Abstract
Left-right (L-R) asymmetries in neuroanatomy exist throughout the animal kingdom, with implications for function and behavior. The molecular mechanisms that control formation of such asymmetries are beginning to be understood. Significant progress has been made by studying the zebrafish parapineal organ, a group of neurons on the left side of the epithalamus. Parapineal cells arise from the medially located pineal complex anlage and migrate to the left side of the brain. We have found that Fgf8a regulates a fate decision among anterior pineal complex progenitors that occurs just prior to the initiation of leftward migration. Cell fate analysis shows that in the absence of Fgf8a a subset of cells in the anterior pineal complex anlage differentiate as cone photoreceptors rather than parapineal neurons. Fgf8a acts permissively to promote parapineal fate in conjunction with the transcription factor Tbx2b, but might also block cone photoreceptor fate. We conclude that this subset of anterior pineal complex precursors, which normally become parapineal cells, are bipotential and require Fgf8a to maintain parapineal identity and/or prevent cone identity.
Keywords: Specification, Determination, Differentiation, Epithalamus, Zebrafish, Neurons
INTRODUCTION
The presence of brain asymmetries is widespread among vertebrates (Geschwind and Levitsky, 1968; Kohl et al., 2011; de Schotten et al., 2011). Although direct experimental evidence is scarce, such L-R differences are thought to increase the capacity and speed of multi-task performance (Rogers, 2000; Nowicka and Tacikowski, 2011). Only recently has there been advancement in understanding the molecular mechanisms governing the formation of lateralized brain structures. The most progress has been made in studies of the zebrafish epithalamus, a forebrain region that exhibits robust molecular and anatomical asymmetries (Taylor et al., 2010; Roussigné et al., 2012). The epithalamus consists of a medially located pineal complex and the paired habenular nuclei flanking either side of the midline (Borg et al., 1983; Butler and Hodos, 1996; Amo et al., 2010). The pineal complex can be further subdivided into a pineal organ and a structure called the parapineal organ that is usually located on the left side of the brain (Borg et al., 1983). The pineal organ, being directly photoreceptive in zebrafish, comprises rod and cone photoreceptors as well as associated projection neurons (Masai et al., 1997). The parapineal organ is a cluster of 10-12 neurons that migrate leftward from the anterior midline of the pineal anlage to lie just posterior to the developing left habenula (Concha and Wilson, 2001; Snelson et al., 2008b). The emergence of the parapineal organ strongly correlates with the induction of anatomical and molecular asymmetries within the habenular nuclei. The full elaboration of these asymmetries requires the parapineal organ; when the parapineal organ is ablated or in mutants in which a left-sided parapineal organ does not form, the habenular nuclei develop more symmetrically with the left nucleus more closely resembling the right in gene expression and neuropil density (Concha and Wilson, 2001; Concha et al., 2003; Gamse et al., 2003; Snelson et al., 2008b; Regan et al., 2009).
Although an increasing body of work has characterized the development of the pineal organ and the habenular nuclei in zebrafish (Masai et al., 1997; Cau and Wilson, 2003; Cau et al., 2008; Quillien et al., 2011), comparatively little attention has been lent to the formation of parapineal neurons. Previous work suggests that parapineal cells are likely to be specified prior to the 15-somite stage from a group of precursors in the anterior pineal complex anlage (Masai et al., 1997; Snelson et al., 2008a). Parapineal cell migration begins by ∼30 hours post-fertilization (hpf) as cells travel in a chain-like fashion towards the left side of the epithalamus. Prior to migration, parapineal precursors are intermingled with and indistinguishable from the surrounding pineal cells in the anterior pineal complex anlage (Concha et al., 2003; Snelson et al., 2008b). In addition to the parapineal organ, the pineal complex anlage gives rise to the projection neurons, rod photoreceptors and cone photoreceptors of the pineal organ (Masai et al., 1997). All pineal complex cell types undergo their final mitotic division in a similar time period between 15 and 24 hpf (Cau et al., 2008; Snelson et al., 2008b). The generation of the proper numbers of the different cell types (parapineal, pineal rod photoreceptor, pineal cone photoreceptor and pineal projection neuron) from the pineal complex anlage requires input from the Notch and Bmp pathways as well as the activity of at least two different transcription factors, T-box containing transcription factor 2b (Tbx2b) and the homeodomain-containing protein Floating head (Flh) (Masai et al., 1997; Cau et al., 2008; Snelson et al., 2008a; Snelson et al., 2008b; Quillien et al., 2011). However, additional genes are likely to be involved in cell specification in the pineal complex anlage, as a small number of parapineal and pineal neurons do form in tbx2b; flh double mutants (Snelson et al., 2008a).
One candidate for pineal and/or parapineal cell specification is the Fgf signaling pathway. Fgf ligands and receptors are expressed in the epithalamus of zebrafish and other vertebrates (Crossley and Martin, 1995; Crossley et al., 1996; Reifers et al., 1998; Reifers et al., 2000; Echevarría et al., 2003). Previous work has shown that Fgf8a can promote migration of the parapineal organ away from the dorsal midline of the pineal complex anlage (Regan et al., 2009). However, a role for Fgf signaling in controlling cell fates within the pineal complex anlage remains to be examined.
Fgfs have well-documented roles as morphogens in the regional patterning of the vertebrate fore- and hindbrain (Sansom and Livesey, 2009; Nakamura et al., 2008). To investigate whether a similar role exists for Fgf in the epithalamus, we performed gain- and loss-of-function experiments in zebrafish. We find that Fgf signaling is required for promoting parapineal cell fate by preventing their incorrect differentiation as cone photoreceptors. Cell fate analysis suggests that a subset of cells in the anterior pineal complex anlage, which give rise to the parapineal organ in wild-type larvae, instead produce cone photoreceptors in fgf8a mutants. Epistasis analysis with Tbx2b reveals that both genes are required for parapineal cells to form but only fgf8a is required to prevent their differentiation as cone photoreceptors. We conclude that, unlike its typical morphogenic role in brain patterning, Fgf signaling acts on bipotential anterior pineal complex precursors to govern a decision between parapineal and cone cell fate.
MATERIALS AND METHODS
Zebrafish
Zebrafish were raised at 28.5°C on a 14/10 hour light/dark cycle and staged according to hpf. The following fish lines were used: AB* (Walker, 1999), fgf8ax15 (Kwon and Riley, 2009), tbx2bc144 (Snelson et al., 2008b), Tg[foxd3:GFP]zf104 (Gilmour et al., 2002), Tg[hsp70:fgf8a]b1193 (Hans et al., 2007) and Tg[flhBAC:kaede]vu376. The Tg[flhBAC:kaede]vu376 transgenic line was generated as previously described (Shin et al., 2003). The first 18 codons of the flh coding sequence in the BAC #101I13 (Yan et al., 1998) were fused to the kaede coding sequence (Ando et al., 2002) using published BAC recombineering methods (Lee et al., 2001). Recombined BAC was injected into one-cell-stage embryos, which were raised to adulthood and screened for germline transmission of the transgene.
In situ hybridization
Whole-mount RNA in situ hybridization was performed as described previously (Gamse et al., 2003), using reagents from Roche Applied Bioscience. Hybridized probes were detected using alkaline phosphatase-conjugated antibodies (Roche) and visualized by 4-nitro blue tetrazolium (NBT; Roche) and 5-bromo-4-chloro-3-indolyl-phosphate (BCIP; Roche) staining for single labeling, or NBT/BCIP followed by iodonitrotetrazolium (INT) and BCIP staining for double labeling. Information on the probes is in supplementary material Table S1.
Cloning
sox1a was cloned by PCR from total cDNA from 26 hpf AB* zebrafish embryos using Phusion polymerase (Finnzymes) and the following primers: 5′-CACCACTGGCTACAGGAGCGAAAA-3′; 5′-CAGAAACGCTGTCAGGATCA-3′. PCR product was purified with a Mini Elute Gel Purification Kit (Qiagen) and ligated into pENTR-D/Topo vector (Invitrogen).
Cryosectioning
After whole-mount in situ hybridization, embryos were embedded in 1.5% agarose, 5% sucrose media. Blocks containing embedded embryos were excised, equilibrated overnight at 4°C in 30% sucrose, and frozen using 2-methylbutane in liquid nitrogen. Frozen blocks were sectioned with a Leica CM1850 cryostat at a thickness of 10-12 μm.
Antibody labeling
Embryos and larvae were fixed overnight at 4°C in 4% paraformaldehyde with 0.3 mM CaCl2, 4% sucrose in 1×PBS, rehydrated with three 5-minute washes in 1×PBSTx (1×PBS with 0.01% Triton X-100) and four 20-minute washes with distilled H2O, and blocked in 1×PBSTx with 10% sheep serum and 1 mg/ml BSA. Antibodies were incubated overnight at 4°C and washed off with four 20-minute washes in 1×PBSTx. Details on primary and secondary antibodies are listed in supplementary material Table S1. Confocal images were taken with a Zeiss LSM 510 microscope and processed using Improvision Velocity software.
