Abstract
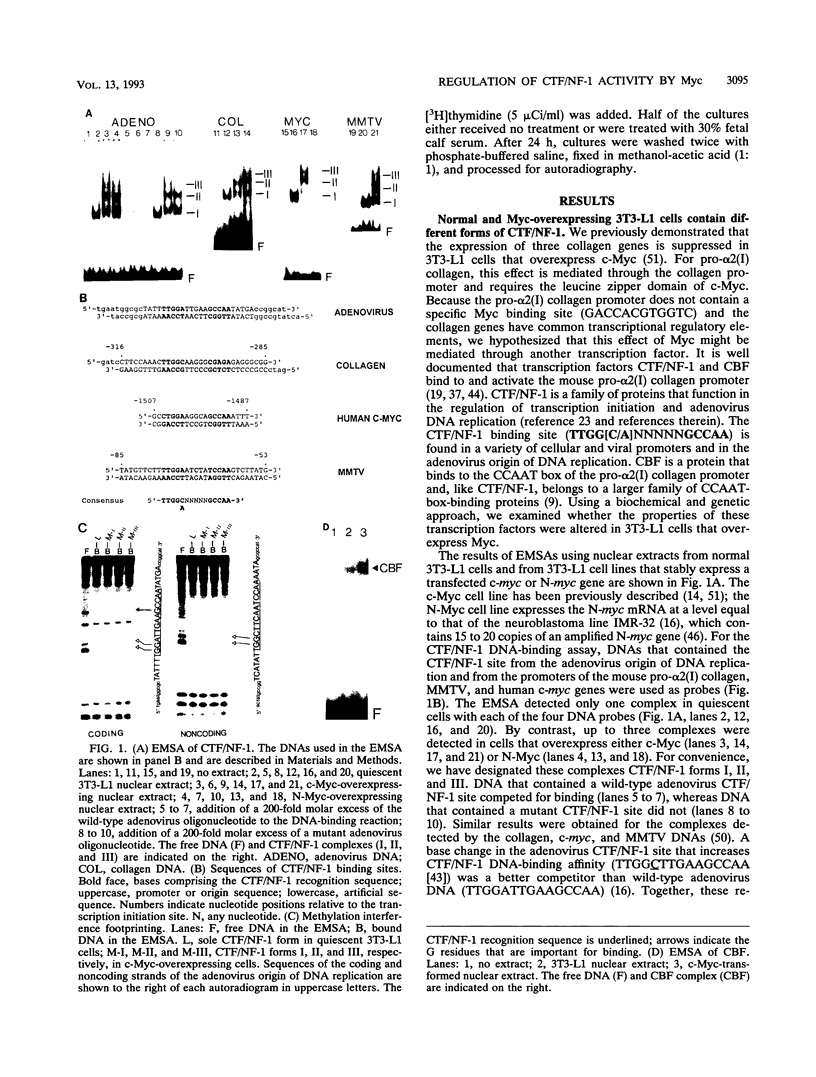
Overexpression of Myc in cells can suppress the transcription of specific genes. Because several of these genes have common transcriptional regulatory elements, we investigated the possibility that this effect of Myc is mediated through a specific transcription factor. In vitro DNA-binding assays detect only one form of CCAAT transcription factor/nuclear factor 1 (CTF/NF-1) in quiescent 3T3-L1 cells. By contrast, quiescent 3T3-L1 cells that stably overexpress either c-Myc or N-Myc contain at least three forms of CTF/NF-1. Biochemical characterization of the various CTF/NF-1 forms showed that they have the same native molecular weight but differ in charge density. The more negatively charged CTF/NF-1 forms present in Myc-overexpressing cells are converted into that found in normal cells by treatment with acid phosphatase, suggesting that they represent a more phosphorylated form of the CTF/NF-1 protein. The various CTF/NF-1 forms have a similar DNA-binding affinity. Transfection experiments demonstrated that transcription from CTF/NF-1-dependent promoters is specifically suppressed in cells that stably overexpress c-Myc. This effect requires CTF/NF-1 binding. CTF/NF-1-dependent promoter activity is also suppressed in 3T3-L1 cells during active growth (relative to the quiescent state). Interestingly, actively growing 3T3-L1 cells contain forms of CTF/NF-1 similar to those in quiescent cells that stably overexpress c-Myc. Thus, the CTF/NF-1 forms present in cells that express high amounts of c-Myc correlate with a lower transcription rate of CTF/NF-1-dependent promoters in vivo. Our results provide a basis for the suppression of specific gene transcription by c-Myc.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson G. M., Freytag S. O. Synergistic activation of a human promoter in vivo by transcription factor Sp1. Mol Cell Biol. 1991 Apr;11(4):1935–1943. doi: 10.1128/mcb.11.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres A. C., van der Valk M. A., Schönenberger C. A., Flückiger F., LeMeur M., Gerlinger P., Groner B. Ha-ras and c-myc oncogene expression interferes with morphological and functional differentiation of mammary epithelial cells in single and double transgenic mice. Genes Dev. 1988 Nov;2(11):1486–1495. doi: 10.1101/gad.2.11.1486. [DOI] [PubMed] [Google Scholar]
- Bernards R., Dessain S. K., Weinberg R. A. N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell. 1986 Dec 5;47(5):667–674. doi: 10.1016/0092-8674(86)90509-x. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. Molecular themes in oncogenesis. Cell. 1991 Jan 25;64(2):235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwood E. M., Eisenman R. N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991 Mar 8;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- Buetti E., Kühnel B. Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J Mol Biol. 1986 Aug 5;190(3):379–389. doi: 10.1016/0022-2836(86)90009-4. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M., Schirm S., Bishop J. M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991 Jan;10(1):133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag S. O., Dang C. V., Lee W. M. Definition of the activities and properties of c-myc required to inhibit cell differentiation. Cell Growth Differ. 1990 Jul;1(7):339–343. [PubMed] [Google Scholar]
