemical studies with purified receptor proteins, it was deduced that SHRs would consist of discrete regions important for hormone and DNA binding (52). Limited proteolysis and antibody binding identified a third distinct region, termed the “immunoactive” domain (53, 54). The cloning of the ERα and GR cDNA in the mid-1980s and subsequent deletion mapping studies refined this domain organization into the picture we have today (Fig. 2A). SHRs are modular proteins that consist of a highly variable N-terminal domain (NTD; also called A/B domain); a 66-amino acid core DNA binding domain (DBD; or C domain); a flexible hinge domain of 40 to 60 amino acids (D domain); a ligand binding domain (LBD) of about 265 amino acids (E domain); and in some receptors a variable stretch of amino acids at the very C terminus referred to as the F domain (Fig. 2A) (24). Two activation domains, termed AF1 and AF2, are located within the NTD and LBD, respectively, and are responsible for mediating the transcriptional activity of SHRs. The two AF can act in an independent manner, although usually full receptor activity requires synergistic cooperation between AF1 and AF2. This cooperativity can be mediated by direct receptor N/C-terminal interactions or the binding of coregulatory proteins to either activation function separately or simultaneously to both AF1 and AF2.
Figure 2.
SHR domain structure and function. A, Schematic representation of the domain organization of SHRs, showing the LBD and DBD and the structurally distinct NTD. Regions of the receptor protein important for ligand binding, DNA binding, dimerization, and transactivation are indicated below the protein. The position of the NLS and the CTE of the core DBD are also shown. Sites of potential posttranslational modification are indicated above the protein: including, acetylation (Ac), phosphorylation (P), methylation (Me) and sumoylation (SUMO-1; small ubiquitin-like modifier 1). B, Crystal structures for the LBD of the ERα bound with agonist (estradiol) (pdb 1ERE) or a selective ER modulator (4-hydroxhtamoxifen) (pdb 3ERT). The ligand binding pocket is indicated, as is the position of helices 1 and 12 and the AF2 surface (blue oval). C, Structures of the ERα-DBD (ER: pdb 1HCQ), which binds to a near palindromic DNA sequence as a homodimer. The presences of the P- and D-box amino acid residues are indicated. D, The crystal structure for PPARγ-RXRα heterodimer complex (pdp 3E00) on a direct repeat response element, with the half-sites separated by 1 bp. The PPARγ structure is in yellow, and RXR is in blue; the hinge region of PPAR is indicated.
A. Ligand binding domain (LBD) (AF2)
1. Hormone binding
In the absence of hormone, SHRs are present in a heterocomplex with heat shock proteins and immunophilins (Fig. 1). This complex has been particularly well studied for the GR and consists of a dimer of hsp90, together with one copy of hsp70, p23, and the immunophilins FKBP51 and -52; the latter have been proposed to link SHRs to the motor protein dynein and the microtubule retrograde transport network (reviewed in Refs. 55 and 56). The binding site for hsp90 on the GR has been mapped to a seven-amino acid sequence at the N-terminal end of the LBD (residues 529 to 535 for human GR), but the exact sequence of amino acids was found not to be critical for this interaction (57). Importantly, the binding of the hsp90 heterocomplex was essential for maintaining the GR in a state primed to bind hormone because reducing levels of hsp90 in yeast cells inhibited receptor action (58). In addition to a role in hormone binding, interactions with hsp90 are important for cycling of SHRs on DNA/chromatin and for receptor turnover (56, 59) (Fig. 1).
The primary role of the LBD is to bind specific steroid ligands, which become buried in the hydrophobic interior. Selectivity of different hormones is based upon the size of the ligand binding pocket and interactions between specific amino acids and the A and D rings of the steroid molecule (reviewed in Ref. 60). From analysis of the amino acid sequences of SHR-like proteins from different organisms, including invertebrates such as molluscs and annelids, it has been proposed that the ancestral gene for SHRs was ER-like and bound estrogens (reviewed in Ref. 61).
The binding of hormone has dramatic consequences for individual receptor structure-function relationships. Structures of the LBD for each class of SHR bound to either agonists or antagonist are available. The LBD is made up of 11 α-helices and a variable number of β-strands; for example, the ERα-LBD has 11 helices and two β-strands (Fig. 2B) (reviewed in Ref. 60). The agonist-bound receptors share a globular folded structure that has been described as a three-layer helical sandwich, with the hormone buried within the ligand binding pocket and sealed in by the conformational change in helix 12 (note that convention names the helices 1 to 12 after the first structures solved; in the SHR subfamily, helix 2 is absent or disordered). The reorientation of helix 12 in response to hormone binding, together with residues in helices 3, 4, and 5 make up the AF2 surface (62–64). This surface pocket has a hydrophobic interior and typically binds coactivator proteins containing the peptide motif LxxLL (where L is leucine and x is any amino acid) that forms a α-helix, which buries the leucine residues within the groove (62). In addition to the hydrophobic surface, the LxxLL helix is orientated by a lysine residue in helix 4 and a glutamic acid in helix 12, which together form a “charge clamp” (63, 65–67).
Extensive analysis of ERα and ERβ LBD revealed the importance of residues outside the ligand binding pocket that contributed to receptor-specific ligand interactions and allosteric communication with the coactivator binding surface (64). In particular, the position of helix 11 was implicated in ER subtype-selective agonist, partial agonist, or antagonist responses to a particular ligand. An allosteric network linking the ligand binding pocket and the AF2 surface was also elegantly demonstrated through a mutation screen with the GR. A number of mutations were identified that reduced the requirement for hsp90 in hormone binding by the receptor: two of the residues, tyrosine 598 (numbering for human GR) in helices 5–6 and methionine 752 (helix 12) contribute to the AF2 surface, whereas a third residue, methionine 604 (helices 5–6) is part of the ligand binding pocket, which provides a structural basis for the allosteric regulation of ligand and coregulatory protein binding to the LBD (68). This study also revealed a further role for hsp90 in connecting this allosteric network.
The structure of the LBD of ER and AR bound to antagonists or SRMs have also been solved (e.g., Fig. 2B). In these cases, it is interesting that the overall fold of the LBD remains similar to that of the agonist-bound receptor, but there are changes within the ligand binding pocket or in the position or flexibility of helix 12. In the case of the ER SRMs, tamoxifen (Fig. 2B) and raloxifene, displacement of helix 12 prevents coregulatory proteins binding to the AF2 surface (60, 69, 70). The agonist activity of tamoxifen in certain cells has been shown to depend on AF1 because removal of the NTD of the receptor abrogates the activation of a target gene by this ligand (71–73).
Interestingly, other surfaces on the AR-LBD have been shown to bind small molecules that interfered with the binding of coregulatory proteins to the AF2 site (74). It is tempting to speculate that this site, termed “BF3,” is an auxiliary protein-protein binding site or that it could be exploited as an allosteric drug binding surface.
2. Dimerization interfaces
Sequences within the LBD have also been identified as dimerization interfaces. ER-LBD dimerization involves amino acids in helix 10, but the dimerization interface for the GR-LBD comprises residues in helices 1, 3, and 5 (43, 75–77). However, whether the presence of a dimerization site within the LBD is common to all SHRs is less clear. In the case of the AR, this is a subject of debate, and it is noteworthy that in the crystal structures of the AR-LBD, only monomers were observed. A recent comparison of the AR-LBD with the GR sequence and structure identified residues in helices 1, 3, and 5 as a potential dimerization interface (78). The involvement of different regions of the LBD in dimerization suggests that this interaction could be differentially regulated allosterically by hormone binding, which in turn could lead to cell- or gene-selective responses through the exposure of different surfaces for coregulatory protein binding.
3. N/C-terminal interactions
Interestingly, whereas other SHRs interact with coactivator proteins via LxxLL motifs, which bind in the hydrophobic groove of AF2, the AR-LBD preferentially binds the AR-NTD and coactivators with more bulky hydrophobic residues in the sequence F/WxxLF/W/Y (65–67, 79–81). This AR-N/C terminal interaction results in stabilization of the receptor protein, a reduction in the dissociation of hormone and selective gene activation (82–84). There is also growing evidence that N/C-terminal domain interactions occur for other SHRs, for example ERα (85, 86), PR (87, 88), and the MR (89).
The structural basis for the AR-LBD/AF2 binding preference for bulky hydrophobic aromatic residues over leucine has been reported (65–67). X-ray crystallography studies showed that the AR-NTD FxxLF motif forms a charge clamp with glutamic acid 897 in helix 12 and lysine 720 at the C-terminal end of helix 3, and the bulky hydrophobic side chains fit better into the surface pocket on the LBD. In contrast, an LxxLL motif peptide fails to make hydrogen bond contacts with the glutamic acid residue in helix 12 and makes fewer hydrophobic contacts with the surface of the LBD (65–67).
It can be concluded from the above studies that the binding of hormone acts as an allosteric switch to regulate SHR-DNA and SHR-protein interactions, including interdomain interactions and/or dimerization.
B. Hinge region
The binding of steroid leads to exposure of the nuclear localization signal (RKxK/RK), within the hinge domain, and translocation of the receptor-steroid complex into the nucleus and subsequent DNA binding (Fig. 1). The hinge region is a 40- to 60-residue linker sequence between the DBD and the LBD, which shows little amino acid homology between different SHRs. The hinge region contains the C-terminal extension (CTE) of the DBD, the nuclear localization signal (NLS), and sites for posttranslational modifications (Fig. 2A).
The CTE is defined as the sequence immediately C-terminal of the conserved second zinc finger of the core DBD. In the case of nonsteroid receptors, for example in the VDR and the thyroid hormone receptor β (NR1A2,), the CTE forms an α-helix and is important for heterodimerization with RXR and minor groove DNA contacts (reviewed in Ref. 90). In the original structures of the GR and ER and subsequently the AR DBD, the CTE appeared disordered, although mutational studies highlighted a role in DNA binding (91–94). Interestingly, recent structural analysis of a number of GR-DBD-DNA complexes observed a helical conformation for the CTE (95). Electron densities were also observed for the first seven to eight residues of the PR-CTE, which revealed interactions with both the minor groove and the core PR-DBD (96, 97). Mutational analysis demonstrated that the PR-CTE was necessary for recognizing nucleotides flanking the DNA response element (96). More recently, the hinge region of the PR was found to regulate the response of the receptor on different promoters. This involved cytoplasmic/nuclear shuttling and retention in response to acetylation of lysines in the NLS and phosphorylation of serines in the NTD (98).
Structural and functional analysis of the AR-hinge region (residues 629 to 634) showed that the NLS interacted with the armadillo repeats 2, 3, and 4 of the NLS receptor protein α-importin (99). An extended receptor peptide (amino acids 617 to 634) adopted a β-turn-like structure when bound to α-importin, which was unique to the AR NLS (99). Recent mutational analysis of this basic motif in the AR demonstrated a number of functional interrelationships, including nuclear localization, DNA binding, intranuclear dynamics, and transcriptional activity (100). The lysines in this motif are also subject to acetylation (101–103) and methylation (104).
Collectively, the above-mentioned studies strongly suggest that the CTE can adopt different conformations depending upon whether the receptor is bound to DNA, protein partners, or free in solution (99, 105, 106). Furthermore, because the CTE itself appears to be an intrinsically disordered (ID) region, it suggests the possibility that CTE interactions are involved in mediating allosteric coupling with the NTD and LBD (discussed in Sections V.B.2. and V.I).
C. DNA binding domain (DBD)
On the basis of amino acid sequence homology and DNA binding specificity, SHRs can be classified as members of the ER subfamily (ERα, β, and ERRα, β, and γ) and the GR subfamily (GR, MR, PR, and AR) (25). The core DBD of SHRs consists of approximately 66 amino acids and involves the coordination of two zinc ions by eight cysteine residues (90, 107). SHRs typically bind in vitro to palindromic or palindromic-like DNA sequences as homodimers. The consensus sequences for ER have the half-site 5′-AGGTCA-3′, and for GR, PR, MR, and AR the half-site is 5′-AGAACA-3′; each half-site is separated by a three-nucleotide spacer, completing the 15-bp receptor response element (90, 107). Early pioneering mutagenesis and “domain swapping” experiments identified a sequence of three amino acids, termed the P-box, within the first zinc-finger module as being important for DNA response element recognition and binding. In the case of the ER, these residues are glutamic acid, glycine, and alanine, whereas the corresponding amino acids in the GR subfamily are glycine, serine, and valine (108, 109). In the second zinc-finger module, a five-amino acid sequence, the D-box, was found to mediate dimerization of the receptor on DNA (90, 107).
The first SHR structures to be solved were the GR-DBD by nuclear magnetic resonance (NMR) spectroscopy (91) and the solution and crystal structures of the ERα-DBD (92, 93). These studies revealed that the DBD folded into a compact globular structure containing two principal α-helices perpendicular to each. The recognition helix fitted into the major groove of DNA and contained the P-box residues (Fig. 2C), which made direct and water-mediated hydrogen bonds with the nucleotide sequence. In addition, there are a number of interactions between amino acid side chains and the phosphate backbone of the DNA. The second helix acts to stabilize the recognition helix in place. The five amino acids of the D-box form an exposed loop structure (Fig. 2C) and mediate the “head-to-head” binding of the receptor monomers on the DNA response element.
Although the overall folding of the DBD is very similar for all receptors studied, there are some differences that are thought to be functionally important. For example, the structure of the AR-DBD forms a more closely packed dimerization interface than that observed for the GR-DBD, and it has been proposed that this, in part, is the basis for AR binding to androgen response elements, a subset of DNA response elements that are selective for this receptor (94). Furthermore, whereas the main determinants for DNA binding reside within the P-box residues, it has become apparent that the region immediately adjacent to the DBD, termed the CTE, also has a role to play in stabilizing DNA binding and/or response element selection by SHRs through contacts with the minor groove of DNA (96, 110).
A recent structural analysis has highlighted the allosteric properties of DNA binding. Receptor binding sites associated with glucocorticoid-regulated genes were shown to influence the composition of coregulatory complexes recruited by the GR (95). The binding of the GR-DBD to different response elements was found to be asymmetric, and a region of the DBD, the “lever arm,” was found to adopt different conformations depending on the DNA architecture (95). The lever arm consists of the amino acids Glu450-Gly-Gln-His-Asn-Tyr455, which are conserved in the PR- and MR-DBD, but shows three and four amino acid changes in the AR and ER, respectively. The histidine at position 453 adopted one of two possible conformations in the DBD dimer: in the upstream monomer the histidine was packed in the DBD core, whereas for the downstream monomer the residue was “flipped out” and conformationally flexible (95). The glutamic acid and the tyrosine participated in contacting DNA; both these residues are found in AR, MR, and PR, but only the tyrosine is found at the same position in the ERα/β. It remains to be investigated more fully to determine whether the lever arm is a common property of the SHR-DBD and to determine the mechanism(s) of allosteric control to the GR-AF1 or -AF2 activities. These studies reveal the structural dynamics imparted by DNA recognition and binding and highlight the potential regulatory role of different DNA sequences or binding partners, which could lead to cell or promoter selective SHR function.