Heat shock conditions
Embryo clutches containing both heterozygous Tg[hsp70I:fgf8a]b1943 transgenic embryos and their non-transgenic siblings were placed in a 2-ml tube (35-40 embryos/tube). Pre-warmed egg water containing 0.3% PTU was added at a volume of 2 ml per tube. Tubes were incubated in a 37°C water bath, then moved into a dish in a 28.5°C incubator. Expression of fgf8a was induced by single heat shock treatment at 24 hpf for 30 minutes. We also tried multiple short heat shocks (15 minutes) at 37°C between the 18-somite stage and 30 hpf, or continuous heat shock for 6 or 15 hours at lower temperatures (30°C or 32°C). All such treatments resulted in embryo death.
Caged fluorescein injection, uncaging and detection
One-cell stage Tg[foxd3:GFP]zf104; Tg[flhBAC:kaede]vu376 double transgenic embryos were injected with 0.5 nl of a 1% 2,3-dimethyl-2,3-dinitrobutane (DMNB)-caged fluorescein-dextran solution. Caged fluorescein dextran synthesis, injection, uncaging and detection were performed as previously described (Clanton et al., 2011).
Morpholino injection
Embryos were injected at the one-cell stage. The following morpholinos were used in this study: tbx2b splice blocking morpholino, 5′-AAAATATGGGTACATACCTTGTCGT-3′ (Snelson et al., 2008b); flh MO, 5′-AATCTGCATGGCGTCTGTTTAGTCC-3′.
Inhibitor treatments
For in situ hybridizations and whole-mount antibody labeling, we incubated embryos in their chorions in 12 μM SU5402 (Calbiochem and Tocris) dissolved in 0.3% dimethyl sulfoxide (DMSO) in egg water in 0.003% n-phenylthiourea (1×PTU; Sigma-Aldrich). Control embryos were treated with 0.3% DMSO in parallel with their SU5402-treated siblings. To hinder parapineal migration, embryos in their chorions were treated with 6 μM SU5402 dissolved in 0.3% DMSO in egg water in 1×PTU in the dark from 24 to 30 hpf. Control embryos were treated with 0.15% DMSO in egg water in 1×PTU in the dark. For cell fate analysis, dechorionated embryos (which are more sensitive to SU5402) were treated with 8 μM SU5402 and control embryos were treated with 0.2% DMSO.
Measuring parapineal cell migration
Tg[foxd3:GFP]zf104 and sox1a co-labeled embryos were imaged using a Leica TCSSP5 confocal microscope under a 40× oil immersion objective. Confocal images were analyzed using Volocity software. Parapineal cell distance was calculated by using the line function in the xyz view to measure from the center of a sox1a-positive cell to the middle of the pineal organ.
RESULTS
Fgf ligands, receptor, and a target gene are expressed in the pineal complex anlage
Previous work showed that fgf8a and fgf17, two ligands in the Fgf family, are expressed in the epithalamus (Reifers et al., 1998; Reifers et al., 2000; Itoh, 2007; Jovelin et al., 2007; Regan et al., 2009). However, the expression of these ligands has not been analyzed relative to markers of the pineal complex anlage at stages beyond 24 hpf. To precisely describe expression at high resolution by confocal microscopy, we used fluorescent in situ hybridization (FISH) in combination with immunofluorescence. The transgenes Tg[foxd3:GFP]zf104 (Gilmour et al., 2002) and Tg[flhBAC:kaede]vu376 together label all cells of the pineal complex anlage and its derivatives, the pineal organ and parapineal organ. We confirmed that expression of the Tg[flhBAC:kaede]vu376 transgene is faithful to endogenous flh expression by co-labeling for Kaede and flh (supplementary material Fig. S1A,B). Transcripts for fgf17 and the Fgf signaling target erm (ets related molecule; etv5a – Zebrafish Information Network) are not abundant enough for our FISH protocol (Clay and Ramakrishnan, 2005) and were thus detected with two-color chromogenic in situ hybridization.
At 24 hpf, fgf8a expression encompasses the anterior-most two to three cell diameters of pineal complex (Fig. 1A,A′). At 30 hpf, fgf8a continues to be expressed in the anterior pineal complex, including parapineal cells that are beginning to separate from the pineal complex anlage (Fig. 1B,B′). At this time, significant expression of fgf8a is also present in cells found directly to the left and right sides of the anterior pineal complex, where habenular precursor cells are located (Concha et al., 2003) (Fig. 1B). By 36hpf, fgf8a is mostly excluded from the migrating parapineal organ (Fig. 1C,C′).
Fig. 1.
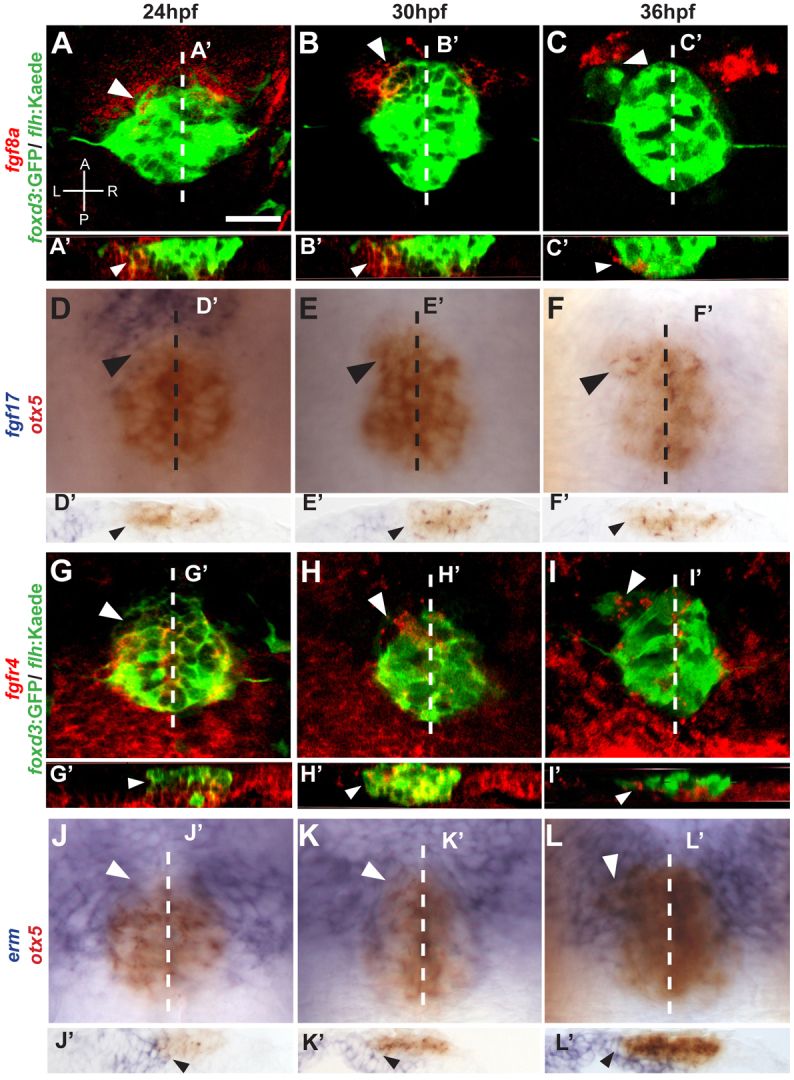
Fgf components are expressed in the epithalamus during parapineal formation in zebrafish. (A-C) fgf8a (red) is expressed in the anterior pineal complex (white arrowheads) of foxd3:GFP/flhBAC:Kaede (green)-expressing embryos during parapineal development. A, anterior; L, left; P, posterior; R, right. (A′-C′) Optical cross sections at the level of the dashed lines in A-C, respectively. fgf8a is expressed in the ventral region of the anterior pineal complex anlage (white arrowheads). (D-F) Dorsal views of in situ hybridizations of fgf17 (blue) relative to the pineal complex (otx5, red). fgf17 is expressed in the anterior pineal complex (black arrowheads) at 24 hpf, but is almost gone by 30 and 36 hpf. Dashed line indicates plane of sectioning. (D′-F′) Cryosections at the levels of the dashed lines in D-F, respectively. fgf17 expression is evident in the region ventral to the anterior pineal complex anlage (black arrows) at 24 hpf. (G-I). Confocal projections show that fgfr4 (red) is expressed within the pineal complex anlage including the anterior portion (white arrowheads). (G′-I′) Optical cross sections at the level of the dashed lines in G-I, respectively, showing fgfr4 expresseion in the anterior pineal complex anlage where parapineal precursors are located (white arrowheads). (J-L) Dorsal views of in situ hybridization of erm (blue) relative to the pineal complex anlage (otx5; red). erm expression shows that Fgf signaling activity is high in the anterior pineal complex (white arrowheads). White dashed lines indicate the plane of sectioning. (J′-L′) Cryosections at the level of the dashed lines in J-L, respectively, of in situ hybridizations of erm. erm is detected in the anterior pineal complex anlage at all stages (black arrowheads). Scale bar: 25 μm.