- Freytag S. O. Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol. 1988 Apr;8(4):1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag S. O., Geddes T. J. Reciprocal regulation of adipogenesis by Myc and C/EBP alpha. Science. 1992 Apr 17;256(5055):379–382. doi: 10.1126/science.256.5055.379. [DOI] [PubMed] [Google Scholar]
- Grignani F., Lombardi L., Inghirami G., Sternas L., Cechova K., Dalla-Favera R. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. EMBO J. 1990 Dec;9(12):3913–3922. doi: 10.1002/j.1460-2075.1990.tb07612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P., Leavitt J., Muscat G., Ng S. Y., Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatamochi A., Golumbek P. T., Van Schaftingen E., de Crombrugghe B. A CCAAT DNA binding factor consisting of two different components that are both required for DNA binding. J Biol Chem. 1988 Apr 25;263(12):5940–5947. [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Inghirami G., Grignani F., Sternas L., Lombardi L., Knowles D. M., Dalla-Favera R. Down-regulation of LFA-1 adhesion receptors by C-myc oncogene in human B lymphoblastoid cells. Science. 1990 Nov 2;250(4981):682–686. doi: 10.1126/science.2237417. [DOI] [PubMed] [Google Scholar]
- Jackson S. P., MacDonald J. J., Lees-Miller S., Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990 Oct 5;63(1):155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Kaddurah-Daouk R., Greene J. M., Baldwin A. S., Jr, Kingston R. E. Activation and repression of mammalian gene expression by the c-myc protein. Genes Dev. 1987 Jun;1(4):347–357. doi: 10.1101/gad.1.4.347. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Barrett J., Villa-Garcia M., Dang C. V. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990 Nov;10(11):5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G. J., Lee W. M., Chen L. L., Dang C. V. Max: functional domains and interaction with c-Myc. Genes Dev. 1992 Jan;6(1):81–92. doi: 10.1101/gad.6.1.81. [DOI] [PubMed] [Google Scholar]
- Kingston R. E., Baldwin A. S., Jr, Sharp P. A. Regulation of heat shock protein 70 gene expression by c-myc. Nature. 1984 Nov 15;312(5991):280–282. doi: 10.1038/312280a0. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Rustgi A. K., Schievella A. R., Bernards R. Suppression of MHC class I gene expression by N-myc through enhancer inactivation. EMBO J. 1989 Nov;8(11):3351–3355. doi: 10.1002/j.1460-2075.1989.tb08497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Eisenman R. N. New light on Myc and Myb. Part I. Myc. Genes Dev. 1990 Dec;4(12A):2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Gander I., Rogge L., Winnacker E. L. A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res. 1988 May 25;16(10):4419–4435. doi: 10.1093/nar/16.