Recent advances in genome-wide analysis of SHR-regulated genes have dramatically increased our ability to identify hormone-regulated gene networks and have allowed for the identification of SHR binding regions (SBRs) throughout the genome. The outcome of these studies has been the revelation that receptor binding sites can have degenerate sequences and different architecture of half-sites (palindromes, inverted palindromes, and repeats), although typically a high-affinity half-site matching the consensus sequence is always present (111–117). The other outcomes from these chromatin immunoprecipitation (ChIP), or ChIP-seq, studies are the observations that: 1) there is a high preponderance of other transcription factor binding motifs associated with receptor response elements (e.g., Foxo A1, ETS1, GATA2); and 2) in some cases the SBR can be more than 10 kb away from the transcription start site of the regulated gene (111–113, 116, 117), although other studies reported SBR within 1.5 kb of the transcription start site (112, 115). Of particular interest was the comparison of genomic binding sites for the GR across four mammalian species, which revealed that sequence conservation for individual sites was greater between species than for receptor binding sites within the same species (116, 118). The latter observation emphasizes the potential importance of DNA architecture of SBR for gene selective regulation by the GR.
D. N-terminal domain (NTD) (AF1)
The NTD of SHRs is highly variable in both amino acid sequence and length, with the MR, PR-B isoform, and AR having NTD of greater than 500 amino acids, the largest of any members of the nuclear receptor superfamily (reviewed in Ref. 50). In contrast to the DBD and LBD, there is little amino acid sequence homology between the different classes of SHR-NTD. However, comparison of the primary sequence for a given receptor across different species does reveal regions of conserved residues (50). Such conserved sequences often colocalize with regions delineated as necessary for receptor-dependent gene regulation, which in turn are involved in protein-protein interactions (49, 50). Deletion of the LBD of SHRs results in a protein that is constitutively active in reporter gene assays, and thus the function of AF1 has been described as hormone-independent (see Ref. 50 and references therein). A significant number of coregulatory proteins have been identified binding to AF1, including chromatin modifying enzymes, basal transcription factors, coactivators, and corepressors (reviewed in Ref. 50). In some cases, the same binding partner has been described as interacting with both AF1 and AF2 through distinct receptor-interacting domains (119, 120).
Deletion studies have defined the AF1 of the ERα (72, 121) and GR (122) to 100 and 200 amino acid regions of the respective NTD. The location and nature of the GR-AF1 was further refined to a 58-residue core domain (amino acids 187 to 244; GR-AF1core) (123). This AF1core domain retained 60 to 70% of the activity of the full-length AF1. Point mutational studies revealed an important role for hydrophobic amino acids in AF1core activity (124, 125), whereas mutations that reduced the overall acidity of the AF1 domain led to progressive impairment of transactivation (126). It was speculated that these residues were important structurally and defined the solvent exposed surface of the transactivation domain. It is significant, therefore, that mutating key glutamic acid, phenylalanine, or tryptophan residues in the corresponding enh2 domain (amino acids 108 to 317) of the rat GR impaired transactivation activity, but not the ability to repress transcription through protein-protein interactions with the transcription factor activator protein 1 (127).
Although the term “AF1” is used to describe the transactivation function of the SHR-NTD in some receptors, the activity is highly modular and can map to multiple sequences within the NTD. Deletion of the AR-NTD results in a transcriptionally weak protein, providing evidence for the main transactivation function being located within the NTD (128, 129). The AR-AF1 is modular in nature, and regions important for transactivation have been characterized by deletion analysis (128, 129), by the use of fusion proteins (129), and by point mutation (130, 131). These studies identified two overlapping regions, amino acids 101 to 370 and 360 to 485, as being critical for receptor-dependent transactivation, and highly conserved hydrophobic amino acids within this region have been shown to be important for activity and protein-protein interactions.
Similarly, multiple regions have been identified within the PR-NTD (132) and MR-NTD (133–135) that, when deleted, impaired receptor-dependent transactivation. The AF1 domain or related SHR-NTD activities have been shown to function in a variety of mammalian cell types and in the budding yeast Saccharomyces cerevisae and when fused with a heterologous DNA binding domain. A striking exception to the later observation is the isolated PR-AF1 domain, which requires the PR-DBD to be functionally active, emphasizing the importance of intradomain communication (132). When taken together, these studies illustrate that different surfaces, even within a single SHR-NTD, can be involved in activation, or indeed repression, of transcription. Furthermore, the strength of the respective SHR-NTD in activating transcription is also variable. A clear correlation between AF1 activity (and inversely AF2 activity) and the length of the NTD has been demonstrated using GalDBD-NTD fusion proteins, with the longer AR- and PR-NTD showing the highest activity in a reporter gene assay (66).
III. Structural Analysis of Full-Length Nuclear Receptors
Significant insight has been provided by x-ray crystallography and/or NMR spectroscopy of the isolated DBD and LBD. However, the presence of significant regions of intrinsic disorder (Section V) and overall protein size have hampered attempts to determine the three-dimensional structure of a full-length SHR.
In contrast, a recent x-ray structure of a complex containing the full-length peroxisome proliferator-activated receptor (PPAR) γ and RXRα nonsteroid nuclear receptors has been published (105) (Fig. 2D), whereas solution studies, using small angle x-rays and fluorescence resonance transfer, have described the “shape” of a number of nonsteroid nuclear receptor complexes (136). Isothermal calorimetry and hydrogen deuterium exchange (HDX) experiments have elegantly revealed interdomain and interreceptor communication for the thyroid hormone receptor (137) and the VDR (138) heterodimer complexes with RXR, respectively.
In the crystal structure of PPARγ and RXRα bound to a DR1 DNA response element (5′-AGGTCAnAGGTCA-3′), the complex is asymmetrical, and the overall conformation of the individual LBD and DBD were remarkably similar to the structures of the isolated domains and those described above for SHRs (Fig. 2D) (105). In the crystal complex, the PPAR monomer adopted a “closed” conformation with extensive interactions between the LBD and the PPAR-DBD and the RXR-LBD, Hinge, and DBD (105). In contrast, the RXR monomer had an “open” conformation with the hinge region extended, creating a surface for PPAR binding. The hinge region of the PPAR also made contact with the DNA and contained two α-helices, whereas the RXR hinge region was more flexible and lacked secondary structure (Fig. 2D) (105). The functional importance of the interaction of the PPAR-LBD with the DBD was supported by the introduction of a F347A mutation. This residue is not directly in contact with either the ligand binding pocket or the AF2 region, but impaired DNA binding and transcriptional activation (105). The NTD of both receptors, although relatively short at 110 and 134 amino acids, respectively, were not observed in the crystal structure of the complex and were highly dynamic as determined by HDX experiments.
However, these different conformations of the hinge region or the closed complex of PPAR were not observed in the solution structures of a number of class II receptors bound with RXR to different response elements (136). Although these studies lack atomic resolution, they resolved as a homogenous complex and generally all show the receptors as an elongated shape in an open conformation, with the hinge regions in extended conformations, permitting the ordering of the LBD over the 5′ half-site of the DNA element (136). Furthermore, the coactivator protein Med1 bound with a 1:1 stoichiometry with the receptor heterodimer complex and only bound to the RXR partner (136). This is in contrast to other studies that suggest binding of coregulatory protein fragments to both receptors in similar receptor complexes, although binding to one partner could be modulated by the DNA response element (138).
Zhang et al. (138) have used HDX to investigate the consequences for receptor conformation of ligand binding, DNA response element binding, and coregulatory protein interactions using the VDR-RXRα heterocomplex as a model. Binding of either ligand alone caused changes in HDX within the cognate receptor LBD and the LBD of the receptor partner. As might be expected, a number of these changes mapped to regions of the receptors involved in heterodimerization (138). However, there were also perturbations at distant sites; for example, 1,25-vitamin D3 binding caused changes in helix 3 of the RXR partner, suggesting allosteric interreceptor communication. Most striking was the destabilization of the VDR-DBD by binding of either 1,25-vitamin D3 or 9-cis-retnoic acid (138). DNA response element recognition and binding also resulted in changes in HDX of both receptors; there was strong protection from solvent exchange for the VDR-DBD/CTE, consistent with more contacts with the DNA (138). DNA binding, and significantly the architecture of the DNA response element, also led to changes in the LBD of both receptors: the regions affected were the dimerization interface and AF2 surface. A 1:1 complex was formed between a fragment of the steroid receptor coactivator-1 (SRC-1) (NCoA1) and VDR-RXR: with VDR binding to the NR3 box and RXR to NR1 box (p160 family of coactivators, NCoA1, 2, and 3, have three LxxLL motifs termed NR box 1 to 3). The nature of the DNA response element was shown to alter the conformation of the AF2 regions such that there was a reduction in VDR and enhancement of RXR interactions with SRC-1 (138).
Taken together, the structural analysis of full-length or two-domain receptor proteins, in complex with different DNA response elements, illustrates the complexity of multiple inter/intradomain interactions and the possible mechanisms of allosteric regulation imparted upon ligand, DNA, or coregulatory protein binding. Furthermore, the above-mentioned structural studies have highlighted the growing importance of the hinge domain in regulating receptor conformation and complex “shape.” In light of these studies with class II nuclear receptors, which form heterodimeric complexes with RXR, it will be vital to solve the structures for SHR complexes containing at least two domains. Because these receptors bind to palindromic-like DNA sequences as homodimers, such studies are required to determine what conformation(s) are adopted by the hinge regions of each monomer and to identify the intra/interdomain communications, which would highlight the potential mechanisms of allosteric regulation in response to hormone and DNA binding.
IV. Steroid Hormone Receptors Function in a Ligand-, Cell-, and Promoter-Specific Manner
Allosteric regulation is a property that arises from the modular structure of SHRs. The consequence of this is that multiple surfaces can engage in protein-protein interactions (i.e., AF1, AF2, lever arm, and the CTE), and accessibility of these sites is regulated by the binding of hormone and the DNA architecture. Thus, the means of gene- and tissue-specific regulation reside within a single transcription factor. The cellular context (SHR protein levels and coregulatory protein levels) and chromatin environment will also influence the SHR transcriptional activity, but in this review we emphasize the features within the receptor protein that underpin this regulation.
A. Selective utilization of AF1 and AF2: cell culture studies
Initial studies mapping the functional domains of the ERα led to the observation of cell and promoter (gene) discriminating functions for the AF1 and AF2 surfaces. Strikingly, using receptor constructs lacking the NTD or the LBD, it has been shown that AF1 and AF2 function in a cell type-specific manner. For example, AF1 was active in CEF (chicken embryonic fibroblast) cells, HepG2 liver cells, and the breast cancer cell lines T47D and MCF-7, but not in HeLa cells (42, 73, 139). Furthermore, dependence on AF1 appeared to correlate with the more differentiated breast and prostate cancer cell lines, whereas AF2-dependent activity showed the opposite trend (73).
The activation functions of ERα also showed promoter selectivity, with AF2 being active on more complex promoters and having the ability to synergize with both AF1 and other transcription factors (71, 139). In contrast, in CEF permissible cells, AF dependence did not distinguish between “complex” and “simple” (response element + TATA-box) promoters and failed to act cooperatively with other transcription factors (139).
Evidence for differential utilization of the GR-AF1 and -AF2 domains comes from studies using the human osteosarcoma cell line U2OS expressing wild-type or variants of the GR with mutations in either AF1 (E219K/F220L/W234R) or AF2 (E773R) (95, 140, 141). After expression profiling to identify GR-regulated genes in U20S cells, a panel of nine genes were selected for investigation. On the basis of expression in response to the synthetic glucocorticoid dexamethasone, the genes were classified as: AF1-dependent (IGFBP); AF2-dependent (PDGF, SGK, ladinin1, SDPR, MSK2, IRF8, and GILZ); AF1- and AF2-dependent (hIAP); or intriguingly, immune to mutation in either AF1 or AF2 (I6PK) (140, 141).
More recently, differential phosphorylation of the human GR-AF1 at specific serine residues has been correlated with gene expression. There is an increase in phosphorylation of the GR on serines 203, 211, and 226 within the AF1 domain in response to hormone binding (142, 143). GR phosphorylated on serines 211 and 226 was recruited, albeit with different kinetics, to receptor binding sites in the TAT, SULT and GILZ genes in response to hormone. In contrast, the receptor phosphorylated on serine 203 was not observed at these glucocorticoid-regulated genes (143).
In the case of the AR, a dependence upon the N/C-terminal interaction was found to distinguish different sets of AR-regulated genes, such that mutations disrupting the interaction impaired transcription of PSA, probasin, and C3(1) genes, but not slp or MMTV (83, 84). Collectively, these studies illustrate the importance of different regions of SHRs (AF1/NTD, AF2/LBD) for target gene expression and provide a starting point for evaluating the mechanisms for this selectivity, which is likely to involve specific protein-protein interactions and posttranslational modifications.
B. Selective utilization of AF1 and AF2: in vivo studies
The above-mentioned in vitro studies have been instrumental in highlighting the potential for differential use of SHR activation functions, but compelling in vivo evidence has only recently become available. In vivo it has been shown that ERα-AF1 was not required for vasoprotective actions of estradiol but was important for ERα function in reproductive tissues (144). A similar transgenic mouse model has been used to investigate the role of ERα activation functions in bone: comparing the consequences of a complete ERα gene deletion with either a deletion of the ERα-NTD(AF1) or a deletion impairing AF2 function in ovariectomized animals treated with estradiol (17). ERα is necessary for maintaining bone density and structure, and it was demonstrated that AF2 was important for both cortical and trabecular bone; crucially the requirement for AF1 was more tissue specific, having a role in trabecular, but not cortical bone (17). Together, these in vivo studies could show that AF1 activity was important for estradiol action in the uterus, but not for the increase in liver weight in response to hormone treatment (17, 144). The significance of these findings lies not just in providing evidence for tissue/cell-specific selectivity of AF1 and AF2, but also in illustrating the possibility of therapeutically targeting these receptor functions to achieve tissue restricted effects. Thus, blocking ERα-AF1 to treat uterine cancer should not impact upon ERα activity in maintaining cortical bone.
The above-mentioned studies demonstrate the potential for different SHR surfaces to be used in a cell- and promoter-specific manner. Further experiments are clearly needed to extend these observations and to determine the molecular mechanisms regulating the display and function of different SHR surfaces.