Like fgf8a, fgf17 is expressed in the anterior-most one-third of the pineal complex (Fig. 1D,D′). However, at 30 and 36 hpf, fgf17 expression is almost undetectable in the epithalamus (Fig. 1E-F′). In summary, fgf17 and fgf8a are both expressed in anterior pineal complex anlage where parapineal cells are present at 24 hpf (Concha et al., 2003), but only fgf8a persists later.
Once secreted, Fgf ligands can bind to and activate any of four Fgf receptors (Böttcher and Niehrs, 2005). fgfr1, fgfr2 and fgfr3 are not highly expressed in the vicinity of the pineal complex between 24 and 36 hpf (Ota et al., 2010) (data not shown). We did detect fgfr4 expression in the pineal complex between 24 and 36 hpf, in agreement with a previous report (Regan et al., 2009). At 24 hpf, fgfr4 transcript is in both the anterior and posterior pineal complex anlage (Fig. 1G), with highest expression in the most ventral cells (Fig. 1G′). By 30 hpf and continuing to 36 hpf, fgfr4 expression is found throughout the pineal complex including the parapineal precursor cells (Fig. 1H-I′).
Fgf signaling is necessary and sufficient to induce expression of the gene erm, making it a convenient readout of Fgf signaling (Roehl and Nusslein-Volhard, 2001). The expression of erm in the pineal complex anlage is very robust at the anterior-most aspect, where parapineal precursors are located, indicating that these cells are responding to high levels of Fgf signaling at 24, 30 and 36 hpf (Fig. 1J-L′).
Attenuating Fgf signaling disrupts parapineal formation
To quantify the effect of Fgf8a loss on parapineal development, we examined the expression of growth factor inhibited 1.2 (gfi-1.2; gfi1ab – Zebrafish Information Network) and sex determining Y box 1a (sox1a) in fgf8ax15mutants, which express a null allele of fgf8a (Kwon and Riley, 2009).
In wild-type (WT) embryos at 52 or 96 hpf, the parapineal is a distinct cluster of 9-12 gfi-1.2-expressing cells (average 9.3±0.3, n=23) located to the left side of the pineal organ (Fig. 2A,E; supplementary material Table S2). gfi-1.2 expression correlates strongly with the cluster of Tg[foxd3:GFP]zf104-positive cells to the left of the pineal organ, indicating that gfi-1.2 is present in all parapineal cells (Dufourcq et al., 2004). At 52 and 96 hpf, fgf8ax15 mutants exhibit an almost 60% reduction in the number of gfi-1.2-expressing cells (4.0±0.4, n=27) (Fig. 2B,E; supplementary material Table S2). The remaining gfi-1.2-expressing cells are located within the pineal organ and do not form a distinct parapineal organ, as previously reported (Regan et al., 2009).
Fig. 2.
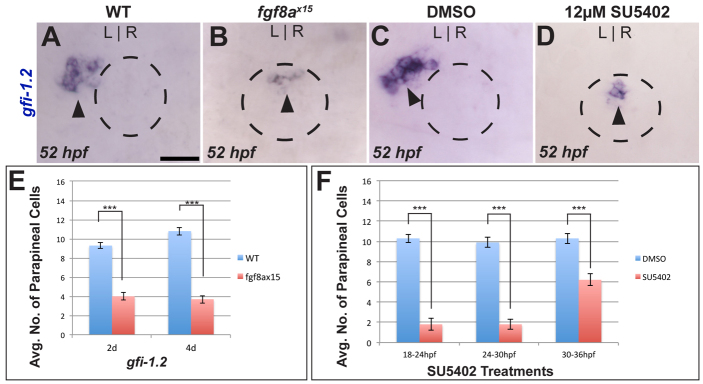
Loss of Fgf signaling results in fewer parapineal cells. (A,B) Dorsal views of WT and fgf8ax15 mutant zebrafish embryos labeled with gfi-1.2 (parapineal cells; black arrowheads). (C,D) Dorsal views showing expression of gfi-1.2 (black arrowheads) in WT larvae at 52 hpf that were treated between 24 and 30 hpf with either DMSO or SU5402. In A-D, dashed line encircles the pineal complex anlage. (E) fgf8ax15 mutants displayed a significant reduction in parapineal cell number compared with WT (***P=<0.0005 by t-test). (F) Reductions in parapineal cell numbers were observed in embryos treated with SU5402, particularly for 18-24 hpf and 24-30 hpf treatments (***P<0.0005 by t-test). Error bars represent s.e.m. Scale bar: 25 μm.
Because gfi1.2 is not expressed until 45 hpf, we analyzed expression of a novel early parapineal marker, sox1a. In WT embryos, the expression of sox1a in the epithalamus is first evident in a few cells of the anterior pineal complex at 26 hpf (supplementary material Fig. S2A). By 28 hpf, sox1a is expressed in a cluster of cells spanning the midline of the anterior pineal complex (supplementary material Fig. S2B), similar to the placement of parapineal cell precursors at 24 hpf (Concha et al., 2003). The sox1a-expressing cells are found to the left of the midline at 36 hpf; by 48 hpf, sox1a is co-expressed with Tg[foxd3:GFP]zf104 in a group of cells to the left of the pineal complex, similar to gfi-1.2 (supplementary material Fig. S2C,D). Thus, we conclude that sox1a is expressed in parapineal cells just prior to migration. At 32 hpf, fgf8ax15 mutants have fewer than half the number of sox1a-positive cells as WT embryos (3.0±0.5, n=17, and 7.5±0.3, n=16, respectively) (supplementary material Fig. S2E-G). Reduced expression of the early parapineal marker sox1a in fgf8ax15 mutants supports the idea that Fgf signaling is required for specification or differentiation of parapineal fate.
Expression of fgf8a is found in the epithalamus from 20 hpf to 72 hpf (Fig. 1; data not shown). Previous work had shown that Fgf signaling is needed between 24 and 44 hpf for parapineal migration (Regan et al., 2009). To establish a temporal requirement for Fgf signaling in production of parapineal cells, we used SU5402, a small molecule that blocks Ras/mitogen-activated protein kinase (MAPK) activation by Fgf receptors (Mohammadi et al., 1997), to abrogate Fgf signaling at different intervals between 18 and 36 hpf. Although all treatment regimens reduced the number of parapineal cells, blocking Fgf activity between 18 and 30 hpf proved to be the most effective in reducing parapineal cell number (Fig. 2F; supplementary material Table S3). Embryos treated with SU5402 from 18 to 24 hpf or from 24 to 30 hpf had ∼80% fewer gfi-1.2-expressing cells compared with control embryos. Blocking Fgf signaling from 30 to 36 hpf results in a ∼40% reduction in parapineal cell number compared with control embryos (Fig. 2F; supplementary material Table S3), suggesting that the timing of parapineal specification is slightly later in some embryos.
In an effort to identify precisely when Fgf is required for parapineal development, we performed an array of 2-hour treatments between 18 and 30 hpf. Inhibiting Fgf signaling in these short intervals resulted in significantly reduced parapineal cell numbers (supplementary material Fig. S3A-C and Table S3). However, the reduction is less severe than the 6-hour treatments (Fig. 2F; supplementary material Table S3). All together, these data suggest that Fgf signaling is required over a broad time frame from 18 to 30 hpf to ensure formation of the correct number of parapineal cells. This encompasses the period prior to parapineal migration.
It is possible that reduced parapineal cell number in fgf8a mutants is secondary to failure of parapineal migration, i.e. parapineal cells might need to move away from the midline to properly differentiate. However, we found that parapineal migration can be uncoupled from differentiation. To block parapineal migration, we used a low dose of SU5402. Treating embryos with 6 μM SU5402 from 24 to 30 hpf resulted in reduced migration of parapineal cells, but did not significantly alter parapineal cell number (supplementary material Fig. S4A,D and Table S4). These data suggest that parapineal differentiation is not dependent on migration.
Conditional expression of Fgf8a is not sufficient to induce parapineal cells
As Fgf8a is necessary for parapineal cell generation, we next tested whether Fgf8a is sufficient to produce additional parapineal cells. We conditionally overexpressed Fgf8a using the Tg[hsp70l:fgf8a]b1193 transgenic line, in which fgf8a transcription is induced by elevated temperature (Hans et al., 2007). The efficacy of fgf8a overexpression via the hsp70l:fgf8a transgene to activate Fgf signaling was confirmed by in situ hybridization for fgf8a and erm transcripts (data not shown).