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Miksicek R., Borgmeyer U., Nowock J. Interaction of the TGGCA-binding protein with upstream sequences is required for efficient transcription of mouse mammary tumor virus. EMBO J. 1987 May;6(5):1355–1360. doi: 10.1002/j.1460-2075.1987.tb02375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink S., Härtig E., Jennewein P., Doppler W., Cato A. C. A mammary cell-specific enhancer in mouse mammary tumor virus DNA is composed of multiple regulatory elements including binding sites for CTF/NFI and a novel transcription factor, mammary cell-activating factor. Mol Cell Biol. 1992 Nov;12(11):4906–4918. doi: 10.1128/mcb.12.11.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikarinen J., Hatamochi A., de Crombrugghe B. Separate binding sites for nuclear factor 1 and a CCAAT DNA binding factor in the mouse alpha 2(I) collagen promoter. J Biol Chem. 1987 Aug 15;262(23):11064–11070. [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Land H. Negative autoregulation of c-myc transcription. EMBO J. 1990 Apr;9(4):1113–1121. doi: 10.1002/j.1460-2075.1990.tb08217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn L. J., Brooks M. W., Laufer E. M., Littlewood T. D., Morgenstern J. P., Evan G. I., Lee W. M., Land H. Domains of human c-myc protein required for autosuppression and cooperation with ras oncogenes are overlapping. Mol Cell Biol. 1990 Sep;10(9):4961–4966. doi: 10.1128/mcb.10.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G. C., Lawe D., Ziff E. B. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991 May 3;65(3):395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Ziff E. B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., O'Neill E. A., Wides R. J., Kelly T. J. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol Cell Biol. 1987 Feb;7(2):875–886. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P., Karsenty G., Roberts A. B., Roche N. S., Sporn M. B., de Crombrugghe B. A nuclear factor 1 binding site mediates the transcriptional activation of a type I collagen promoter by transforming growth factor-beta. Cell. 1988 Feb 12;52(3):405–414. doi: 10.1016/s0092-8674(88)80033-3. [DOI] [PubMed] [Google Scholar]
- Schwab M., Alitalo K., Klempnauer K. H., Varmus H. E., Bishop J. M., Gilbert F., Brodeur G., Goldstein M., Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983 Sep 15;305(5931):245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Hennighausen L., Battey J., Leder P. Chromatin structure and protein binding in the putative regulatory region of the c-myc gene in Burkitt lymphoma. Cell. 1984 Jun;37(2):381–391. doi: 10.1016/0092-8674(84)90368-4. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990 Feb;64(2):543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg R., Noordermeer I. A., Krüse-Wolters M., Ruiter D. J., Schrier P. I. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. 1988 Apr;7(4):1023–1029. doi: 10.1002/j.1460-2075.1988.tb02909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. S., Geddes T. J., Pogulis R. J., de Crombrugghe B., Freytag S. O. Transcriptional suppression of cellular gene expression by c-Myc. Mol Cell Biol. 1991 Apr;11(4):2291–2295. doi: 10.1128/mcb.11.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]