V. Intrinsically Disordered Structure and Steroid Hormone Receptor Action
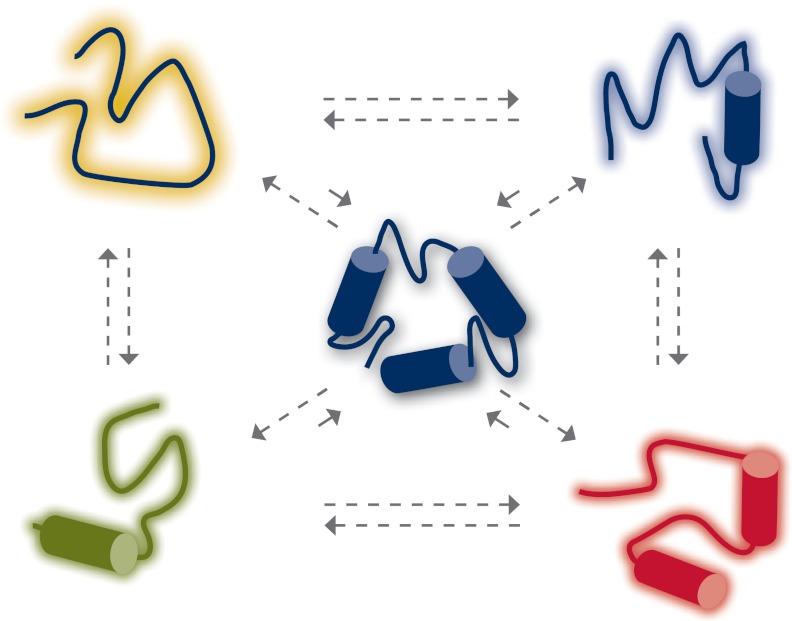
The traditional view in biology is that the specific function of a given protein is determined by its unique three-dimensional structure, the so-called “lock and key” hypothesis. However, in recent years, it has become quite evident that many biologically important proteins possess large stretches of amino acid sequences that do not adopt a well-defined three-dimensional structure (145–150). These unstructured proteins/protein regions have been termed “intrinsically disordered” and exist as dynamic ensembles of interconverting conformers that are capable of undergoing disorder/order transition under specific physiological conditions (Fig. 3) (151–154). However, in contrast to structured or ordered proteins whose conformation is relatively stable with occasional cooperative conformational switches, they do not automatically adopt a classical fully folded and well-defined functional structure and typically undergo conformational changes (155–159). Both random coil-like and collapsed (partially folded or molten globule-like and premolten globule-like) conformations with poorly packed side chains are the features of ID proteins/regions (Fig. 3) (160, 161).
Figure 3.
ID proteins/domains exist as an ensemble of conformers, which collectively appear to be unstructured. Each conformer is in a reversible equilibrium with each other. Except for a very small fraction, which may be relatively well ordered (shown in center), all other conformers possess the characteristics of random coil or molten globule-like structures.
In recent years, it has become evident that eukaryotic genomes are highly enriched in ID proteins, which appear to promote molecular recognition (162). Based on biophysical and computational analyses, ID regions/domains are prevalent in a majority of transcription factor proteins, including the hinge and NTD of SHRs (49, 50, 163–166). Characterization of the conformational propensities and function of such nonglobular protein sequences represents a major challenge.
A. What does it mean to be intrinsically disordered?
The complex and highly coordinated interactions of proteins play a fundamental role in the control of cellular physiology, where different functions can be achieved via recognition of specific and unique identification sequences frequently found inside ID regions (145–150, 154). It has been predicted that a number of signaling proteins (∼65%) possess long ID regions/domains, which play a critical role in cell cycle control, transcriptional and translational regulation, and signal transduction; this number is predicted to be much higher (∼75%) for cancer-associated proteins (165, 167). The significance of such ID domains/regions in signaling molecules is that their conformational flexibility creates large interaction surfaces that allow macromolecular interactions with high specificity and low affinity through coupled binding and folding, an important property of signaling proteins promoting molecular recognition that provide specific, but reversible, interactions with target molecules (151–154). The function-associated conformational changes and disorder-to-order transitions may be brought about by alterations in environmental or cellular conditions. The ID nature of proteins also provides a possible mechanism for their actions to be regulated through posttranslational modifications, such as kinase-dependent phosphorylation (153, 180). Furthermore, sites of other posttranslational modifications such as acetylation, hydroxylation, ubiquitination, methylation, and sites of regulatory proteolytic attack are frequently associated with ID regions (168). The functional importance of conformational changes and disorder-to-order transitions in ID proteins stems from a large decrease in conformation entropy, which in turn can uncouple specificity from binding strength. This enables highly specific interactions to be easily reversible, which is beneficial for proteins involved in signaling and transcriptional regulation.
B. Coupled folding and binding of the steroid hormone receptor NTD
The primary amino acid sequences of the NTD of the SHR, which contain AF1, are much less conserved than are DBD and LBD regions (50, 60, 169–171). Despite having poor sequence homology, these regions are highly enriched in charged amino acids and have low hydrophobicity, which is a signature for ID (49, 50). Studies from several laboratories have confirmed the ID nature of several SHR-NTD or AF1 domains using circular dichroism-, NMR-, Fourier transformed infrared-, and fluorescence emission- spectroscopies and proteomic methods (135, 172–176). Table 1 summarizes the secondary structure content for different SHR-NTD and AF1 activities under different experimental conditions. From these studies, it can be concluded that the NTD typically lack stable secondary structure but have the propensity to form α-helical conformation. Interestingly, the AF1b region of the MR-NTD appears to be more stably folded than the rest of the domain and to have predominantly β-strand secondary structure (135). SHR-NTD therefore exist as an ensemble of conformations having more or less stable secondary and tertiary structures (Fig. 3).
Table 1.
Measured secondary structure in nuclear receptor NTD and or AF1 domains
| Receptor | Conditions | Secondary structure elements |
Method | Refs. | ||||
|---|---|---|---|---|---|---|---|---|
| α-helix | β-strand | β-turn | Bend | Coil | ||||
| AR-AF1 | Buffer | 13 | 20 | 32 | 36 | CD | 176 | |
| Buffer | 16 | 24 | 17 | 19 | 24 | FTIR | 188 | |
| TFE | 40 | 15 | 20 | 25 | CD | 176 | ||
| TMAO | 37 | 17 | 11 | 11 | 24 | FTIR | 188 | |
| + TFIIF | 35 | 15 | 14 | 17 | 21 | FTIR | 188 | |
| AR-NTD | Buffer | 14 | 27 | 24 | 34 | CD | 242 | |
| TFE | 58 | 12 | 9 | 22 | CD | 242 | ||
| GR-AF1 | Buffer | 26.7 | 11.6 | 12.3 | 10 | 39.4 | FTIR | 243 |
| Buffer | 26 | 13 | 61 | CD | 180 | |||
| + TBP | 42.1 | 11.8 | 12.4 | 9.3 | 24.4 | FTIR | 243 | |
| P-serine | 45 | 19 | 36 | CD | 180 | |||
| MR-AF1a | Buffer | 11 | 29 | 25 | 36 | CD | 135 | |
| TFE | 63 | 5 | 10 | 22 | CD | 135 | ||
| MR-MD | Buffer | 13 | 22 | 24 | 40 | CD | 135 | |
| TFE | 66 | 13 | 7 | 15 | CD | 135 | ||
| MR-AF1b | Buffer | 19 | 32 | 31 | 19 | CD | 135 | |
| TFE | 13 | 31 | 23 | 33 | CD | 135 | ||
| MR-NTD | Buffer | 20 | 21 | 24 | 35 | CD | 135 | |
| TFE | 79 | 2 | 3 | 15 | CD | 135 | ||
| PR-AnDBD | Buffer | 30 | CD | 97 | ||||
| + JPD2 | 58 | 11 | 17 | CD | 97 | |||
| EcR-NTD | Buffer | 8 | 31 | 23 | 89 | CD | 244 | |
| dHR38-NTD | Buffer | 11.1 | 11.7 | 10.3 | 66.6 | CD | 245 | |
| TFE | 72.3 | 4.4 | 8.7 | 17.8 | CD | 245 | ||
JPD2, Jun-protein dimerization 2; P-serine, phosphorylated serine(s); TMAO, trimethylamine N oxide; EcR, ecdysone receptor; CD, circular dichroism; FTIR, Fourier transform infrared spectroscopy.
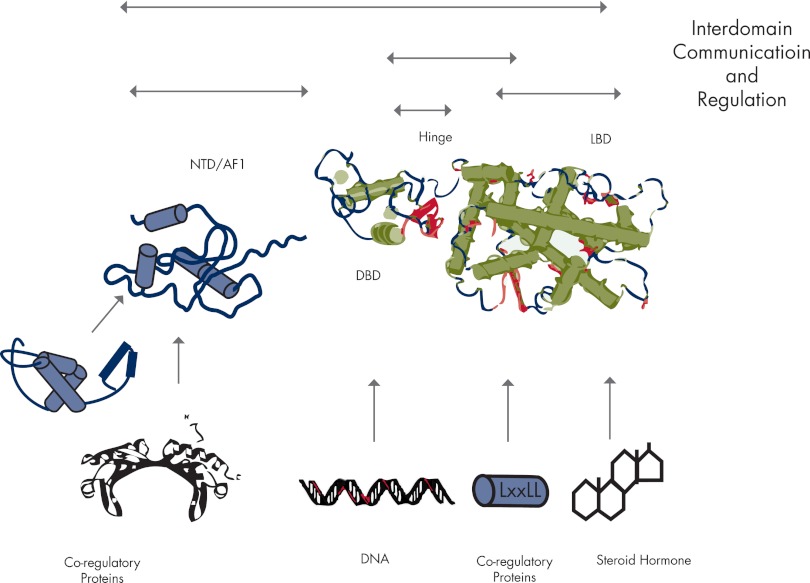
It is well known that the SHR AF1/NTD generally work in conjunction with other coregulatory proteins and by multiple mechanisms (177–182). This raises the question: what is the structural basis of the functional activity of the ID NTD/AF1 in the context of full-length SHR action? As discussed in Section III, studies carried out so far, including analysis of the crystal structure of the full-length PPARγ and RXRα (105), suggest that the AF1 may be unstructured even in full-length receptor. Furthermore, in the case of the GR, PR, and AR, the NTD is ID within the context of a two-domain fragment containing the entire NTD and DBD (174, 183, 184). It can be hypothesized that one of the reasons why the holo-receptor is associated with several chaperone proteins in the cytosol, before ligand binding, is protecting this large ID region from degradation. However, available data indicate that all the known chaperone proteins bind to the LBD of the receptor (see Section II.A), except in the case of the AR-NTD interactions with the C terminus of the Hsp70-interacting protein (246) and the co-chaperone protein Bag1L (185, 186). That raises another question: if these proteins bind to the LBD, how do they protect NTD structure? The answer to this question may lie within the dynamic structure of the SHR protein that allows sensing of the intracellular environment and efficient intramolecular domain interactions, which in turn may induce structure in the otherwise ID NTD. In fact, this hypothesis is supported by the finding that certain chemical chaperones known to protect/stabilize protein structure are capable of inducing a compact structure in the AF1/NTD of the AR, GR, and MR (135, 176, 187–190) (Table 1 and Fig. 4) and the growing evidence for N/C-terminal interactions among SHRs (see Section II.A.3). Because structural flexibility has advantages for the assembly of large multiprotein complexes, once the ligand-bound receptor enters the nucleus, the NTD/AF1 may be again unstructured until it encounters a specific binding partner(s) including site-specific DNA and/or coregulatory proteins.
Figure 4.
Folding of the SHR-NTD. The NTD exists has an ensemble of conformations, having more or less stable structure (middle molecule). A more stably folded conformation of the NTD can be induced or selected by small molecules (osmolytes), posttranslational modification (e.g., phosphorylation), DNA binding, and interactions with coregulatory proteins. In these models of NTD folding, the more stable structure is shown to be α-helical by the solid cylinders (blue).
Regulatory regions of many signaling proteins including SHRs are known to form an assembly of protein complexes in a rapid and well-coordinated manner for efficient and target-specific regulation of gene expression (49, 119, 120). It has been proposed that the ID nature of the NTD/AF1 allows rapid and reversible adoption of various structural configurations controlled by allosteric modulations through inter- and intramolecular communication and selective responses to cellular environments. Because ID domains (e.g., SHR-NTD/AF1) can exist as a large collection of highly dynamic and rapidly interconverting conformations (Fig. 3), which may vary at a given time depending upon cellular crowding under physiological conditions, the ID nature of the NTD/AF1 allows it to rapidly sample the cellular environment until partner binding proteins of appropriate concentration and affinity are found. Then, either by induced fit or selective binding of a particular conformer, a high-affinity NTD/AF1:coregulatory protein interaction occurs (Fig. 4). During the course of gene regulation, SHRs interact with various coregulatory proteins, site-specific DNA, and small molecule steroid ligands (Fig. 1). These interactions occur at precisely defined locations within the SHR protein, but their effects are sometimes propagated to distal regions/domains, triggering highly specific responses.
Generally, under physiological conditions, proteins must have specific structure to carry out their proper functions. The GR-AF1 domain was the first region of any nuclear receptor to be characterized as being ID (172, 173). It was further reported that this ID region can be induced to adopt a helical structure when incubated in the presence of trifluoroethanol (TFE) (Table 1). At that time, it was hypothesized that the GR-AF1 might not require an ordered conformation and act through the so-called “acid blob” concept (172). However, several mutagenesis studies suggest that negative charges per se are not sufficient for AF1 activity; indeed, key hydrophobic amino acids may be crucial for AF1 activity (124, 125). The pattern of proteolytic degradation products in cell-free extracts suggested that the GR-AF1 might be structured in vivo (126). Furthermore, helix-destroying mutations of amino acids in three hypothetical helices of the GR-AF1 strongly reduced GR transcriptional activity (172, 191). Thus, it could be hypothesized that the GR-AF1 may be structured in vivo, at least when directly involved in transcriptional activation. Subsequent studies with other SHRs such as AR, PR, ER, and MR showed similar trends (135, 174–176, 189). In recent years, it has been shown that the NTD/AF1 of SHRs undergo coupled binding/folding events. It has been well established that like many other transcription factors, during transcription regulation the NTD/AF1 shifts to a conformational space in which more structured conformers exist (Table 1 and Fig. 4).
The crowded conditions inside the cell have been suggested to cause ID proteins to fold into an ordered three-dimensional structure. Molecular crowding in cells may include small molecule solutes and macromolecules including proteins, DNA, and RNA. Because SHRs are known to function in a cell/tissue-specific manner and AF1-mediated transcriptional activity is particularly sensitive to cellular environment (see Section IV), it can be hypothesized that one of the reasons for cell-specific effects of AF1 may be influenced by binding/folding events governed by molecular crowding in specific cells. Several mechanisms important in inducing ordered conformations of the NTD/AF1 under physiological conditions have been described, which in turn may facilitate receptor interaction with specific coactivators and subsequent transcriptional activity.