Based on the temporal requirement for Fgf signaling determined by our SU5402 treatments, we induced fgf8a expression at 24 hpf (see Materials and methods for details) and examined gfi1.2 expression at 52 hpf. We detected no significant difference in parapineal cell number in hsp70l:fgf8a transgenic embryos relative to their non-transgenic siblings (Fig. 3A-C; supplementary material Table S5). We also tested whether Fgf8a overexpression could rescue the parapineal defects seen in fgf8ax15 mutants. We analyzed the expression of sox1a at 36 hpf and gfi-1.2 at 52 hpf in mutants with and without the hsp70l:fgf8a transgene. When heat shocked at 24 hpf, there was no rescue of parapineal cells in mutants (supplementary material Fig. S5 and Table S5), suggesting that we cannot restore parapineal number in fgf8ax15 mutants by the addition of exogenous Fgf8a.
Fig. 3.
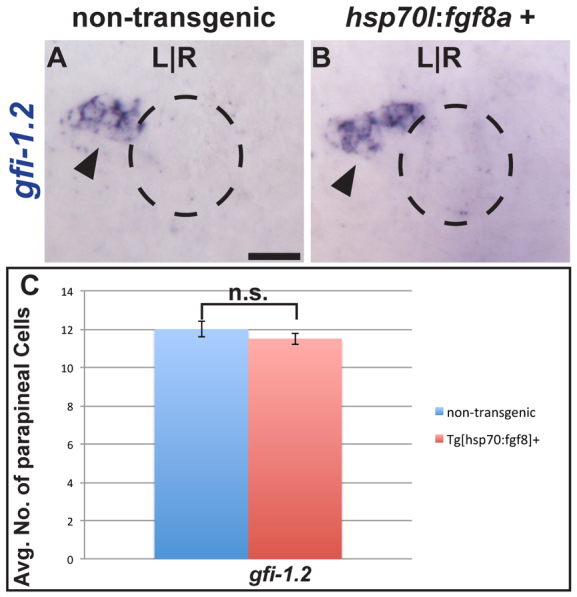
Fgf8a overexpression is not sufficient to induce supernumerary parapineal cells. (A,B) Dorsal view of non-transgenic and Tg[hps70:fgf8a]b193 zebrafish larvae at 52 hpf that were induced for 30 minutes at 24 hpf and labeled with gfi-1.2 (black arrowheads). Dashed line encircles the pineal complex anlage. (C) Graph quantifying the number of gfi-1.2-expressing cells. Error bars represent s.e.m. n.s., not significant. Scale bar: 25 μm.
Reduced Fgf activity leads to a selective increase in cone cell number
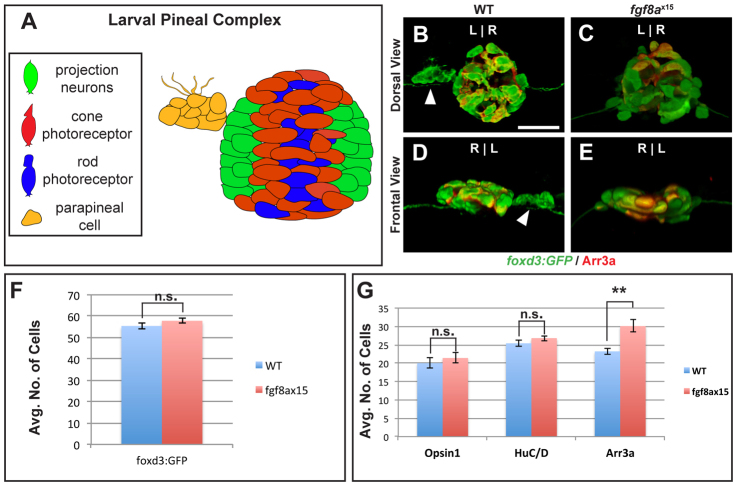
The deficit of parapineal cells seen in fgf8ax15 mutants and SU5402-treated embryos could result from alteration of cell fate or reductions in the total number of pineal complex cells. To distinguish between these possibilities, we counted the total number of pineal complex cells, as well as the number of each cell type. The pineal complex is composed of rod photoreceptors (labeled by Opsin-1), cone photoreceptors (labeled by Arrestin 3a), projection neurons (labeled by HuC; Elavl3 – Zebrafish Information Network) and parapineal cells (Fig. 4A) (Butler and Hodos, 1996; Masai et al., 1997; Concha et al., 2000; Cau et al., 2008; Snelson et al., 2008b). With the exception of rod cells, all pineal complex cells express Tg[foxd3:GFP]zf104 at 52 hpf. Thus, by counting cells that express either Tg[foxd3:GFP]zf104 or Opsin-1, we can quantify the total number of pineal complex cells.
Fig. 4.
fgf8ax15 mutants have a selective increase in cone photoreceptor number compared with WT larvae. (A) Schematic of the pineal complex of the larval zebrafish at 52 hpf. (B-E) Dorsal (B,C) and frontal (D,E) views of WT and fgf8ax15 mutants labeled with foxd3:GFP (pineal complex) and Arr3a (cone photoreceptors) at 52 hpf. fgf8ax15 mutants lack a distinct parapineal organ compared with WT (white arrowheads). L, left; R, right. (F,G) fgf8ax15 and WT embryos have equivalent numbers of foxd3:GFP expressing cells (F), projection neurons (HuC/D) and rod photoreceptors (Opsin1) (G). However, fgf8ax15 mutants have more cone photoreceptors (Arr3a) in the pineal complex than WT (G) (**P<0.005 by t-test). Error bars represent s.e.m. Scale bar: 25 μm.
The morphology of the pineal organ is largely unaltered in fgf8ax15 mutants compared with WT siblings (Fig. 4B-E). However, fgf8ax15 mutant larvae do not have a distinct parapineal organ (Fig. 4C,E). Despite the lack of a parapineal organ, the total number of Tg[foxd3:GFP]zf104-positive cells in fgf8ax15 mutants is not significantly different from that observed in WT larvae at 52 hpf (Fig. 4F; supplementary material Table S6) or 96 hpf (supplementary material Fig. S7A-C and Table S6).
The number of rods and projection neurons was unchanged in fgf8a mutants (Fig. 4G; supplementary material Fig. S6 and Table S6). However, the number of cone photoreceptors, labeled by Arrestin 3a (Arr3a; also known as Zpr1 and Fret43) (Larison and Bremiller, 1990; Masai et al., 1997; Ile et al., 2010) and Tg[foxd3:GFP]zf104, is significantly different in fgf8ax15 mutants and WT. At 52 hpf, WT larvae had an average of 23.2±0.8 (n=10) cells expressing Arr3a (Fig. 4G). Arr3a expression was only observed in the pineal organ and was never seen in the parapineal of WT (Fig. 4B,D). fgf8ax15 mutants have an average of 30.1±1.7 (n=12) cone cells (Fig. 4G). The average increase of approximately seven cone cells in fgf8ax15 mutants relative to WT is comparable to the decrease in gfi-1.2-expressing cells (Fig. 2E). The increase in cone number in fgf8ax15 mutant larvae persists at 96 hpf (supplementary material Fig. S7A-C and Table S6). A similar phenotype to fgf8ax15 mutants was detected in WT embryos treated from 24 to 30 hpf with 12 μM SU5402 (supplementary material Fig. S7D-F and Table S3). Taken together, these data show that the total number of pineal complex cells is unchanged in fgf8ax15 mutants compared with their WT siblings, arguing against a role for Fgf8a in governing cell proliferation or cell survival. However, fgf8ax15 mutants and 12 μM SU5402-treated larvae exhibit an increase in one cell type, cone photoreceptors, mirroring the decrease in the number of mature parapineal cells.
As mentioned above, we could not rescue parapineal cell number by overexpressing Fgf8a in fgf8a mutants. Similarly, we did not see a rescue of cone cell number by overexpressing Fgf8a via the Tg[hsp70l:fgf8a]b1193 transgenic line (supplementary material Fig. S5F and Table S5), and the number of Tg[foxd3:GFP] remains unchanged between hsp70l:fgf8-negative fgf8ax15 mutants and hsp70l:fgf8-positive mutants, indicating that the overall pineal complex cell number is not governed by Fgf8a (supplementary material Fig. S5F and Table S5).
Parapineal precursors give rise to cone cells in fgf8ax15 mutants
Because the number of cone cells is increased and the number of parapineal cells is decreased in fgf8ax15, we hypothesized that cells that would normally become parapineal cells were differentiating instead as cone photoreceptors. Fate-mapping data for the pineal complex indicated that by 24 hpf, parapineal precursor cells reside in the anterior one-third of the pineal complex anlage intermingled with precursors of the pineal organ (Concha et al., 2003). To conduct cell fate analysis of parapineal precursor cells, we photoactivated caged fluorescein dextran using a laser microbeam in 10-15 cells of the anterior pineal complex anlage at 24 hpf in fgf8ax15 mutants or WT siblings and allowed them to develop until 52 hpf (Fig. 5A). To mark pineal complex cells, we used the Tg[foxd3:GFP]zf104 and Tg[flhBAC:kaede]vu376 transgenes. In WT embryos, fluorescein-labeled parapineal cells were clearly detectable at 52 hpf to the left of the pineal organ (Fig. 5B,B′). As the anterior pineal anlage does not give rise exclusively to parapineal cells, some cells of the pineal organ were also labeled (Fig. 5B,B′). In WT, only 27%±5.4 (n=14) of fluorescein-labeled cells became cone photoreceptors (Fig. 5D). However, in fgf8ax15 mutants, cone photoreceptors made up almost twice as many of the fluorescein-labeled cells (48%±4.7, n=14) as in WT siblings (Fig. 5C-D).
Fig. 5.
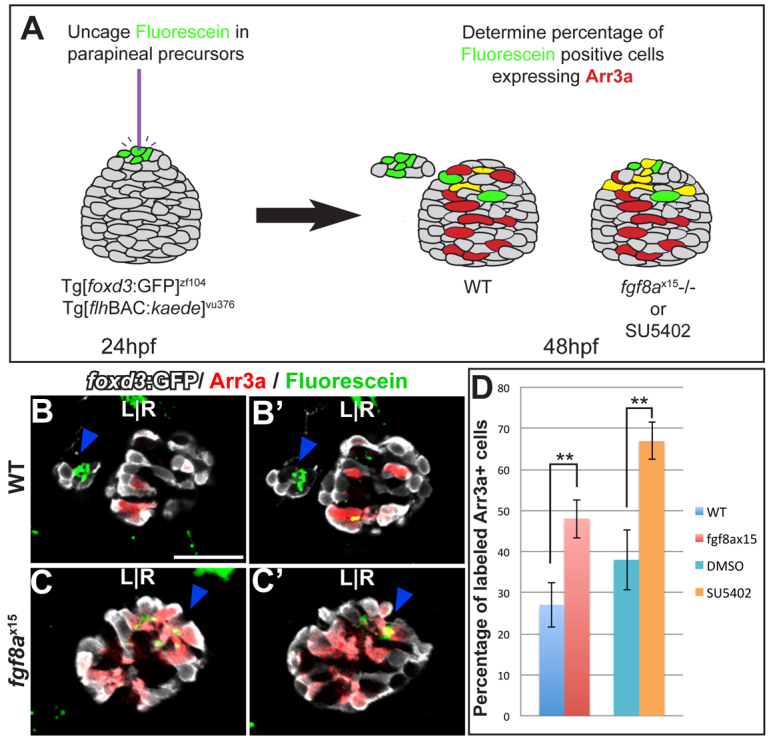
Cell fate analysis of the anterior pineal complex indicates that parapineal precursors adopt a cone photoreceptor fate in fgf8ax15 mutants. (A) Fate analysis scheme. Green, fluorescein-positive cells; red, Arr3a-positive cells; yellow, cells expressing both fluorescein and Arr3a. (B-C′) Dorsal views of serial confocal sections in the pineal complex of WT (B,B′) or fgf8ax15 (C,C′) zebrafish embryos at 52 hpf, after labeling of the anterior pineal complex anlage at 24 hpf. In WT, clusters of labeled parapineal cells are evident (blue arrowheads). In fgf8a mutants, many of the cells that were labeled in the anterior pineal complex anlage at 24 hpf become cone photoreceptors (blue arrowheads). (D) When Fgf signaling is reduced, a significantly higher percentage of labeled cells become cone photoreceptors compared with controls (**P<0.005 by t-test). Error bars represent s.e.m. Scale bar: 30 μm.
A possible explanation for the increase in labeled cone photoreceptors in our cell fate analysis experiments is that in fgf8ax15 mutants, parapineal precursor cells might not reside in the anterior pineal anlage. To exclude this possibility, we performed cell fate analysis of the anterior pineal complex anlage using embryos treated with SU5402 between 24 and 30 hpf (Fig. 5D). In SU5402-treated embryos 67%±4.5 (n=9) of fluorescein-labeled cells became cone cells. However, only 38%±7.4 (n=7) of labeled cells in control embryos gave rise to cone cells, similar to fgf8ax15 mutants.
We conclude that the additional cone cells in fgf8ax15 mutants are derived from parapineal precursor cells that have incorrectly adopted a cone photoreceptor fate. To reflect more accurately the bipotentiality of these cells, we will refer to them as ‘anterior pineal complex precursor cells’ from now on.
Flh is not responsible for altering anterior pineal complex precursor fate in fgf8a mutants
The transcription factor Flh is required for neurogenesis in the developing pineal organ but not in the parapineal organ (Masai et al., 1997; Cau and Wilson, 2003; Snelson et al., 2008a). One explanation for the excess cone photoreceptors at the expense of parapineal cells in fgf8ax15 mutants is increased Flh activity. If so, depletion of flh in fgf8ax15 mutants would suppress the parapineal-to-cone fate change. However, we find no change in parapineal cell number in fgf8a mutants with reduced flh activity (Fig. 6A-C; supplementary material Table S6), indicating that the switch from parapineal to cone fate in fgf8ax15 mutants is not a result of increased Flh activity. Importantly, this underscores that the supernumerary cone cells in fgf8ax15 mutants are likely to arise from a population of cells that are not affected by Flh depletion, with the most likely source being the anterior pineal complex precursors.
Fig. 6.
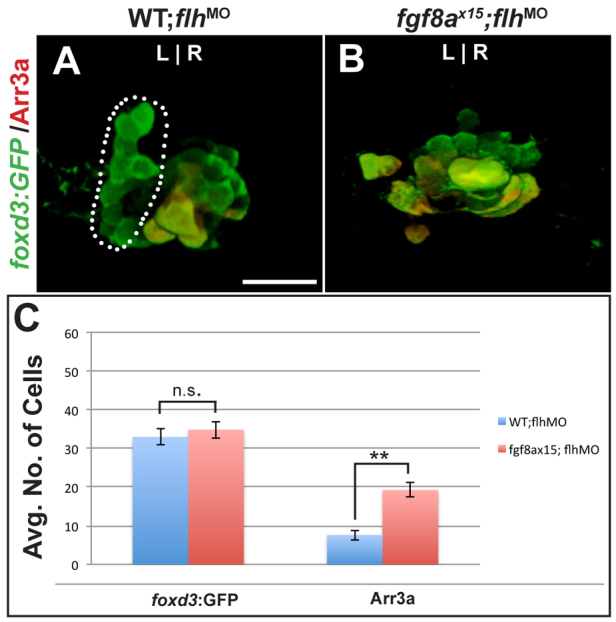
Ectopic cone photoreceptors persist in flh-depleted fgf8ax15 mutants. (A,B) Dorsal views of 96 hpf WT and fgf8ax15 mutant zebrafish embryos injected with flh morpholino (flhMO) labeled with foxd3:GFP (pineal complex) and Arr3a (cone photoreceptors). In WT injected with flhMO, a full-sized parapineal organ (dotted line) is evident, but pineal cells are reduced in number. In flhMO-injected fgf8ax15 mutants, no parapineal organ is visible. (C) The number of foxd3:GFP-expressing cells is unchanged between fgf8ax15;flhMO and WT;flhMO. However, the number of cone photoreceptors increased in fgf8ax15;flhMO relative to WT;flhMO larvae (**P<0.005 by t-test). Error bars represent s.e.m. n.s., not significant. Scale bar: 25 μm.
Fgf8a and Tbx2b act during different steps of parapineal development
Previously, the transcription factor Tbx2b was implicated in parapineal specification (Snelson et al., 2008b). Indeed, tbx2bc144 mutants and fgf8ax15 mutants exhibit a strikingly similar phenotype with reduction in the number of gfi-1.2-expressing cells and failure of the remaining cells to migrate towards the left side of the brain. In chick nasal mesenchyme, Fgf8 can activate the expression of tbx2, an ortholog of tbx2b (Firnberg and Neubüser, 2002). Therefore, we tested whether a similar regulatory relationship exists between Fgf signaling and Tbx2b in parapineal formation. We blocked Fgf signaling by SU5402 treatment from 24 to 30 hpf and examined tbx2b expression at 30 hpf. We see no change in tbx2b transcript level in the epithalamus of SU5402-treated embryos compared with controls (supplementary material Fig. S8A,B). Likewise, we looked at expression of erm, a target gene of Fgf signaling in tbx2bc144 mutants. At 30 hpf, erm expression is similar in tbx2bc144 mutants and WT controls, but different from that seen in fgf8ax15 mutants (supplementary material Fig. S8C-E). These data indicate that in the epithalamus, Fgf8a does not induce tbx2b expression nor is Tbx2b required for Fgf activity.