1. Role of small molecules (osmolytes)
Functionally folded structures of most macromolecules are sensitive to changes in cellular environmental conditions. The native functional activities and stability of macromolecules are fine-tuned through accumulation of high concentrations of small organic molecules known as osmolytes or chemical chaperones. Cells regulate many biological processes such as protein folding and protein-protein interactions via accumulation of specific osmolytes (190, 192–195). In recent years, the mechanism of osmolyte compatibility and osmolyte-induced stability has attracted considerable attention. Osmolytes that occur naturally represent different chemical classes such as amino acids (proline and glycine), methylamines (betaine and trimethylamine-N-oxide), and polyols and sugars (sorbitol and trehalose) (190). Because osmolytes predominantly affect the protein backbone, the balance between osmolyte-backbone interactions and amino acid side chain-solvent interactions determines protein folding (192–195). In ID proteins, the hydrophobic amino acids are insufficient, relative to the charged side chains, to cause spontaneous folding (156, 188, 205–207). Addition of the solvophobic effect of a profolding osmolyte can tip the energy balance in the favor of folding because the backbone collapses to avoid solvent interactions with the osmolyte (192–195). The quantity of osmolyte required depends on both its inherent solvophobic properties in the interaction with the peptide backbone and the free energy balance provided by the sum of all backbone-osmolyte interactions and the sum of all amino acid side chain-solvent interactions (192–195).
The ID NTD of SHRs, or isolated AF1 regions, can be induced to fold spontaneously to native, functional forms by addition of certain organic osmolytes to the solvent (135, 173, 176, 196, 197) (Table 1 and Fig. 4). Incubation with several osmolytes causes the ID AF1 domain of the GR to fold into a form that can bind strongly to specific coregulatory proteins that are critical for the transcriptional activity of the receptor (196). The folding curves were cooperative and typical of a protein that folds spontaneously via the classic “two-state” model. This strongly suggests that a natural conformation had been reached (196). Osmolytes also reduced limited proteolysis of the GR-AF1 polypeptide and resulted in the tryptophan and tyrosine residues becoming less solvent exposed, which is consistent with folding or increased conformational stability of the receptor domain (196). Similarly, the ID AF1 domain of the AR has been shown to have the propensity to form helical structures in the presence of the osmolyte, trimethylamine N-oxide (188), and to adopt a more folded conformation that was resistant to limited proteolysis (176, 189) (Table 1). Furthermore, this osmolyte-induced conformation in the AR-AF1 domain significantly facilitates its interaction with a critical coregulatory protein (188, 189). Trimethylamine N-oxide has also been shown to stabilize a more folded conformation for regions of the MR-NTD and, in the case of central 139 amino acids, increase binding of a number of coactivator and corepressor proteins (Table 1) (135).
Recent studies suggest that trehalose-induced folding of the GR-NTD facilitates binding to a glucocorticoid response element (GRE) in the context of a two-domain GR polypeptide containing the entire NTD plus the DBD, termed GR500 (190). It has been previously reported that GR500 is capable of binding to GRE and can stimulate AF1-mediated GR activity similar to that observed with the hormone-bound full-length receptor. Taken together, the above results suggest that osmolyte-induced folding of the NTD or AF1 of SHRs may be important for the interaction of the receptor protein with both coregulatory proteins and response element DNA.
2. Binding of DNA and protein partners
According to the classic model of SHR action (Fig. 1), receptor bound to specific hormone response element (HRE) by virtue of high-affinity interactions modifies chromatin structure and/or contacts the multiprotein transcription machinery to regulate transcription from a target promoter (59, 95, 119, 120, 230).
a. DNA architecture and SHR-NTD folding.
In recent years, it has become evident that SHR:HRE binding also influences the three-dimensional configuration of the receptor (95). Consequently, in an HRE-specific manner, through conformational rearrangements, the surfaces of the SHRs are modified such that various critical ancillary factors can bind (95). Furthermore, DNA-binding can also be a trigger for an active intramolecular communication that can change the conformation of the SHRs in a site-specific manner (199). Because transcriptional control of a specific gene depends upon the interactions of the SHR with DNA/chromatin, the exact DNA sequences and architecture of the response elements in the regulatory regions of the gene could help determine the hormone response. Specific sequences within estrogen response element (ERE) have been shown to modulate the conformation of ER (200, 201), and conformational changes resulted in altered recruitment of coactivator proteins to the ER:ERE complex (199). This provides an intellectually satisfying rationale for the varied transcriptional effects of specific sequences found in the SHR genomic binding sites controlling specific genes. Furthermore, as discussed in Section III, recent HDX experiments demonstrated that binding of DNA response elements led to changes in the dimerization and AF2 surfaces of each partner in a heterodimer receptor complex (138). However, it also raises the question: what are the regions/domains of the receptor that may be most prone to these conformational changes due to HRE binding? Several studies have shown that dramatic folding effects have been seen on the AF1/NTD of SHRs when DNA binding takes place (174, 183, 184, 201). Moderate conformational changes have also been observed in other domains, namely, the DBD (95) and LBD (138), due to DBD:HRE interactions. Studies with the GR have shown that stoichiometric binding to a consensus GRE by the two-domain GR500 polypeptide led to the formation of secondary/tertiary structure in the NTD, suggesting that considerable binding energy may have been devoted to intramolecular rearrangement in the otherwise ID NTD (183). Similar studies on the two-domain PR fragment indicated that DNA binding resulted in the stabilization of structure in the ID NTD of both the A and B forms of the receptor (174). It is important to note here that when expressed as recombinant protein, even in these two-domain fragments, the NTD remains mostly unstructured until it binds to specific HRE sequences (183).
This interdomain communication also works in reverse. The presence of the NTD was found to reduce the binding affinity of AR-NTD-DBD relative to AR-DBD alone (184). The NTD had to be covalently attached to the DBD, supporting a role for intramolecular contacts, and did not significantly change the protein-nucleotide contacts.
Together, these results suggest that one of the reasons why sequence specific DNA binding has such a profound effect on the function of SHRs may be because the ID NTD can gain a functionally active structure such that it can bind appropriate coregulatory proteins when HRE:DBD interactions occur. Consequently, coregulatory proteins that bind to the SHRs are excluded or included in the transcription complex by virtue of the receptor surfaces available, which in turn are a consequence of site-specific DNA binding and DNA architecture (Fig. 4). Thus, part of the functional structure of the SHR necessary for positive regulation of transcription depends on receptor-DNA binding. This structure may differ from the conformation at genes negatively regulated by SHRs, where the receptor is not necessarily bound to DNA—for example, receptor interactions with other transcription factors to repress the expression of target genes. At such sites, the SHR is tethered to DNA indirectly via binding to a heterologous transcription factor. For example, the effect of estrogen at the pS2 gene is mediated through a cross talk between the ER, ERE, and an activator protein 1 response element (202). More extensive analyses are needed to reveal all the structure-function variations in the SHR and its NTD/AF1 in order to understand receptor-mediated transcription in various contexts.
b. Coregulatory protein binding and SHR-NTD folding.
Several coregulatory binding partner proteins are involved in the action of the SHR on target gene transcription (49, 119, 203, 204). There are several examples in which binding sites within the ID regions of many transcription factors contain molecular recognition features, which consist of short stretches of amino acid sequences that undergo a disorder-to-order transition and are stabilized by binding to a partner protein(s) that act as coregulators (153, 156, 188, 205–207). Thermodynamics predicts that if one or more conformers of an ID protein/region bind specific proteins with high affinity, interaction of the ID region with that partner at appropriate concentrations may cause its structure to stabilize (208, 209). It has been predicted that these segments may have advantages for cell signaling by allowing the decoupling of partner binding specificity and affinity, due to which the strength and duration of signaling events can evolve (208, 209).
Coregulatory proteins are known to modulate the transcriptional activity of the SHRs through multiple mechanisms. The SHR:coregulator complexes can act to modify chromatin, influence RNA polymerase II phosphorylation, and bind mediators and proteins of the basal transcription complex. Many of the known key SHR coregulators bind to both the NTD/AF1 and AF2, using distinct receptor binding sites (210–213). In recent years, the mechanism of action and the structure of AF2:coregulator interaction and its effects on the regulation of SHR target genes have been explained in depth (5, 45, 46, 63, 231). However, how the choice of AF1:coregulator interaction is made and its functional significance in gene regulation by SHRs remain poorly understood. There are examples in which the ID regions of a transcription factor protein take full shape upon interaction with protein binding partner(s) through an induced-fit model of folding (154, 214). Based on this model, it has been hypothesized that NTD/AF1 is not fully structured in vivo until it binds to one or more coregulatory protein(s), and this induced conformation or limited set of conformations in NTD/AF1 is a prerequisite for its interaction with specific sets of other coregulatory proteins in a cell- and promoter-specific manner.
The NTD/AF1 of SHRs is known to recruit proteins from the basal transcriptional machinery. For example, the TATA box binding protein (TBP) can directly bind to the NTD/AF1 domain of several SHRs (135, 175, 182), and RAP74, a subunit of the transcription factor IIF (TFIIF) complex, interacts with the AR AF1 domain (179, 215). Significantly, the TBP binding induces secondary/tertiary structure formation in the GR-AF1 (182) (Table 1) and ERα-NTD (175). Using several biophysical and partial proteolytic digestion experiments, the GR-AF1 has been shown to assume a three-dimensional fold with significant helical content upon interaction with TBP (182). The TBP binding-induced folding in the GR-AF1 significantly enhances its interaction with SRC-1, and subsequent AF1-mediated, GRE-driven promoter-reporter activity (182). Similar studies have been reported for the ER-NTD (175). TBP has a central role in the basal transcription machinery, and it is interesting to note that it directly binds to the NTD of the ERα but failed to bind ERβ-NTD (175). This difference in TBP binding implies differential recruitment of target proteins by the NTD of ERα and ERβ. The affinity of the ERα and GR-AF1/NTD:TBP interaction was determined to be in the micromolar range, as assessed by surface plasmon resonance spectroscopy (175, 182). Based on these results, it has been proposed that the interaction between the AF1/NTD and TBP may proceed in a two-step manner with initial very fast, low-affinity association, followed by a slow, folding event and tighter association (175, 182). The initial association may occur by electrostatic interactions between the acidic residues of highly negatively charged AF1/NTD and the positively charged TBP. However, this initial unstable protein complex subsequently may convert into a more stable form by the folding of the ID AF1/NTD and the formation of specific contacts between the two proteins (216–218).
Furthermore, the dissociation of this binding interaction suggested a complex behavior, with a rapid dissociation for AF1/NTD molecules that did not undergo proper folding and a slower dissociation for those molecules that did fold successfully upon physical interaction with the TBP (175). Such a two-step binding mechanism is consistent with the change in protein conformation that accompanies the AF1/NTD:TBP interaction. Based on the binding and consequent folding of the AF1/NTD, it can be hypothesized that the interaction between SHR-NTD/AF1 domains and protein(s) from basal transcription machinery may be a unified mechanism, through which these ID AF1/NTD acquire a functionally active conformation under physiological conditions. In this conformation, the NTD/AF1 may be able to create favorable protein interaction surfaces for binding with other coregulatory proteins. The exclusion of certain other binding partners cannot be ruled out. It could thus be hypothesized that a complex and dynamic binding pattern for the NTD of SHRs occurs to achieve transcriptional activation, where the NTD/AF1 region must be able to obtain different conformations dependent on the binding partner(s) (Fig. 4). Induced folding of the AR-AF1 domain due to its interaction with RAP74 and facilitation of its interaction with SRC-1 lends further support to this hypothesis (176, 188, 189). Together, these results may provide a potential mechanism through which SHR AF1 domains may regulate the expression of specific genes, information essential to an understanding of how the hormone signals are passed from the receptor to target genes. However, a clearer picture will likely emerge when the functionally folded three-dimensional structures of the NTD/AF1 bound to different coregulatory proteins are solved and can be compared.
In addition to DNA binding and dimerization, the DBD has also been highlighted as a site for protein-protein interactions and may act as a hub for the transmission of allosteric signals as a result of receptor binding of both hormone and DNA. Significantly, binding of coregulatory proteins to the PR-DBD CTE has been described, which results in allosteric regulation and folding of the PR-NTD (97, 106, 219) (Table 1).
3. Posttranslational modifications and SHR-NTD folding
Posttranslational modification is an important phenomenon that controls the functions of transcription factors in cells by regulating their DNA-binding affinity, interaction with components of the transcription initiation complex, and the shuttling between the cellular compartments (220). The SHRs are the target for a plethora of posttranslational modifications, including phosphorylation, acetylation, methylation, and sumoylation, and these modifications have a complex effect on SHR signaling (221). In some cases, one type of posttranslational modification can influence another type (198, 222). Acetylation of the lysine residues in the AR hinge domain plays an important role in AR-mediated gene regulation, and mutation of these residues can impair phosphorylation of serine 94 in the NTD (103, 198). Similarly, methylation of these residues enhances AR-dependent transactivation by increasing N/C-terminal interactions and recruitment of the receptor to target genes (104, 223). The interplay of phosphorylation and sumoylation in the regulation of ERR-NTD transcriptional activity has also been observed, and these modifications form a conserved phosphosumoyl switch that exists within a larger synergy control motif (224).
Recent studies have suggested that signaling via phosphorylation-regulated protein-protein interaction often involves ID regions, and these regions have a much higher frequency of known phosphorylation sites than ordered regions, suggesting a strong preference for locating phosphorylation sites in the ID regions (153). One of the main reasons for such propensity is to facilitate extensive formation of hydrogen bonding between the backbones and/or side chains that can occur through disorder-order transition within the ID region. The formation of these hydrogen bonds would be difficult if the sites of phosphorylation were located within ordered regions (153, 209). In terms of structural consequences of site-specific phosphorylation, both disorder-to-order and order-to-disorder conformational transitions have been observed to follow the phosphorylation event, and these conformational changes often affect protein function (153). Lack of well-defined structure is generally associated with specific sites of enzyme-catalyzed phosphorylation and ubiquitination sites (209), which facilitates the inclusion or exclusion of specific binding partners in the complex (209). Because a significant part of the binding energy is required to fold an ID region, high specificity coupled with low affinity provides the basis for easily reversible interactions (209).