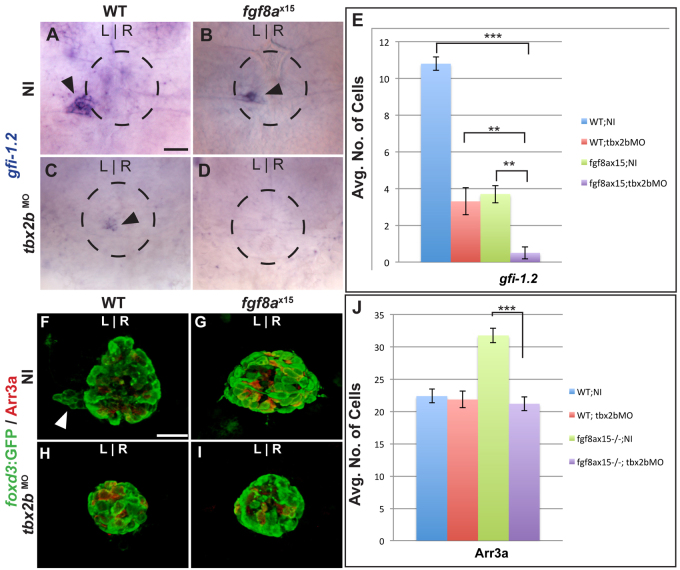
If two genes have a similar mutant phenotype, and the double mutant has an additive phenotype, then the two genes act in independent pathways. To determine whether loss of both Fgf8a and Tbx2b leads to an additive phenotype with respect to parapineal cell number, we analyzed gfi-1.2 expression in larvae lacking the function of both Fgf8a and Tbx2b at 96 hpf. To knock down Tbx2b, we injected a tbx2b morpholino (tbx2bMO) into WT and fgf8ax15 mutants. We find that the larvae of fgf8ax15 mutants injected with tbx2bMO (fgf8ax15;tbx2bMO) had significantly fewer gfi-1.2-expressing cells than either WT;tbx2bMO or NI;fgf8ax15 embryos (Fig. 7D,E; supplementary material Table S2), demonstrating that the loss of both Fgf8a and Tbx2b creates an additive defect with respect to parapineal cell number.
Fig. 7.
Fgf8a and Tbx2b act additively in the formation of parapineal cells, whereas Fgf8a acts downstream of Tbx2b to prevent the formation of cone photoreceptors. (A-D) Dorsal views of uninjected (NI) WT (A), uninjected fgf8ax15 mutant zebrafish larvae (B), WT injected with tbx2bMO (C) and fgf8ax15 mutant embryos injected with tbx2bMO (D) at 96 hpf, labeled for gfi-1.2 to mark parapineal cells (arrowheads). (E) Many fewer parapineal cells are detected when tbx2b and fgf8a are simultaneously reduced than when either is singly depleted (**P<0.005, ***P<0.0005). (F-I) Dorsal views of confocal slices of WT;NI (F), fgf8ax15;NI (G), WT;tbx2bMO (H) and fgf8ax15 ;tbx2bMO (I) larvae labeled with foxd3:GFP to mark the entire pineal complex and Arr3a to mark cone photoreceptors. (J) Tbx2b depletion reduces the number of cone photoreceptors to a level indistinguishable from non-injected WT and tbx2b morphants (***P<0.0005). Error bars represent s.e.m. Scale bars: 25 μm.
We also examined the phenotype resulting from simultaneous depletion of Tbx2b and Fgf8a with respect to cone photoreceptor number. Previous work suggested that although tbx2b mutants had a deficit in gfi-1.2-positive cells, cone photoreceptor number was unchanged (Snelson et al., 2008b). As tbx2b and fgf8a mutants had different phenotypes with respect to cone photoreceptor number, we performed epistasis testing. fgf8ax15;tbx2bMO larvae (21.2±1.0; n=12) had approximately ten fewer cone cells than fgf8ax15 uninjected (fgf8ax15;NI) larvae (31.8±1.3; n=12) and were indistinguishable from WT;NI (22.4±1.1; n=11) and WT; tbx2bMO (21.8±1.1; n=6) larvae (Fig. 7F-J). These data indicate that regarding cone cell number, the depletion of tbx2b is epistatic to mutation of fgf8a. As we do not detect an increase in any differentiated cell type, we speculate that anterior pineal complex cells remain in an undifferentiated state following tbx2b loss of function.
DISCUSSION
We have shown that Fgf signaling is required during a fate decision by a subset of anterior pineal complex cells in order for them to form a left-sided parapineal organ. In fgf8ax15 mutants, there is a significant decrease in the number of parapineal neurons, accompanied by an increase in the number of cone photoreceptors in the pineal organ. In the absence of Fgf8a, anterior pineal complex precursor cells adopt a cone photoreceptor fate. Fgf8a acts in parallel to, but downstream of, Tbx2b to prevent differentiation of anterior pineal complex precursors as cone photoreceptors. Therefore, we propose that cells in the epithalamus are specified by Tbx2b as bipotential anterior pineal complex precursors that can go on to form parapineal cells or cone photoreceptors. They require Fgf signaling, and perhaps Tbx2b, to differentiate as parapineal cells, and might also require Fgf signaling to prevent differentiation as cone cells (Fig. 8).
Fig. 8.
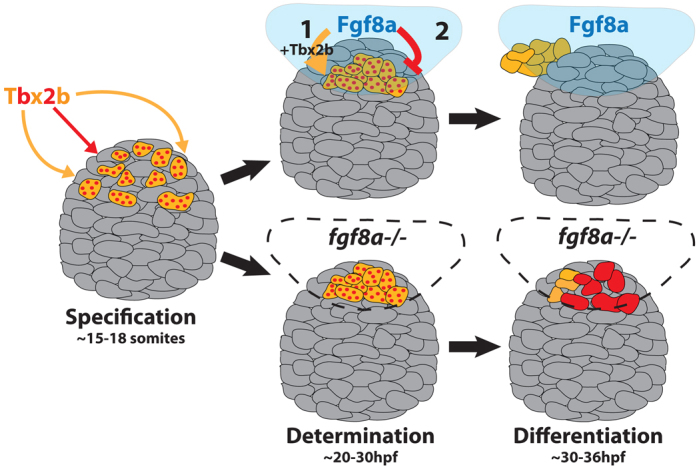
Model: Fgf8a acts on bipotential parapineal/cone cells to promote parapineal fate and/or inhibit cone photoreceptor fate. The transcription factor Tbx2b specifies cells in the anterior region of the pineal complex anlage (gray) as bipotential parapineal/cone precursors (red-dotted orange cells). Fgf8a acts on the bipotential cells (1) in parallel with Tbx2b (orange arrow), to determine them as unipotential parapineal cells (solid orange cells) and (2) by itself (red arrow) to suppress the cone photoreceptor (red cell) differentiation program. In the absence of Fgf8a (lower diagrams), the putative bipotential parapineal/cone precursors lack the proper signal to differentiate as parapineal cells. Consequently, many of the bipotential precursor cells may adopt a cone cell fate instead, resulting in only a few parapineal cells.
There are several ways that Fgf8a could govern cell fate within the anterior pineal complex. One possibility is that the cell fate changes seen in the anterior pineal complex of Fgf-deficient larvae is secondary to perturbed parapineal cell migration. A second possibility is that Fgf acts directly on anterior pineal complex precursors to bias them with parapineal fate. We favor the latter hypothesis for several reasons. First, preventing parapineal cell migration is insufficient to reduce the number of parapineal cells (supplementary material Fig. S4A-C). Also, axin1 and oep mutants often exhibit a significant delay in parapineal migration, but produce no fewer gfi-1.2-expressing cells than WT (Carl et al., 2007; Gamse et al., 2002). Finally, sox1a is expressed specifically in anterior pineal complex precursor cells prior to obvious parapineal cell migration. At 32 hpf, there are far fewer sox1a-expressing cells in fgf8ax15 mutants compared with WT. Therefore, we propose that Fgf has independent roles in parapineal fate differentiation and migration.