Like many other transcription factors, the SHRs are phosphoproteins, and it has been suggested that kinase-mediated phosphorylation plays an important role in the regulation of their activities (225). Phosphorylation of specific residues within the NTD/AF1 of the GR and ER regulates the transcriptional activity of these receptors (226–228). There are also reports suggesting that phosphorylation may affect SHR stability and thus alter receptor activity (180). Analysis of known phosphorylated residues within the AR-, ERα-, GR-, and PR-NTD with predicted ID reveals a strong correlation (Table 2). In the case of the ERβ and the GR, site-specific phosphorylation has been associated with conformational changes in the ID AF1 region (180, 229). Based on a simulation modeling approach, it was proposed that phosphorylation of serine 211 in the human GR-AF1 may result in the formation of more compact structure in the surrounding peptide within the AF1 (amino acids 187–244), which may expose novel protein surface for cofactor interaction (228). Furthermore, it was shown that common surfaces within GR affected by phosphorylation may be responsible for influencing the regulation of selective genes (228). More recent studies show that when GR-AF1 is phosphorylated at serine 211, using the p38 MAPK, secondary/tertiary structure is formed in the AF1 domain (Table 1), suggesting that under physiological conditions, site-specific phosphorylation may play a crucial role in allowing the ID AF1 domain of the GR to adopt functionally active conformation(s) (180). Furthermore, it was found that the resulting structurally modified forms of AF1 facilitated interactions with critical coregulatory proteins (CBP, TBP, and SRC-1), and possibly additional factors, resulting in the assembly of multiprotein complexes involved in GR-mediated regulation of transcription (180).
Table 2.
Location of known phosphorylated residues in regions of predicted intrinsic disorder
| Receptor | Residue | Predictor of intrinsic disorder |
|||
|---|---|---|---|---|---|
| PONDR | RONN | GlobPlot | Fold index | ||
| AR-NTD | S16 | D | D | D | |
| S81 | D | D | D | D | |
| S94 | D | D | D | D | |
| S210 | D | D | D | D | |
| S256 | D | Border | |||
| S308 | D | D | |||
| S424 | D | D | D | ||
| S515 | |||||
| ERα-NTD | S106 | D | D | D | |
| S118 | D | Border | D | ||
| S167 | D | D | Border | D | |
| GR-NTD | S134 | D | D | D | Border |
| S203 | D | D | D | D | |
| S211 | D | D | D | D | |
| S226 | D | ||||
| S404 | D | D | D | ||
| PR-NTD | S20 | D | D | D | |
| S25 | D | D | D | ||
| S81 | D | D | D | ||
| S102 | D | D | D | Border | |
| S130 | D | D | D | ||
| S162 | D | D | |||
| S190 | D | D | |||
| S213 | D | D | Border | D | |
| S294 | D (Border) | D | |||
| S345 | D | D | D | D | |
| S400 | D | D | |||
| T430 | D | D | D | ||
| S554 | D | D | D | ||
These findings demonstrate a mechanism through which the ID NTD of SHRs may adopt a functionally active conformation under physiological conditions in response to phosphorylation. More generally, it can be concluded that the SHR-NTD may be structured in vivo, at least when phosphorylated and/or directly involved in interactions with DNA and binding partner proteins (Fig. 4).
VI. Intrinsic Disorder Can Optimize Allosteric Coupling
Allosteric coupling is a common phenomenon associated with proteins with modular structures (230) and can be defined by propagation of conformational perturbation in one domain/region of the protein due to signals passed from distant sites in the same molecule. As discussed above, due to their modular structures and regulation through specific ligands interacting with different domains (i.e., coregulatory proteins for NTD and LBD, DNA for DBD, and steroid hormone for LBD), SHRs are tightly regulated through allosteric coupling (230, 231). Each of these interactions can act as signals that modulate interactions with other molecules and/or establish intramolecular and interdomain communications (Fig. 5). As a result, SHRs can switch from one functional state to another by selective stabilization of different conformations, and the ID NTD/AF1 may be the region that undergoes most significant structural rearrangement and so plays the major role in the transmission of information to distal sites.
Figure 5.
Interdomain communication and allosteric regulator of SHRs. Various inter- and intramolecular events may allosterically regulate the structure and functions of the ID NTD/AF1 domain. Arrows indicate the flow of signals from one region/domain to another, throughout the SHR protein. For example, binding of different SRM in the ligand binding pocket can pass the signal to the surface of the LBD and dynamically reorient AF2 conformation and other parts of the domain. Signals are then passed to the hinge region, resulting in conformational rearrangements in the hinge, transferring to the DBD and eventually to ID NTD/AF1. In a similar fashion, HRE-DBD binding passes signals to influence the structure of NTD/AF1 and/or the AF2 surface. Direct binding of a coregulatory protein to NTD/AF1, site-specific phosphorylation, and possibly other posttranslational modifications, and even ID NTD/AF1 flanking sequences within the NTD can be avenues for allosteric coupling involving ID AF1 and other receptor domains.
In the above description (Fig. 5) allosteric coupling is proposed to induce an ordered conformation in the ID domain through domain-domain interactions, and structure formation in these ID domains/regions is linked to ligand binding either in the ID domain itself or in other domains of the molecule. Thus, the disorder/order transition is coupled to long-range allosteric communication within the molecule and is therefore important for its function (232). Significantly, it has recently been argued that an additional functional consequence of ID is to maximize allosteric interactions (232). This has led to a new model whereby allosteric coupling does not necessarily require a conformational link between the two sites, but is rather a consequence of the “energetic balance” and overall stability of the protein (232). In this manner, SHRs sample various conformations and are primed to respond to changes in different intracellular compartments. It is interesting therefore that the removal of the NTD-flanking sequences on both sides of the GR-AF1, by deleting amino acids, that produces a peptide with AF1 immediately upstream from the DBD leads to acquisition of an ordered conformation in AF1. In this construct, interaction of AF1 with CBP/p300, but not SRC-1, is significantly increased and concomitantly, the GRE-mediated AF1 activity (233). Thus, proximity to the DBD or loss of flanking sequences enhances certain folding properties of AF1 and subsequent activity, suggesting that these flanking sequences play an inhibitory role in regulating GR-AF1 structure and activity (233). Unlike, the two-domain GR fragment (GR500), binding of this peptide to a GRE fails to induce any further structure formation in AF1 (233). These position-specific structural effects suggest that AF1 structure (or propensity for structure formation) is strongly influenced by the amino acids that surround it as well as by influences from the DBD:GRE interaction. Strikingly, these findings also support the process of allosteric coupling through an intrinsic disorder mechanism, and the NTD amino acid sequences that flank AF1 must influence these signaling processes (232). These results also provide a mechanistic explanation for why certain N-terminally truncated GR, and possibly other SHRs, splice variants differ in their activities (10).
Allosteric mechanisms have evolved in many transcription factor proteins, and this flexibility may allow SHRs to mediate diverse genetic responses directly by modifying their malleable surfaces for specific interactions. Identification of different ligand binding events and subsequent allosteric coupling is likely to play an important role in understanding the diverse target function of SHRs and could be critical in developing improved therapeutic interventions in several pathological conditions associated with SHRs. In cells, SHR function in a very dynamic fashion, existing as an ensemble of conformations that can sample the cellular environment in a rapid manner, and subsets of these conformations can be selected and stabilized by hormone and DNA binding.
VII. Structural Dynamics and the Design of Selective Small Molecule Modulators of Steroid Hormone Receptor Function
It is generally accepted that receptor function is dependent upon ligand-induced conformational changes (i.e., helix 12 orientation), together with the cellular complement and levels of coregulatory proteins. As discussed in Section V, it seems certain that the DNA architecture of receptor binding sites and the folding of the AF1/NTD domain will also contribute to cell-specific receptor actions. SHRs are well-validated drug targets and are prognostic indicators in a number of pathophysiological conditions, including inflammation, hormone-dependent cancers, osteoporosis, and cardiovascular disease (see Section I). Classical antagonists of SHRs act as competitive inhibitors of the natural hormone by binding to the ligand binding pocket and inducing a conformation that is unable to bind coactivator proteins, or retains the receptor complex in the cytosol, and/or targets the receptor for degradation. However, there is increasing interest in the development and characterization of compounds that exhibit mixed agonist/antagonist properties depending on the cellular context; these are best described as “selective receptor modulators.” The “Holy Grail” for hormone therapies is to be able to target SHR signaling in a receptor- and tissue-selective manner to maximize therapeutic benefits and to minimize deleterious outcomes in other receptor target tissues.
The general problem with traditional antagonists or SRMs is the development of resistance and/or off-target side effects. It therefore makes sense to investigate the possibility of identifying inhibitors that act outside of the ligand binding pocket, which could complement or replace existing SRMs. A number of groups have screened different classes of molecules (natural products, biologics, or synthetic compounds) for such inhibitors, and a number of exciting and surprising outcomes have been reported. For example, there are several examples where SRM binding influences the conformation of the receptor (234, 235), and these conformational changes collectively determine the specific biological activity. Thus, a number of peptide probes that recognize different conformations of a SHR bound to different ligands have been identified. Although these peptide libraries have proved their worth in characterizing ligand-binding profiles, initially for the ERα and ERβ, they also open the possibility for developing selective receptor peptide inhibitors (for example, Refs. 236–238) that will selectively disrupt receptor-SRM-coregulatory protein complexes in a tissue-specific manner.
The potential for allosteric inhibitors of receptor function was emphasized by the finding that a number of small molecules (e.g., flufenamic acid, 3,3′,5-triiodothyroacetic acid) disrupted protein-protein interactions involving AR-AF2 and inhibited AR-dependent transactivation (74). The surprise came when x-ray structures revealed these molecules bound to a hydrophobic surface of the LBD, termed BF3 (involving residues in helix 1, helices 3 to 5, a loop, and helix 9), which is distinct from the ligand binding pocket, and crucially the AF2 region (74). This finding, together with the information from peptide binding studies, suggests that other surfaces of the LBD may participate in protein-protein interactions and could therefore be novel drug targets. Allosteric regulation could also stabilize a specific conformer that reduces the affinity of the second binding site for its ligand, and this dynamic could be explored as a novel approach to structure-based SRM design to produce differential responses.
In a separate study, a number of hits were identified from a screen of two small-molecule libraries that inhibited ER-dependent DNA binding (239). One of these molecules, theophylline,8-[(benzylthio)methyl]-(7Cl,8Cl), selectively inhibited ERα-dependent transcription and acted outside of the ligand binding pocket. Similarly, two compounds, pyrvinium pamoate and a natural product, harmol hydrochloride, have been described that inhibit AR activity, independent of hormone binding (240). Harmol appeared to disrupt DNA binding, whereas pyrvinium acted at a subsequent step in receptor signaling, possibly disrupting protein-protein interactions (240). Although the binding sites on the ER or AR for these small molecules have yet to be identified, they illustrate the possibilities for developing inhibitors that target other domains of SHRs.
The structural flexibility of the SHR-AF1/NTD may have appeared to be a rather unattractive target for small-molecule inhibitors of receptor function. However, the uniqueness and functional importance of the NTD makes it a worthwhile, even if technically challenging, drug target. It is therefore highly significant that a small molecule, identified in a screen of compounds isolated from a marine sponge, was recently shown to be highly effective and specific at inhibiting AR activity in vitro and in vivo (241). The compound, termed EPI-001, is a bisphenol A ester derivative (BADGE), and it interacted with the AR-AF1 domain, differentially disrupted protein-protein interactions, and reduced binding to androgen-regulated genes (241). Overall, this study provides “proof-of-principle” that small-molecule inhibitors can be identified that target the structurally plastic SHR-NTD.
Traditionally, it has been difficult to design or identify small molecules that target protein-protein interfaces, given the hydrophobic nature and relatively large surface area of such sites. However, the above-mentioned studies would challenge this preconception and open up the potential to develop tissue-specific modulators of SHR action that would complement or replace exiting therapeutics.
VIII. Conclusions and Future Challenges
The last 25 yr have seen tremendous progress in our understanding of the structure-function relationships of members of the nuclear receptor superfamily, including SHRs. This has included the isolation of receptor cDNA; the solving, at atomic resolution, of the structures of the isolated DBD and LBD; the identification of binding partners; and a clearer appreciation of the role of ID and allosteric coupling in receptor action. The future will prove to be just as challenging and exciting, and we may reasonably expect progress in a number of areas.
Solving the three-dimensional structure of a full-length SHR has proved a major technical challenge. However, our increased understanding of the functional and structural properties of the ID NTD, together with the allosteric coupling through hormone and DNA binding, opens up the possibility of investigating the conformation of a SHR-DNA-coregulatory protein(s) complex. In the absence of a structure for a full-length SHR complex, it will be important at the very least to obtain structural information, perhaps through small angle x-rays or DHX experiments, for a two-domain SHR in the presence and absence of DNA binding site and protein binding partners. Such studies will be invaluable if we are to further understand the structural basis for SHR-dependent gene regulation and allosteric control. Such information will also be vital to our understanding of the impact of point mutations in receptor-dependent diseases and in the design of novel inhibitors and SRM ligands to modulate tissue-specific SHR action.
In conclusion, SHRs are highly modular proteins, and this property has been exploited in the regulation of these molecules by the binding of different ligands (hormone, DNA, coregulatory proteins). The result is the exquisite control of normal physiology by steroid hormones and the serious consequences that arise through misregulation in the pathophysiology of certain cancers, inflammatory disease, and cardiovascular disorders. It has become increasingly clear that the NTD, containing the major transactivation function AF1, is naturally ID, and that this structural plasticity is functionally important for SHR activity. The SHR-NTD exists as an ensemble of conformers, having more or less structure, which prime this region of the receptor to rapidly respond to changes in the intracellular environment through hormone binding and posttranslation modifications. It is interesting that the NTD itself is often highly modular, with multiple regions contributing to receptor-dependent gene regulation. This seems particularly the case for the SHRs with the largest NTD: AR, PR, and MR. The consequences of this are unclear at present, but it may be significant that the length of the NTD has been correlated with AF1 “strength” in a reporter gene assay. A picture is emerging where the induced folding and/or stabilization of a functional conformation in the NTD is achieved through coregulatory protein binding and/or allosteric coupling with the DBD and DNA response element binding, and we are beginning to understand how different surfaces within the SHR protein may be created and used to control target gene expression.
Acknowledgments
Work in the authors' laboratories is supported by National Institutes of Health Grant R01DK058829 (to R.K.) and The Chief Scientist Office, Scottish Government, Prostate Cancer Charity (United Kingdom) and The Kosterlitz Center, University of Aberdeen (to I.J.M.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AF
- Activation function
- AR
- androgen receptor
- CTE
- C-terminal extension
- DBD
- DNA binding domain
- ER
- estrogen receptor
- ERE
- ER element
- ERR
- estrogen-related receptor
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response element
- HDX
- hydrogen deuterium exchange
- HRE
- hormone response element
- ID
- intrinsically disordered
- LBD
- ligand binding domain
- MR
- mineralocorticoid receptor
- NLS
- nuclear localization signal
- NMR
- nuclear magnetic resonance
- NTD
- N-terminal domain
- PPAR
- peroxisome proliferator-activated receptor
- PR
- progesterone receptor
- RXR
- retinoid X receptor
- SBR
- SHR binding region
- SHR
- steroid hormone receptor
- SRC-1
- steroid receptor coactivator-1
- SRM
- SHR modulator
- TBP
- TATA box binding protein
- TFE
- trifluoroethanol
- TFIIF
- transcription factor IIF
- VDR
- vitamin D3 receptor.
References
- 1. Gronemeyer H, Gustafsson JA, Laudet V. 2004. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3:950–964 [DOI] [PubMed] [Google Scholar]
- 2. Yang X, Lamia KA, Evans RM. 2007. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol 72:387–394 [DOI] [PubMed] [Google Scholar]
- 3. Beck IM, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, De Bosscher K. 2009. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev 30:830–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McEwan IJ. 2009. Nuclear receptors: one big family. Methods Mol Biol 505:3–18 [DOI] [PubMed] [Google Scholar]
- 5. Stanisiæ V, Lonard DM, O'Malley BW. 2010. Modulation of steroid hormone receptor activity. Prog Brain Res 181:153–176 [DOI] [PubMed] [Google Scholar]
- 6. Briet M, Schiffrin EL. 2010. Aldosterone: effects on the kidney and cardiovascular system. Nat Rev Nephrol 6:261–273 [DOI] [PubMed] [Google Scholar]
- 7. Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O'Malley BW. 2010. Global characterization of transcriptional impact of the SRC-3 coregulator. Mol Endocrinol 24:859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vicent GP, Nacht AS, Zaurín R, Ballaré C, Clausell J, Beato M. 2010. Minireview: role of kinases and chromatin remodeling in progesterone signaling to chromatin. Mol Endocrinol 24:2088–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao C, Dahlman-Wright K, Gustafsson JÅ. 2010. Estrogen signaling via estrogen receptor β. J Biol Chem 285:39575–39579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oakley RH, Cidlowski JA. 2011. Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem 286:3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. 2011. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145:224–241 [DOI] [PubMed] [Google Scholar]
- 12. Umetani M, Domoto H, Gormley AK, Yuhanna IS, Cummins CL, Javitt NB, Korach KS, Shaul PW, Mangelsdorf DJ. 2007. 27-Hydroxycholesterol is an endogenous SERM that inhibits the cardiovascular effects of estrogen. Nat Med 13:1185–1192 [DOI] [PubMed] [Google Scholar]
- 13. Connell JM, MacKenzie SM, Freel EM, Fraser R, Davies E. 2008. A lifetime of aldosterone excess: long-term consequences of altered regulation of aldosterone production for cardiovascular function. Endocr Rev 29:133–154 [DOI] [PubMed] [Google Scholar]
- 14. Bhasin S, Jasuja R. 2009. Selective androgen receptor modulators as function promoting therapies. Curr Opin Clin Nutr Metab Care 12:232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Funder JW, Reincke M. 2010. Aldosterone: a cardiovascular risk factor? Biochim Biophys Acta 1802:1188–1192 [DOI] [PubMed] [Google Scholar]
- 16. Konduri SD, Medisetty R, Liu W, Kaipparettu BA, Srivastava P, Brauch H, Fritz P, Swetzig WM, Gardner AE, Khan SA, Das GM. 2010. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci USA 107:15081–15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Börjesson AE, Windahl SH, Lagerquist MK, Engdahl C, Frenkel B, Movérare-Skrtic S, Sjögren K, Kindblom JM, Stubelius A, Islander U, Antal MC, Krust A, Chambon P, Ohlsson C. 2011. Roles of transactivating functions 1 and 2 of estrogen receptor-α in bone. Proc Natl Acad Sci USA 108:6288–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McEwan IJ. 2012. Intrinsic disorder in the androgen receptor: identification, characterisation and drugability. Mol BioSyst 8:82–90 [DOI] [PubMed] [Google Scholar]
- 19. Zennaro MC, Lombès M. 2004. Mineralocorticoid resistance. Trends Endocrinol Metab 15:264–270 [DOI] [PubMed] [Google Scholar]
- 20. Boonyaratanakornkit V, Edwards DP. 2004. Receptor mechanisms of rapid extranuclear signalling initiated by steroid hormones. Essays Biochem 40:105–120 [DOI] [PubMed] [Google Scholar]
- 21. Gellersen B, Fernandes MS, Brosens JJ. 2009. Non-genomic progesterone actions in female reproduction. Hum Reprod Update 15:119–138 [DOI] [PubMed] [Google Scholar]
- 22. McEwen BS. 2010. Stress, sex, and neural adaptation to a changing environment: mechanisms of neuronal remodeling. Ann NY Acad Sci 1204 Suppl:E38–E59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Groeneweg FL, Karst H, de Kloet ER, Joëls M. 2011. Rapid non-genomic effects of corticosteroids and their role in the central stress response. J Endocrinol 209:153–167 [DOI] [PubMed] [Google Scholar]
- 24. Evans RM. 1988. The steroid and thyroid hormone receptor superfamily. Science 240:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Escriva H, Bertrand S, Laudet V. 2004. The evolution of the nuclear receptor superfamily. Essays Biochem 40:11–26 [DOI] [PubMed] [Google Scholar]
- 26. Weinberger C, Hollenberg SM, Ong ES, Harmon JM, Brower ST, Cidlowski J, Thompson EB, Rosenfeld MG, Evans RM. 1985. Identification of human glucocorticoid receptor complementary DNA clones by epitope selection. Science 228:740–742 [DOI] [PubMed] [Google Scholar]
- 27. Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. 1986. Sequence and expression of human estrogen receptor complementary DNA. Science 231:1150–1154 [DOI] [PubMed] [Google Scholar]
- 28. Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. 1986. Human oestrogen receptor cDNA: sequence, expression and homology to v-Erb-A. Nature 320:134–139 [DOI] [PubMed] [Google Scholar]
- 29. Conneely OM, Sullivan WP, Toft DO, Birnbaumer M, Cook RG, Maxwell BL, Zarucki-Schulz T, Greene GL, Schrader WT, O'Malley BW. 1986. Molecular cloning of the chicken progesterone receptor. Science 233:767–770 [DOI] [PubMed] [Google Scholar]
- 30. Jeltsch JM, Krozowski Z, Quirin-Stricker C, Gronemeyer H, Simpson RJ, Garnier JM, Krust A, Jacob F, Chambon P. 1986. Cloning of the chicken progesterone receptor. Proc Natl Acad Sci USA 83:5424–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang CS, Kokontis J, Liao ST. 1988. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science 240:324–326 [DOI] [PubMed] [Google Scholar]
- 32. Lubahn DB, Joseph DR, Sar M, Tan J, Higgs HN, Larson RE, French FS, Wilson EM. 1988. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol Endocrinol 2:1265–1275 [DOI] [PubMed] [Google Scholar]
- 33. Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. 1985. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature 318:635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kornberg RD. 2007. The molecular basis of eucaryotic transcription. Cell Death Differ 14:1989–1997 [DOI] [PubMed] [Google Scholar]
- 35. Malik S, Roeder RG. 2010. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11:761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roeder RG. 2005. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579:909–915 [DOI] [PubMed] [Google Scholar]
- 37. McNally JG, Müller WG, Walker D, Wolford R, Hager GL. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262–1265 [DOI] [PubMed] [Google Scholar]
- 38. Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA. 2001. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat Cell Biol 3:15–23 [DOI] [PubMed] [Google Scholar]
- 39. Nagaich AK, Walker DA, Wolford R, Hager GL. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14:163–174 [DOI] [PubMed] [Google Scholar]
- 40. van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB. 2007. Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol 177:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL. 2005. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol 25:2406–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berry M, Metzger D, Chambon P. 1990. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J 9:2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. 1997. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753–758 [DOI] [PubMed] [Google Scholar]
- 44. Xu J, Wu RC, O'Malley BW. 2009. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer 9:615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bulynko YA, O'Malley BW. 2011. Nuclear receptor coactivators: structural and functional biochemistry. Biochemistry 50:313–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith CL, O'Malley BW. 2004. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev 25:45–71 [DOI] [PubMed] [Google Scholar]
- 47. Ball LJ, Levy N, Zhao X, Griffin C, Tagliaferri M, Cohen I, Ricke WA, Speed TP, Firestone GL, Leitman DC. 2009. Cell type- and estrogen receptor-subtype specific regulation of selective estrogen receptor modulator regulatory elements. Mol Cell Endocrinol 299:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spillman MA, Manning NG, Dye WW, Sartorius CA, Post MD, Harrell JC, Jacobsen BM, Horwitz KB. 2010. Tissue-specific pathways for estrogen regulation of ovarian cancer growth and metastasis. Cancer Res 70:8927–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumar R, Thompson EB. 2003. Transactivation functions of the N-terminal domains of nuclear hormone receptors: protein folding and coactivator interactions. Mol Endocrinol 17:1–10 [DOI] [PubMed] [Google Scholar]
- 50. Lavery DN, McEwan IJ. 2005. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J 391:449–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McEwan IJ, Lavery D, Fischer K, Watt K. 2007. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal 5:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wrange O, Gustafsson JA. 1978. Separation of the hormone- and DNA-binding sites of the hepatic glucocorticoid receptor by means of proteolysis. J Biol Chem 253:856–865 [PubMed] [Google Scholar]
- 53. Carlstedt-Duke J, Okret S, Wrange O, Gustafsson JA. 1982. Immunochemical analysis of the glucocorticoid receptor: identification of a third domain separate from the steroid-binding and DNA-binding domains. Proc Natl Acad Sci USA 79:4260–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carlstedt-Duke J, Strömstedt PE, Wrange O, Bergman T, Gustafsson JA, Jörnvall H. 1987. Domain structure of the glucocorticoid receptor protein. Proc Natl Acad Sci USA 84:4437–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith DF. 1993. Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol 7:1418–1429 [DOI] [PubMed] [Google Scholar]
- 56. Pratt WB, Morishima Y, Osawa Y. 2008. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283:22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaul S, Murphy PJ, Chen J, Brown L, Pratt WB, Simons SS., Jr 2002. Mutations at positions 547–553 of rat glucocorticoid receptors reveal that hsp90 binding requires the presence, but not defined composition, of a seven-amino acid sequence at the amino terminus of the ligand binding domain. J Biol Chem 277:36223–36232 [DOI] [PubMed] [Google Scholar]
- 58. Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. 1990. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348:166–168 [DOI] [PubMed] [Google Scholar]
- 59. Freeman BC, Yamamoto KR. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296:2232–2235 [DOI] [PubMed] [Google Scholar]
- 60. Huang P, Chandra V, Rastinejad F. 2010. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol 72:247–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eick GN, Thornton JW. 2011. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol 334:31–38 [DOI] [PubMed] [Google Scholar]
- 62. Heery DM, Kalkhoven E, Hoare S, Parker MG. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- 63. Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev 12:3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nagy L, Schwabe JW. 2004. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci 29:317–324 [DOI] [PubMed] [Google Scholar]
- 65. Dubbink HJ, Hersmus R, Verma CS, van der Korput HA, Berrevoets CA, van Tol J, Ziel-van der Made AC, Brinkmann AO, Pike AC, Trapman J. 2004. Distinct recognition modes of FXXLF and LXXLL motifs by the androgen receptor. Mol Endocrinol 18:2132–2150 [DOI] [PubMed] [Google Scholar]
- 66. He B, Gampe RT, Jr, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. 2004. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell 16:425–438 [DOI] [PubMed] [Google Scholar]
- 67. Hur E, Pfaff SJ, Payne ES, Grøn H, Buehrer BM, Fletterick RJ. 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol 2:E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD. 2007. A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J Mol Biol 368:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Egea PF, Klaholz BP, Moras D. 2000. Ligand-protein interactions in nuclear receptors of hormones. FEBS Lett 476:62–67 [DOI] [PubMed] [Google Scholar]
- 70. Pike AC, Brzozowski AM, Hubbard RE. 2000. A structural biologist's view of the oestrogen receptor. J Steroid Biochem Mol Biol 74:261–268 [DOI] [PubMed] [Google Scholar]
- 71. Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. 1994. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol 8:21–30 [DOI] [PubMed] [Google Scholar]
- 72. McInerney EM, Katzenellenbogen BS. 1996. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. J Biol Chem 271:24172–24178 [DOI] [PubMed] [Google Scholar]
- 73. Mérot Y, Métivier R, Penot G, Manu D, Saligaut C, Gannon F, Pakdel F, Kah O, Flouriot G. 2004. The relative contribution exerted by AF-1 and AF-2 transactivation functions in estrogen receptor α transcriptional activity depends upon the differentiation stage of the cell. J Biol Chem 279:26184–26191 [DOI] [PubMed] [Google Scholar]
- 74. Estébanez-Perpiñá E, Arnold LA, Arnold AA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ. 2007. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci USA 104:16074–16079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fawell SE, Lees JA, White R, Parker MG. 1990. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell 60:953–962 [DOI] [PubMed] [Google Scholar]
- 76. Savory JG, Préfontaine GG, Lamprecht C, Liao M, Walther RF, Lefebvre YA, Haché RJ. 2001. Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. Mol Cell Biol 21:781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93–105 [DOI] [PubMed] [Google Scholar]
- 78. Centenera MM, Harris JM, Tilley WD, Butler LM. 2008. The contribution of different androgen receptor domains to receptor dimerization and signaling. Mol Endocrinol 22:2373–2382 [DOI] [PubMed] [Google Scholar]
- 79. Langley E, Zhou ZX, Wilson EM. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem 270:29983–29990 [DOI] [PubMed] [Google Scholar]
- 80. Langley E, Kemppainen JA, Wilson EM. 1998. Intermolecular NH2-/carboxyl-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem 273:92–101 [DOI] [PubMed] [Google Scholar]
- 81. Doesburg P, Kuil CW, Berrevoets CA, Steketee K, Faber PW, Mulder E, Brinkmann AO, Trapman J. 1997. Functional in vivo interaction between the amino-terminal, transactivation domain and the ligand binding domain of the androgen receptor. Biochemistry 36:1052–1064 [DOI] [PubMed] [Google Scholar]
- 82. He B, Kemppainen JA, Wilson EM. 2000. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem 275:22986–22994 [DOI] [PubMed] [Google Scholar]
- 83. He B, Lee LW, Minges JT, Wilson EM. 2002. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem 277:25631–25639 [DOI] [PubMed] [Google Scholar]
- 84. Callewaert L, Verrijdt G, Christiaens V, Haelens A, Claessens F. 2003. Dual function of an amino-terminal amphipatic helix in androgen receptor-mediated transactivation through specific and nonspecific response elements. J Biol Chem 278:8212–8218 [DOI] [PubMed] [Google Scholar]
- 85. Métivier R, Penot G, Flouriot G, Pakdel F. 2001. Synergism between ERα transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: requirement for the AF-1 α-helical core and for a direct interaction between the N- and C-terminal domains. Mol Endocrinol 15:1953–1970 [DOI] [PubMed] [Google Scholar]