Fgf signaling governs a binary fate decision in pineal complex precursors
During embryogenesis, secreted ligands, such as Fgfs, often emanate from organizing centers in the brain (Sato et al., 2004; Iwata and Hevner, 2009; Sansom and Livesey, 2009; Suzuki-Hirano and Shimogori, 2009). Epithalamic Fgf signaling (Fgf8a and Fgf17) could play a similar morphogenic role in specifying pineal complex cell types. Fgf8 knockout mice exhibit severe deficits in all epithalamic cell types, including the pineal organ (Martinez-Ferre and Martinez, 2009), precluding the analysis of how different cell fates are specified; in the zebrafish, the pineal complex is largely intact, allowing us to examine the role of Fgf signaling in the generation of pineal complex subtypes. Our data suggest that Fgf signaling does not act as a morphogen in the pineal complex, i.e. intensity and duration of Fgf signaling does not specify different pineal complex cell types. In fgf8a mutants, the total cell number in the pineal complex number is the same as that in WT; rather, anterior pineal complex precursors change cell fates to become cone photoreceptors. Other pineal complex cell types are unaffected by the loss of Fgf8a. This is a surprising result, as examples of Fgf signaling participating in binary cell fate decisions in the developing nervous system are rare (see Minokawa et al., 2001).
Despite broad expression of the Fgf receptor fgfr4 and the Fgf target gene erm throughout the pineal complex, the anterior pineal complex precursors appear to be the only cells that change fate when Fgf signaling is abrogated. This indicates that the response of specified parapineal precursors to Fgf signaling is regulated by other factor(s). Previously, we found that a small number of cells at the anterior dorsal midline of the pineal complex anlage maintain expression of tbx2b but not flh, a transcription factor required for pineal neurogenesis, and proposed that anterior pineal complex precursors were specified by the combination of tbx2b expression and flh exclusion (Snelson et al., 2008a). The limitation of parapineal competency to tbx2b+, flh– cells agrees with our data that overexpression of Fgf8a is unable to induce supernumerary parapineal neurons.
Fgf signaling is required over a broad period of time to ensure parapineal formation, and parapineal formation is sensitive to the precise level of Fgf signaling. This may explain our inability to rescue parapineal cell specification by overexpression of Fgf8a from a heat-shock inducible transgene. More sophisticated conditional methods will be required to carefully modulate the time and place that Fgf signaling is restored and reveal whether Fgf acts in an instructive or a permissive manner during parapineal development.
Fgf signaling might have multiple roles during parapineal development
Previously, Fgf8a was shown to be required for the migration of parapineal cells (Regan et al., 2009). According to the model of Regan et al., Fgf8a promotes migration of parapineal cells away from the midline and, in the absence of epithalamic Nodal signaling, can provide a directional cue for parapineal migration (Regan et al., 2009; Roussigné et al., 2012). However, the authors do not note any change in parapineal cell number in fgf8a mutants. In the paper by Regan and colleagues, the authors used fgf8ati282, a hypomorphic allele that disrupts splicing of ∼70% of fgf8a transcripts (Draper et al., 2001). Therefore, one possibility is that the levels of Fgf signaling required for parapineal migration and differentiation might not be the same. This is supported by our data. By treating embryos with lower doses of SU5402, we were able to stymie parapineal cell migration without affecting parapineal cell number (supplementary material Fig. S4). The fgf8a mutant allele used in our studies, fgf8ax15, is likely to be a null allele resulting from a premature stop codon, which truncates much of the protein (Kwon and Riley, 2009). According to our data, complete loss of Fgf8a results in severe reductions in mature parapineal cells.
Additionally, Fgf acts during early to mid-somitogenesis to correctly establish the midline of the forebrain through a gene regulatory loop of the sine oculis homeobox homologs six3b and six7 (J. Neugebauer and J. Yost, personal communication). Importantly, this early role of Fgf signaling in midline formation does not directly impact cell fate within anterior pineal complex precursors, as larvae deficient in both Six3b and Six7 still form a parapineal organ (Inbal et al., 2007). Thus, Fgf signaling controls the establishment of left-sided Nodal signaling (Neugebauer, 2012), is required for habenular neurogenesis (Regan et al., 2009), governs differentiation and migration of parapineal neurons (Regan et al., 2009) and might impact formation of the Kupffer’s vesicle, a ciliated structure similar to the node in other vertebrates that governs propagation of left-sided Nodal signaling (Albertson and Yelick, 2005). This clearly demonstrates the importance of Fgf signaling in many facets of epithalamic development.
Comparisons can be made between formation of the parapineal organ and migration of the posterior lateral line primordium (pLL) in zebrafish, as both require Fgf signaling (Regan et al., 2009). The pLL deposits small, epithelized ‘rosettes’ of cells called neuromasts (Ma and Raible, 2009). Disruption of Fgf signaling was shown to perturb formation of neuromast rosettes and migration of the pLL (Aman and Piotrowski, 2008; Lecaudey et al., 2008; Nechiporuk and Raible, 2008). Prior to migration, parapineal cells seem to organize into a similar rosette-like formation in the anterior pineal complex anlage (Roussigné et al., 2009) that might facilitate the cell-cell communication needed for migration. Significantly, Fgf signaling in the pLL also controls cell fate within neuromasts, which eventually differentiate into hair cells and supporting cells (Nechiporuk and Raible, 2008). Abrogating Fgf activity only within the neuromast results in hair cell deficits although supporting cells seem to form normally (Nechiporuk and Raible, 2008). This paradigm could offer interesting parallels to parapineal formation as Fgf signaling could be controlling migration and differentiation of parapineal cells independently. Thus, the parapineal organ could serve as a model for studying growth factor signaling during both cell decisions and collective migration in the brain.
Acknowledgments
We would like to thank Bruce Riley, Paul Hargrove, Monte Westerfield and Vince Tropepe for providing fish and reagents used in this study; Gena Gustin and Qiang Guan for fish care; Erin Booton for creating transgenic fish lines; and Sataree Khuansuwan, Simon Wu, Maureen Gannon and Patrick Blader for help in preparing this manuscript. Confocal data was collected on instruments of the VUMC Cell Imaging Shared Resource.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant HD054534 to J.T.G.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.083709/-/DC1
References
- Albertson R. C., Yelick P. C. (2005). Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 283, 310–321 [DOI] [PubMed] [Google Scholar]
- Aman A., Piotrowski T. (2008). Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev. Cell 15, 749–761 [DOI] [PubMed] [Google Scholar]
- Amo R., Aizawa H., Takahoko M., Kobayashi M., Takahashi R., Aoki T., Okamoto H. (2010). Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. J. Neurosci. 30, 1566–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R., Hama H., Yamamoto-Hino M., Mizuno H., Miyawaki A. (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 12651–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg B., Ekstrom P., Van Veen T. (1983). The parapinela organ of teleosts. Acta Zoolog. 64, 211–218 [Google Scholar]
- Böttcher R. T., Niehrs C. (2005). Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 26, 63–77 [DOI] [PubMed] [Google Scholar]
- Butler A. B., Hodos W. (1996). Comparative Vertebrate Neuroanatomy: Evolution and Adaptation. New York, NY: Wiley-Liss; [Google Scholar]
- Carl M., Bianco I. H., Bajoghli B., Aghaallaei N., Czerny T., Wilson S. W. (2007). Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron 55, 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E., Wilson S. W. (2003). Ash1a and Neurogenin1 function downstream of Floating head to regulate epiphysial neurogenesis. Development 130, 2455–2466 [DOI] [PubMed] [Google Scholar]
- Cau E., Quillien A., Blader P. (2008). Notch resolves mixed neural identities in the zebrafish epiphysis. Development 135, 2391–2401 [DOI] [PubMed] [Google Scholar]
- Clanton J. A., Shestopalov I., Chen J. K., Gamse J. T. (2011). Lineage labeling of zebrafish cells with laser uncagable fluorescein dextran. J. Vis. Exp. 50, e2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay H., Ramakrishnan L. (2005). Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish 2, 105–111 [DOI] [PubMed] [Google Scholar]
- Concha M. L., Wilson S. W. (2001). Asymmetry in the epithalamus of vertebrates. J. Anat. 199, 63–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha M. L., Burdine R. D., Russell C., Schier A. F., Wilson S. W. (2000). A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron 28, 399–409 [DOI] [PubMed] [Google Scholar]