- 86. Kraus WL, McInerney EM, Katzenellenbogen BS. 1995. Ligand-dependent, transcriptionally productive association of the amino- and carboxyl-terminal regions of a steroid hormone nuclear receptor. Proc Natl Acad Sci USA 92:12314–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tetel MJ, Giangrande PH, Leonhardt SA, McDonnell DP, Edwards DP. 1999. Hormone-dependent interaction between the amino- and carboxyl-terminal domains of progesterone receptor in vitro and in vivo. Mol Endocrinol 13:910–924 [DOI] [PubMed] [Google Scholar]
- 88. Dong X, Challis JR, Lye SJ. 2004. Intramolecular interactions between the AF3 domain and the C-terminus of the human progesterone receptor are mediated through two LXXLL motifs. J Mol Endocrinol 32:843–857 [DOI] [PubMed] [Google Scholar]
- 89. Pippal JB, Yao Y, Rogerson FM, Fuller PJ. 2009. Structural and functional characterisation of the interdomain interaction in the mineralocorticoid receptor. Mol Endocrinol 23:1360–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Claessens F, Gewirth DT. 2004. DNA recognition by nuclear receptors. Essays Biochem 40:59–72 [DOI] [PubMed] [Google Scholar]
- 91. Härd T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP, Carlstedt-Duke J, Yamamoto KR, Gustafsson JA, Kaptein R. 1990. Solution structure of the glucocorticoid receptor DNA-binding domain. Science 249:157–160 [DOI] [PubMed] [Google Scholar]
- 92. Schwabe JW, Neuhaus D, Rhodes D. 1990. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature 348:458–461 [DOI] [PubMed] [Google Scholar]
- 93. Schwabe JW, Chapman L, Finch JT, Rhodes D. 1993. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 75:567–578 [DOI] [PubMed] [Google Scholar]
- 94. Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. 2004. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci USA 101:4758–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roemer SC, Donham DC, Sherman L, Pon VH, Edwards DP, Churchill ME. 2006. Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements. Mol Endocrinol 20:3042–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wardell SE, Kwok SC, Sherman L, Hodges RS, Edwards DP. 2005. Regulation of the amino-terminal transcription activation domain of progesterone receptor by a cofactor-induced protein folding mechanism. Mol Cell Biol 25:8792–8808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Daniel AR, Gaviglio AL, Czaplicki LM, Hillard CJ, Housa D, Lange CA. 2010. The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol Endocrinol 24:2126–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. 2008. Structural basis for the nuclear import of the human androgen receptor. J Cell Sci 121:957–968 [DOI] [PubMed] [Google Scholar]
- 100. Tanner TM, Denayer S, Geverts B, Van Tilborgh N, Kerkhofs S, Helsen C, Spans L, Dubois V, Houtsmuller AB, Claessens F, Haelens A. 2010. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci 67:1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. 2000. P300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem 275:20853–20860 [DOI] [PubMed] [Google Scholar]
- 102. Fu M, Wang C, Wang J, Zhang X, Sakamaki T, Yeung YG, Chang C, Hopp T, Fuqua SA, Jaffray E, Hay RT, Palvimo JJ, Jänne OA, Pestell RG. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol Cell Biol 22:3373–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Faus H, Haendler B. 2008. Androgen receptor acetylation sites differentially regulate gene control. J Cell Biochem 104:511–524 [DOI] [PubMed] [Google Scholar]
- 104. Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K, Shaheen F, Watt K, McEwan IJ, Wang C, Pestell RG, Robson CN. 2011. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 39:1266–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. 2008. Structure of the intact PPAR-γ-RXR-nuclear receptor complex on DNA. Nature 456:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hill KK, Roemer SC, Jones DN, Churchill ME, Edwards DP. 2009. A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain. J Biol Chem 284:24415–24424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zilliacus J, Wright AP, Carlstedt-Duke J, Gustafsson JA. 1995. Structural determinants of DNA-binding specificity by steroid receptors. Mol Endocrinol 9:389–400 [DOI] [PubMed] [Google Scholar]
- 108. Green S, Kumar V, Theulaz I, Wahli W, Chambon P. 1988. The N-terminal DNA-binding ‘zinc finger’ of the oestrogen and glucocorticoid receptors determines target gene specificity. EMBO J 7:3037–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Green S, Chambon P. 1989. Chimeric receptors used to probe the DNA-binding domain of the estrogen and glucocorticoid receptors. Cancer Res 49:2282s–2285s [PubMed] [Google Scholar]
- 110. Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. 2000. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem 275:12290–12297 [DOI] [PubMed] [Google Scholar]
- 111. Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- 112. Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- 113. Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. 2007. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 21:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27:5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, Mills IG. 2007. New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep 8:871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. 2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. 2007. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. 2008. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA 105:5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Acevedo ML, Kraus WL. 2004. Transcriptional activation by nuclear receptors. Essays Biochem 40:73–88 [DOI] [PubMed] [Google Scholar]
- 120. Rosenfeld MG, Lunyak VV, Glass CK. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20:1405–1428 [DOI] [PubMed] [Google Scholar]
- 121. Metzger D, Ali S, Bornert JM, Chambon P. 1995. Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J Biol Chem 270:9535–9542 [DOI] [PubMed] [Google Scholar]
- 122. Hollenberg SM, Evans RM. 1988. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell 55:899–906 [DOI] [PubMed] [Google Scholar]
- 123. Dahlman-Wright K, Almlöf T, McEwan IJ, Gustafsson JA, Wright AP. 1994. Delineation of a small region within the major transactivation domain of the human glucocorticoid receptor that mediates transactivation of gene expression. Proc Natl Acad Sci USA 91:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Almlöf T, Gustafsson JA, Wright AP. 1997. Role of hydrophobic amino acid clusters in the transactivation activity of the human glucocorticoid receptor. Mol Cell Biol 17:934–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Almlöf T, Wallberg AE, Gustafsson JA, Wright AP. 1998. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor τ 1-core activation domain and target factors. Biochemistry 37:9586–9594 [DOI] [PubMed] [Google Scholar]
- 126. Almlöf T, Wright AP, Gustafsson JA. 1995. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. J Biol Chem 270:17535–17540 [DOI] [PubMed] [Google Scholar]
- 127. Iñiguez-Lluhí JA, Lou DY, Yamamoto KR. 1997. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem 272:4149–4156 [DOI] [PubMed] [Google Scholar]
- 128. Simental JA, Sar M, Lane MV, French FS, Wilson EM. 1991. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem 266:510–518 [PubMed] [Google Scholar]
- 129. Jenster G, van der Korput HA, Trapman J, Brinkmann AO. 1995. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem 270:7341–7346 [DOI] [PubMed] [Google Scholar]
- 130. Chamberlain NL, Whitacre DC, Miesfeld RL. 1996. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem 271:26772–26778 [DOI] [PubMed] [Google Scholar]
- 131. Betney R, McEwan IJ. 2003. Role of conserved hydrophobic amino acids in androgen receptor AF-1 function. J Mol Endocrinol 31:427–439 [DOI] [PubMed] [Google Scholar]
- 132. Takimoto GS, Tung L, Abdel-Hafiz H, Abel MG, Sartorius CA, Richer JK, Jacobsen BM, Bain DL, Horwitz KB. 2003. Functional properties of the N-terminal region of progesterone receptors and their mechanistic relationship to structure. J Steroid Biochem Mol Biol 85:209–219 [DOI] [PubMed] [Google Scholar]
- 133. Govindan MV, Warriar N. 1998. Reconstitution of the N-terminal transcription activation function of human mineralocorticoid receptor in a defective human glucocorticoid receptor. J Biol Chem 273:24439–24447 [DOI] [PubMed] [Google Scholar]
- 134. Fuse H, Kitagawa H, Kato S. 2000. Characterization of transactivational property and coactivator mediation of rat mineralocorticoid receptor activation function-1 (AF-1). Mol Endocrinol 14:889–899 [DOI] [PubMed] [Google Scholar]
- 135. Fischer K, Kelly SM, Watt K, Price NC, McEwan IJ. 2010. Conformation of the mineralocorticoid receptor N-terminal domain: evidence for induced and stable structure. Mol Endocrinol 24:1935–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mély Y, Svergun DI, Moras D. 2011. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol 18:564–570 [DOI] [PubMed] [Google Scholar]
- 137. Putcha BD, Fernandez EJ. 2009. Direct interdomain interactions can mediate allosterism in the thyroid receptor. J Biol Chem 284:22517–22524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR. 2011. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol 18:556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. 1989. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell 59:477–487 [DOI] [PubMed] [Google Scholar]
- 140. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. 2002. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA 99:16701–16706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR. 2003. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA 100:13845–13850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wang Z, Frederick J, Garabedian MJ. 2002. Deciphering the phosphorylation “code” of the glucocorticoid receptor in vivo. J Biol Chem 277:26573–26580 [DOI] [PubMed] [Google Scholar]
- 143. Blind RD, Garabedian MJ. 2008. Differential recruitment of glucocorticoid receptor phospho-isoforms to glucocorticoid-induced genes. J Steroid Biochem Mol Biol 109:150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Billon-Galés A, Fontaine C, Filipe C, Douin-Echinard V, Fouque MJ, Flouriot G, Gourdy P, Lenfant F, Laurell H, Krust A, Chambon P, Arnal JF. 2009. The transactivating function 1 of estrogen receptor α is dispensable for the vasculoprotective actions of 17β-estradiol. Proc Natl Acad Sci USA 106:2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Dunker AK, Uversky VN. 2010. Drugs for ‘protein clouds’: targeting intrinsically disordered transcription factors. Curr Opin Pharmacol 10:782–788 [DOI] [PubMed] [Google Scholar]
- 146. Babu MM, van der Lee R, de Groot NS, Gsponer J. 2011. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol 21:432–440 [DOI] [PubMed] [Google Scholar]
- 147. Nilsson J, Grahn M, Wright AP. 2011. Proteome-wide evidence for enhanced positive Darwinian selection within intrinsically disordered regions in proteins. Genome Biol 12:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P. 2011. A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem 112:3256–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tompa P. 2011. Unstructural biology coming of age. Curr Opin Struct Biol 21:419–425 [DOI] [PubMed] [Google Scholar]
- 150. Uversky VN. 2011. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 43:1090–1103 [DOI] [PubMed] [Google Scholar]
- 151. Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. 2002. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 415:549–553 [DOI] [PubMed] [Google Scholar]
- 152. Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. 2007. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 6:2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Dunker AK, Uversky VN. 2008. Signal transduction via unstructured protein conduits. Nat Chem Biol 4:229–230 [DOI] [PubMed] [Google Scholar]
- 154. Wright PE, Dyson HJ. 2009. Linking folding and binding. Curr Opin Struct Biol 19:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. 2003. Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding. Proc Natl Acad Sci USA 100:5148–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. 2006. Intrinsic disorder in transcription factors. Biochemistry 45:6873–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. 2010. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc Natl Acad Sci USA 107:8183–8188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wang X, Zhang S, Zhang J, Huang X, Xu C, Wang W, Liu Z, Wu J, Shi Y. 2010. A large intrinsically disordered region in SKIP and its disorder-order transition induced by PPIL1 binding revealed by NMR. J Biol Chem 285:4951–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kulkarni P, Rajagopalan K, Yeater D, Getzenberg RH. 2011. Protein folding and the order/disorder paradox. J Cell Biochem 112:1949–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yoon MK, Shin J, Choi G, Choi BS. 2006. Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins 65:856–866 [DOI] [PubMed] [Google Scholar]
- 161. Kjaergaard M, Teilum K, Poulsen FM. 2010. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. Proc Natl Acad Sci USA 107:12535–12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. 2009. Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept Lett 16:1533–1547 [DOI] [PubMed] [Google Scholar]
- 163. Joerger AC, Fersht AR. 2008. Structural biology of the tumor suppressor p53. Annu Rev Biochem 77:557–582 [DOI] [PubMed] [Google Scholar]
- 164. Kumar R, Litwack G. 2009. Structural and functional relationships of the steroid hormone receptors' N-terminal transactivation domain. Steroids 74:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Garza AS, Ahmad N, Kumar R. 2009. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sci 84:189–193 [DOI] [PubMed] [Google Scholar]
- 166. Krasowski MD, Reschly EJ, Ekins S. 2008. Intrinsic disorder in nuclear hormone receptors. J Proteome Res 7:4359–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Uversky VN, Oldfield CJ, Dunker AK. 2008. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 37:215–246 [DOI] [PubMed] [Google Scholar]
- 168. Eisenhaber B, Eisenhaber F. 2007. Posttranslational modifications and subcellular localization signals: indicators of sequence regions without inherent 3D structure? Curr Protein Pept Sci 8:197–203 [DOI] [PubMed] [Google Scholar]
- 169. Kumar R, Thompson EB. 1999. The structure of the nuclear hormone receptors. Steroids 64:310–319 [DOI] [PubMed] [Google Scholar]
- 170. Kumar R, Johnson BH, Thompson EB. 2004. Overview of the structural basis for transcription regulation by nuclear hormone receptors. Essays Biochem 40:27–39 [DOI] [PubMed] [Google Scholar]
- 171. McEwan IJ, Nardulli AM. 2009. Nuclear hormone receptor architecture—form and dynamics: the 2009 FASEB summer conference on dynamic structure of the nuclear hormone receptors. Nucl Recept Signal 7:e011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Dahlman-Wright K, Baumann H, McEwan IJ, Almlöf T, Wright AP, Gustafsson JA, Härd T. 1995. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci USA 92:1699–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Baskakov IV, Kumar R, Srinivasan G, Ji YS, Bolen DW, Thompson EB. 1999. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem 274:10693–10696 [DOI] [PubMed] [Google Scholar]