- Concha M. L., Russell C., Regan J. C., Tawk M., Sidi S., Gilmour D. T., Kapsimali M., Sumoy L., Goldstone K., Amaya E., et al. (2003). Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron 39, 423–438 [DOI] [PubMed] [Google Scholar]
- Crossley P. H., Martin G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439–451 [DOI] [PubMed] [Google Scholar]
- Crossley P. H., Martinez S., Martin G. R. (1996). Midbrain development induced by FGF8 in the chick embryo. Nature 380, 66–68 [DOI] [PubMed] [Google Scholar]
- de Schotten M. T., Dell’Acqua F., Forkel S. J., Simmons A., Vergani F., Murphy D. G., Catani M. (2011). A lateralized brain network for visuospatial attention. Nat. Neurosci. 14, 1245–1246 [DOI] [PubMed] [Google Scholar]
- Draper B. W., Morcos P. A., Kimmel C. B. (2001). Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis 30, 154–156 [DOI] [PubMed] [Google Scholar]
- Dufourcq P., Rastegar S., Strähle U., Blader P. (2004). Parapineal specific expression of gfi1 in the zebrafish epithalamus. Gene Expr. Patterns 4, 53–57 [DOI] [PubMed] [Google Scholar]
- Echevarría D., Vieira C., Gimeno L., Martínez S. (2003). Neuroepithelial secondary organizers and cell fate specification in the developing brain. Brain Res. Brain Res. Rev. 43, 179–191 [DOI] [PubMed] [Google Scholar]
- Firnberg N., Neubüser A. (2002). FGF signaling regulates expression of Tbx2, Erm, Pea3, and Pax3 in the early nasal region. Dev. Biol. 247, 237–250 [DOI] [PubMed] [Google Scholar]
- Gamse J. T., Shen Y.-C., Thisse C., Thisse B., Raymond P. A., Halpern M. E., Liang J. O. (2002). Otx5 regulates genes that show circadian expression in the zebrafish pineal complex. Nat. Genet. 30, 117–121 [DOI] [PubMed] [Google Scholar]
- Gamse J. T., Thisse C., Thisse B., Halpern M. E. (2003). The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development 130, 1059–1068 [DOI] [PubMed] [Google Scholar]
- Geschwind N., Levitsky W. (1968). Human brain: left-right asymmetries in temporal speech region. Science 161, 186–187 [DOI] [PubMed] [Google Scholar]
- Gilmour D. T., Maischein H. M., Nüsslein-Volhard C. (2002). Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron 34, 577–588 [DOI] [PubMed] [Google Scholar]
- Hans S., Christison J., Liu D., Westerfield M. (2007). Fgf-dependent otic induction requires competence provided by Foxi1 and Dlx3b. BMC Dev. Biol. 7, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ile K. E., Kassen S., Cao C., Vihtehlic T., Shah S. D., Mousley C. J., Alb J. G., Jr, Huijbregts R. P., Stearns G. W., Brockerhoff S. E., et al. (2010). Zebrafish class 1 phosphatidylinositol transfer proteins: PITPbeta and double cone cell outer segment integrity in retina. Traffic 11, 1151–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal A., Kim S. H., Shin J., Solnica-Krezel L. (2007). Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron 55, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N. (2007). The Fgf families in humans, mice, and zebrafish: their evolutional processes and roles in development, metabolism, and disease. Biol. Pharm. Bull. 30, 1819–1825 [DOI] [PubMed] [Google Scholar]
- Iwata T., Hevner R. F. (2009). Fibroblast growth factor signaling in development of the cerebral cortex. Dev. Growth Differ. 51, 299–323 [DOI] [PubMed] [Google Scholar]
- Jovelin R., He X., Amores A., Yan Y. L., Shi R., Qin B., Roe B., Cresko W. A., Postlethwait J. H. (2007). Duplication and divergence of fgf8 functions in teleost development and evolution. J. Exp. Zool. B Mol. Dev. Evol. 308, 730–743 [DOI] [PubMed] [Google Scholar]
- Kohl M. M., Shipton O. A., Deacon R. M., Rawlins J. N., Deisseroth K., Paulsen O. (2011). Hemisphere-specific optogenetic stimulation reveals left-right asymmetry of hippocampal plasticity. Nat. Neurosci. 14, 1413–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H. J., Riley B. B. (2009). Mesendodermal signals required for otic induction: Bmp-antagonists cooperate with Fgf and can facilitate formation of ectopic otic tissue. Dev. Dyn. 238, 1582–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larison K. D., Bremiller R. (1990). Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 109, 567–576 [DOI] [PubMed] [Google Scholar]
- Lecaudey V., Cakan-Akdogan G., Norton W. H. J., Gilmour D. (2008). Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development 135, 2695–2705 [DOI] [PubMed] [Google Scholar]
- Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., Court D. L., Jenkins N. A., Copeland N. G. (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65 [DOI] [PubMed] [Google Scholar]
- Ma E. Y., Raible D. W. (2009). Signaling pathways regulating zebrafish lateral line development. Curr. Biol. 19, R381–R386 [DOI] [PubMed] [Google Scholar]
- Martinez-Ferre A., Martinez S. (2009). The development of the thalamic motor learning area is regulated by Fgf8 expression. J. Neurosci. 29, 13389–13400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I., Heisenberg C. P., Barth K. A., Macdonald R., Adamek S., Wilson S. W. (1997). floating head and masterblind regulate neuronal patterning in the roof of the forebrain. Neuron 18, 43–57 [DOI] [PubMed] [Google Scholar]
- Minokawa T., Yagi K., Makabe K. W., Nishida H. (2001). Binary specification of nerve cord and notochord cell fates in ascidian embryos. Development 128, 2007–2017 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997). Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Sato T., Suzuki-Hirano A. (2008). Isthmus organizer for mesencephalon and metencephalon. Dev. Growth Differ. 50, S113–S118 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A., Raible D. W. (2008). FGF-dependent mechanosensory organ patterning in zebrafish. Science 320, 1774–1777 [DOI] [PubMed] [Google Scholar]
- Neugebauer J. Y. J. (2012). FGF signaling maintains a beta-catenin-demarked midline for normal brain asymmetry. Neuron (in press). [Google Scholar]
- Nowicka A., Tacikowski P. (2011). Transcallosal transfer of information and functional asymmetry of the human brain. Laterality 16, 35–74 [DOI] [PubMed] [Google Scholar]
- Ota S., Tonou-Fujimori N., Tonou-Fujimori N., Nakayama Y., Ito Y., Kawamura A., Yamasu K. (2010). FGF receptor gene expression and its regulation by FGF signaling during early zebrafish development. Genesis 48, 707–716 [DOI] [PubMed] [Google Scholar]
- Quillien A., Blanco-Sanchez B., Halluin C., Moore J. C., Lawson N. D., Blader P., Cau E. (2011). BMP signaling orchestrates photoreceptor specification in the zebrafish pineal gland in collaboration with Notch. Development 138, 2293–2302 [DOI] [PubMed] [Google Scholar]
- Regan J. C., Concha M. L., Roussigné M., Russell C., Wilson S. W. (2009). An Fgf8-dependent bistable cell migratory event establishes CNS asymmetry. Neuron 61, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y., Brand M. (1998). Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395 [DOI] [PubMed] [Google Scholar]
- Reifers F., Adams J., Mason I. J., Schulte-Merker S., Brand M. (2000). Overlapping and distinct functions provided by fgf17, a new zebrafish member of the Fgf8/17/18 subgroup of Fgfs. Mech. Dev. 99, 39–49 [DOI] [PubMed] [Google Scholar]
- Roehl H., Nusslein-Volhard C. (2001). Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr. Biol. 11, 503–507 [DOI] [PubMed] [Google Scholar]
- Rogers L. J. (2000). Evolution of hemispheric specialization: advantages and disadvantages. Brain Lang. 73, 236–253 [DOI] [PubMed] [Google Scholar]
- Roussigné M., Bianco I. H., Wilson S. W., Blader P. (2009). Nodal signalling imposes left-right asymmetry upon neurogenesis in the habenular nuclei. Development 136, 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigné M., Blader P., Wilson S. W. (2012). The zebrafish epithalamus clears a path through the complexity of brain lateralization. Dev. Neurobiol. 72, 269–281 [DOI] [PubMed] [Google Scholar]
- Sansom S. N., Livesey F. J. (2009). Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harb. Perspect. Biol. 1, a002519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Joyner A. L., Nakamura H. (2004). How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev. Growth Differ. 46, 487–494 [DOI] [PubMed] [Google Scholar]
- Shin J., Park H. C., Topczewska J. M., Mawdsley D. J., Appel B. (2003). Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Science 25, 7–14 [DOI] [PubMed] [Google Scholar]
- Snelson C. D., Burkart J. T., Gamse J. T. (2008a). Formation of the asymmetric pineal complex in zebrafish requires two independently acting transcription factors. Dev. Dyn. 237, 3538–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelson C. D., Santhakumar K., Halpern M. E., Gamse J. T. (2008b). Tbx2b is required for the development of the parapineal organ. Development 135, 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Hirano A., Shimogori T. (2009). The role of Fgf8 in telencephalic and diencephalic patterning. Semin. Cell Dev. Biol. 20, 719–725 [DOI] [PubMed] [Google Scholar]
- Taylor R. W., Hsieh Y. W., Gamse J. T., Chuang C. F. (2010). Making a difference together: reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development 137, 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- Walker C. (1999). Haploid screens and gamma-ray mutagenesis. Methods Cell Biol. 60, 43–70 [DOI] [PubMed] [Google Scholar]
- Yan Y. L., Talbot W. S., Egan E. S., Postlethwait J. H. (1998). Mutant rescue by BAC clone injection in zebrafish. Genomics 50, 287–289 [DOI] [PubMed] [Google Scholar]