- 174. Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. 2001. The N-terminal region of human progesterone B-receptors: biophysical and biochemical comparison to A-receptors. J Biol Chem 276:23825–23831 [DOI] [PubMed] [Google Scholar]
- 175. Wärnmark A, Wikström A, Wright AP, Gustafsson JA, Härd T. 2001. The N-terminal regions of estrogen receptor α and β are unstructured in vitro and show different TBP binding properties. J Biol Chem 276:45939–45944 [DOI] [PubMed] [Google Scholar]
- 176. Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. 2002. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem 277:20079–20086 [DOI] [PubMed] [Google Scholar]
- 177. Shen HC, Buchanan G, Butler LM, Prescott J, Henderson M, Tilley WD, Coetzee GA. 2005. GRIP1 mediates the interaction between the amino- and carboxyl-termini of the androgen receptor. Biol Chem 386:69–74 [DOI] [PubMed] [Google Scholar]
- 178. Georgiakaki M, Chabbert-Buffet N, Dasen B, Meduri G, Wenk S, Rajhi L, Amazit L, Chauchereau A, Burger CW, Blok LJ, Milgrom E, Lombès M, Guiochon-Mantel A, Loosfelt H. 2006. Ligand-controlled interaction of histone acetyltransferase binding to ORC-1 (HBO1) with the N-terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1-dependent coactivation of transcription. Mol Endocrinol 20:2122–2140 [DOI] [PubMed] [Google Scholar]
- 179. Lavery DN, McEwan IJ. 2008. Functional characterization of the native NH2-terminal transactivation domain of the human androgen receptor: binding kinetics for interactions with TFIIF and SRC-1a. Biochemistry 47:3352–3359 [DOI] [PubMed] [Google Scholar]
- 180. Garza AM, Khan SH, Kumar R. 2010. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol 30:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wardell SE, Narayanan R, Weigel NL, Edwards DP. 2010. Partial agonist activity of the progesterone receptor antagonist RU486 mediated by an amino-terminal domain coactivator and phosphorylation of serine 400. Mol Endocrinol 24:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Khan SH, Ling J, Kumar R. 2011. TBP binding-induced folding of the glucocorticoid receptor AF1 domain facilitates its interaction with steroid receptor coactivator-1. PLoS One 6:e21939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Kumar R, Baskakov IV, Srinivasan G, Bolen DW, Lee JC, Thompson EB. 1999. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem 274:24737–24741 [DOI] [PubMed] [Google Scholar]
- 184. Brodie J, McEwan IJ. 2005. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. J Mol Endocrinol 34:603–615 [DOI] [PubMed] [Google Scholar]
- 185. Cutress RI, Townsend PA, Sharp A, Maison A, Wood L, Lee R, Brimmell M, Mullee MA, Johnson PW, Royle GT, Bateman AC, Packham G. 2003. The nuclear BAG-1 isoform, BAG-1L, enhances oestrogen-dependent transcription. Oncogene 22:4973–4982 [DOI] [PubMed] [Google Scholar]
- 186. Shatkina L, Mink S, Rogatsch H, Klocker H, Langer G, Nestl A, Cato AC. 2003. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol Cell Biol 23:7189–7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kumar R, Lee JC, Bolen DW, Thompson EB. 2001. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem 276:18146–18152 [DOI] [PubMed] [Google Scholar]
- 188. Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. 2004. Induced α-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry 43:3008–3013 [DOI] [PubMed] [Google Scholar]
- 189. Lavery DN, McEwan IJ. 2008. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: a collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry 47:3360–3369 [DOI] [PubMed] [Google Scholar]
- 190. Khan SH, Arnott JA, Kumar R. 2011. Naturally occurring osmolyte, trehalose induces functional conformation in an intrinsically disordered activation domain of glucocorticoid receptor. PLoS One 6:e19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Dahlman-Wright K, McEwan IJ. 1996. Structural studies of mutant glucocorticoid receptor transactivation domains establish a link between transactivation activity in vivo and α-helix-forming potential in vitro. Biochemistry 35:1323–1327 [DOI] [PubMed] [Google Scholar]
- 192. Timasheff SN. 2002. Protein-solvent preferential interactions, protein hydration, and the modulation of biochemical reactions by solvent components. Proc Natl Acad Sci USA 99:9721–9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Street TO, Bolen DW, Rose GD. 2006. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci USA 103:13997–14002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kumar R. 2009. Role of naturally occurring osmolytes in protein folding and stability. Arch Biochem Biophys 491:1–6 [DOI] [PubMed] [Google Scholar]
- 195. Holthauzen LM, Rösgen J, Bolen DW. 2010. Hydrogen bonding progressively strengthens upon transfer of the protein urea-denatured state to water and protecting osmolytes. Biochemistry 49:1310–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kumar R, Serrette JM, Khan SH, Miller AL, Thompson EB. 2007. Effects of different osmolytes on the induced folding of the N-terminal activation domain (AF1) of the glucocorticoid receptor. Arch Biochem Biophys 465:452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kumar R. 2008. Osmolyte-induced folding of an intrinsically disordered activation function subdomain of glucocorticoid receptor. J Recept Signal Transduct Res 28:465–474 [DOI] [PubMed] [Google Scholar]
- 198. Fu M, Rao M, Wu K, Wang C, Zhang X, Hessien M, Yeung YG, Gioeli D, Weber MJ, Pestell RG. 2004. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J Biol Chem 279:29436–29449 [DOI] [PubMed] [Google Scholar]
- 199. Loven MA, Likhite VS, Choi I, Nardulli AM. 2001. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor β conformation. J Biol Chem 276:45282–45288 [DOI] [PubMed] [Google Scholar]
- 200. Wood JR, Greene GL, Nardulli AM. 1998. Estrogen response elements function as allosteric modulators of estrogen receptor conformation. Mol Cell Biol 18:1927–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Wood JR, Likhite VS, Loven MA, Nardulli AM. 2001. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol 15:1114–1126 [DOI] [PubMed] [Google Scholar]
- 202. Barkhem T, Haldosén LA, Gustafsson JA, Nilsson S. 2002. PS2 Gene expression in HepG2 cells: complex regulation through crosstalk between the estrogen receptor α, an estrogen-responsive element, and the activator protein 1 response element. Mol Pharmacol 61:1273–1283 [DOI] [PubMed] [Google Scholar]
- 203. McKenna NJ, Lanz RB, O'Malley BW. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344 [DOI] [PubMed] [Google Scholar]
- 204. Glass CK, Rose DW, Rosenfeld MG. 1997. Nuclear receptor coactivators. Curr Opin Cell Biol 9:222–232 [DOI] [PubMed] [Google Scholar]
- 205. McEwan IJ, Dahlman-Wright K, Ford J, Wright AP. 1996. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: evidence for an induced fit model of transactivation domain folding. Biochemistry 35:9584–9593 [DOI] [PubMed] [Google Scholar]
- 206. Dyson HJ, Wright PE. 2002. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12:54–60 [DOI] [PubMed] [Google Scholar]
- 207. Dunker AK, Silman I, Uversky VN, Sussman JL. 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18:756–764 [DOI] [PubMed] [Google Scholar]
- 208. Iakoucheva LM, Brown CJ, Lawson JD, Obradoviæ Z, Dunker AK. 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323:573–584 [DOI] [PubMed] [Google Scholar]
- 209. Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32:1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Bevan CL, Hoare S, Claessens F, Heery DM, Parker MG. 1999. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol 19:8383–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Benecke A, Chambon P, Gronemeyer H. 2000. Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep 1:151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Onate SA, Boonyaratanakornkit V, Spencer TE, Tsai SY, Tsai MJ, Edwards DP, O'Malley BW. 1998. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem 273:12101–12108 [DOI] [PubMed] [Google Scholar]
- 213. Powell SM, Christiaens V, Voulgaraki D, Waxman J, Claessens F, Bevan CL. 2004. Mechanisms of androgen receptor signalling via steroid receptor coactivator-1 in prostate. Endocr Relat Cancer 11:117–130 [DOI] [PubMed] [Google Scholar]
- 214. Sugase K, Dyson HJ, Wright PE. 2007. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447:1021–1025 [DOI] [PubMed] [Google Scholar]
- 215. Reid J, Murray I, Watt K, Betney R, McEwan IJ. 2002. The androgen receptor interacts with multiple regions of the large subunit of general transcription factor TFIIF. J Biol Chem 277:41247–41253 [DOI] [PubMed] [Google Scholar]
- 216. Shoemaker BA, Portman JJ, Wolynes PG. 2000. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA 97:8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Hermann S, Berndt KD, Wright AP. 2001. How transcriptional activators bind target proteins. J Biol Chem 276:40127–40132 [DOI] [PubMed] [Google Scholar]
- 218. Ferreira ME, Hermann S, Prochasson P, Workman JL, Berndt KD, Wright AP. 2005. Mechanism of transcription factor recruitment by acidic activators. J Biol Chem 280:21779–21784 [DOI] [PubMed] [Google Scholar]
- 219. Wardell SE, Boonyaratanakornkit V, Adelman JS, Aronheim A, Edwards DP. 2002. Jun dimerization protein 2 functions as a progesterone receptor N-terminal domain coactivator. Mol Cell Biol 22:5451–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. 2002. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J Biol Chem 277:42241–42248 [DOI] [PubMed] [Google Scholar]
- 221. Duma D, Jewell CM, Cidlowski JA. 2006. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J Steroid Biochem Mol Biol 102:11–21 [DOI] [PubMed] [Google Scholar]
- 222. Wang C, Tian L, Popov VM, Pestell RG. 2011. Acetylation and nuclear receptor action. J Steroid Biochem Mol Biol 123:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. 2011. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol 25:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Tremblay AM, Wilson BJ, Yang XJ, Giguère V. 2008. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-α and -γ transcriptional activity through a synergy control motif. Mol Endocrinol 22:570–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Weigel NL, Moore NL. 2007. Steroid receptor phosphorylation: a key modulator of multiple receptor functions. Mol Endocrinol 21:2311–2319 [DOI] [PubMed] [Google Scholar]
- 226. Chen D, Riedl T, Washbrook E, Pace PE, Coombes RC, Egly JM, Ali S. 2000. Activation of estrogen receptor α by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell 6:127–137 [PubMed] [Google Scholar]
- 227. Miller AL, Webb MS, Copik AJ, Wang Y, Johnson BH, Kumar R, Thompson EB. 2005. P38 Mitogen-activated protein kinase (MAPK) is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol Endocrinol 19:1569–1583 [DOI] [PubMed] [Google Scholar]
- 228. Chen W, Dang T, Blind RD, Wang Z, Cavasotto CN, Hittelman AB, Rogatsky I, Logan SK, Garabedian MJ. 2008. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol 22:1754–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Chen YX, Du JT, Zhou LX, Liu XH, Zhao YF, Nakanishi H, Li YM. 2006. Alternative O-GlcN acylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor β. Chem Biol 13:937–944 [DOI] [PubMed] [Google Scholar]
- 230. Lefstin JA, Yamamoto KR. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885–888 [DOI] [PubMed] [Google Scholar]
- 231. Nettles KW, Sun J, Radek JT, Sheng S, Rodriguez AL, Katzenellenbogen JA, Katzenellenbogen BS, Greene GL. 2004. Allosteric control of ligand selectivity between estrogen receptors α and β: implications for other nuclear receptors. Mol Cell 13:317–327 [DOI] [PubMed] [Google Scholar]
- 232. Hilser VJ, Thompson EB. 2007. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA 104:8311–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Kumar R, Thompson EB. 2010. Influence of flanking sequences on signaling between the activation function AF1 and DNA-binding domain of the glucocorticoid receptor. Arch Biochem Biophys 496:140–145 [DOI] [PubMed] [Google Scholar]
- 234. McDonnell DP. 2000. Selective estrogen receptor modulators (SERMs): a first step in the development of perfect hormone replacement therapy regimen. J Soc Gynecol Investig 7:S10–S15 [DOI] [PubMed] [Google Scholar]
- 235. McDonnell DP, Chang CY, Norris JD. 2000. Development of peptide antagonists that target estrogen receptor-cofactor interactions. J Steroid Biochem Mol Biol 74:327–335 [DOI] [PubMed] [Google Scholar]
- 236. Paige LA, Christensen DJ, Grøn H, Norris JD, Gottlin EB, Padilla KM, Chang CY, Ballas LM, Hamilton PT, McDonnell DP, Fowlkes DM. 1999. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER α and ER β. Proc Natl Acad Sci USA 96:3999–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Wijayaratne AL, Nagel SC, Paige LA, Christensen DJ, Norris JD, Fowlkes DM, McDonnell DP. 1999. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology 140:5828–5840 [DOI] [PubMed] [Google Scholar]
- 238. Yang J, Chang CY, Safi R, Morgan J, McDonnell DP, Fuller PJ, Clyne CD, Young MJ. 2011. Identification of ligand-selective peptide antagonists of the mineralocorticoid receptor using phage display. Mol Endocrinol 25:32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Mao C, Patterson NM, Cherian MT, Aninye IO, Zhang C, Montoya JB, Cheng J, Putt KS, Hergenrother PJ, Wilson EM, Nardulli AM, Nordeen SK, Shapiro DJ. 2008. A new small molecule inhibitor of estrogen receptor α binding to estrogen response elements blocks estrogen-dependent growth of cancer cells. J Biol Chem 283:12819–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Jones JO, Bolton EC, Huang Y, Feau C, Guy RK, Yamamoto KR, Hann B, Diamond MI. 2009. Non-competitive androgen receptor inhibition in vitro and in vivo. Proc Natl Acad Sci USA 106:7233–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, Watt K, Tam T, Yang YC, Bañuelos CA, Williams DE, McEwan IJ, Wang Y, Sadar MD. 2010. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 17:535–546 [DOI] [PubMed] [Google Scholar]
- 242. Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. 2008. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol 41:301–314 [DOI] [PubMed] [Google Scholar]
- 243. Kumar R, Volk DE, Li J, Lee JC, Gorenstein DG, Thompson EB. 2004. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc Natl Acad Sci USA 101:16425–16430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Nocula-£ugowska M, Rymarczyk G, Lisowski M, Ozyhar A. 2009. Isoform-specific variation in the intrinsic disorder of the ecdysteroid receptor N-terminal domain. Proteins 76:291–308 [DOI] [PubMed] [Google Scholar]
- 245. Dziedzic-Letka A, Rymarczyk G, Kapłon TM, Górecki A, Szamborska-Gbur A, Wojtas M, Dobryszycki P, O¿yhar A. 2011. Intrinsic disorder of Drosophila melanogaster hormone receptor 38 N-terminal domain. Proteins 79:376–392 [DOI] [PubMed] [Google Scholar]
- 246. He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM. 2004. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J Biol Chem 279:30643–30653 [DOI] [PubMed] [Google Scholar]
- 247. Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. 2005. Comparing and combining predictors of mostly disordered proteins. Biochemistry 44:1989–2000 [DOI] [PubMed] [Google Scholar]
- 248. Yang ZR, Thomson R, McNeil P, Esnouf RM. 2005. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 21:3369–3376 [DOI] [PubMed] [Google Scholar]
- 249. Linding R, Russell RB, Neduva V, Gibson TJ. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. 2005. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21:3435–3438 [DOI] [PubMed] [Google Scholar]