Abstract
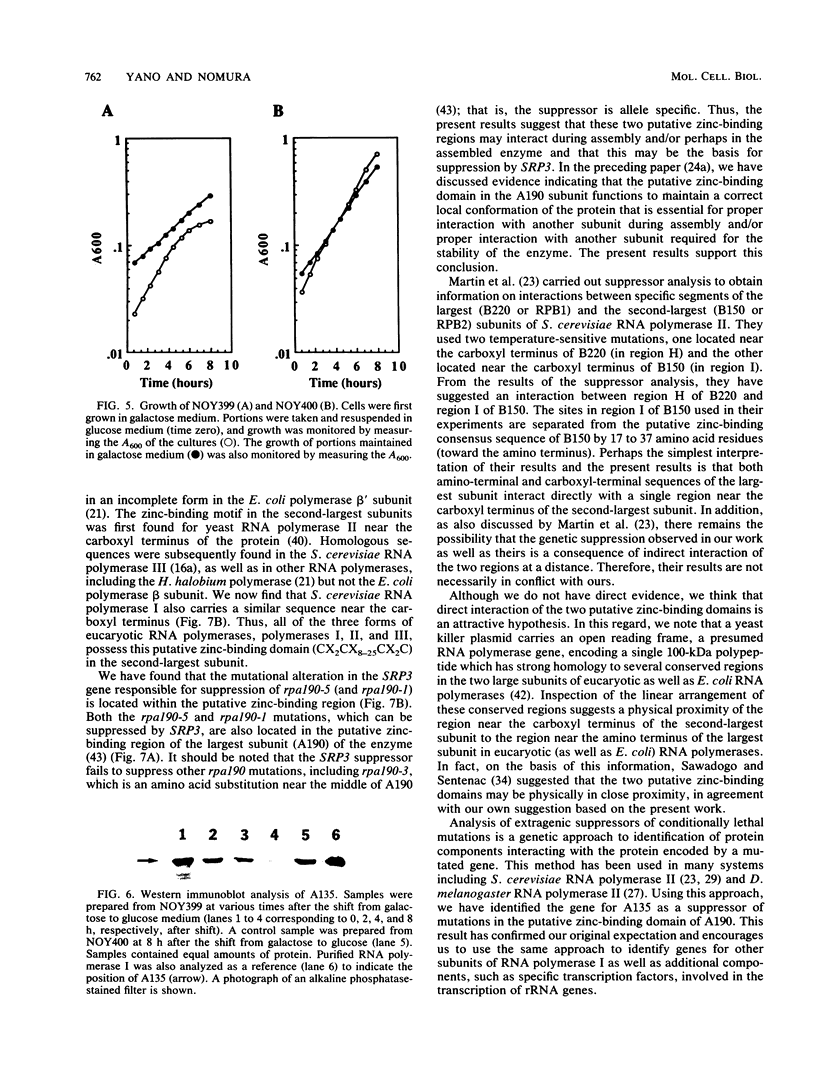
The SRP3-1 mutation is an allele-specific suppressor of temperature-sensitive mutations in the largest subunit (A190) of RNA polymerase I from Saccharomyces cerevisiae. Two mutations known to be suppressed by SRP3-1 are in the putative zinc-binding domain of A190. We have cloned the SRP3 gene by using its suppressor activity and determined its complete nucleotide sequence. We conclude from the following evidence that the SRP3 gene encodes the second-largest subunit (A135) of RNA polymerase I. First, the deduced amino acid sequence of the gene product contains several regions with high homology to the corresponding regions of the second-largest subunits of RNA polymerases of various origins, including those of RNA polymerase II and III from S. cerevisiae. Second, the deduced amino acid sequence contains known amino acid sequences of two tryptic peptides from the A135 subunit of RNA polymerase I purified from S. cerevisiae. Finally, a strain was constructed in which transcription of the SRP3 gene was controlled by the inducible GAL7 promoter. When this strain, which can grow on galactose but not on glucose, was shifted from galactose medium to glucose medium, a large decrease in the cellular concentration of A135 was observed by Western blot analysis. We have also identified the specific amino acid alteration responsible for suppression by SRP3-1 and found that it is located within the putative zinc-binding domain conserved among the second-largest subunits of eucaryotic RNA polymerases. From these results, it is suggested that this putative zinc-binding domain is in physical proximity to and interacts with the putative zinc-binding domain of the A190 subunit.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Auld D. S., Atsuya I. Yeast RNA polymerase I: a eukaryotic zinc metalloenzyme. Biochem Biophys Res Commun. 1976 Mar 22;69(2):548–554. doi: 10.1016/0006-291x(76)90555-6. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Harashima S., Takagi A., Oshima Y. Transformation of protoplasted yeast cells is directly associated with cell fusion. Mol Cell Biol. 1984 Apr;4(4):771–778. doi: 10.1128/mcb.4.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Kikuchi Y., Nogi Y., Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet. 1983;191(1):31–38. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- Hirano T., Konoha G., Toda T., Yanagida M. Essential roles of the RNA polymerase I largest subunit and DNA topoisomerases in the formation of fission yeast nucleolus. J Cell Biol. 1989 Feb;108(2):243–253. doi: 10.1083/jcb.108.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokerst R. S., Weeks J. R., Zehring W. A., Greenleaf A. L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989 Jan;215(2):266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- Kontermann R., Sitzler S., Seifarth W., Petersen G., Bautz E. K. Primary structure and functional aspects of the gene coding for the second-largest subunit of RNA polymerase III of Drosophila. Mol Gen Genet. 1989 Nov;219(3):373–380. doi: 10.1007/BF00259609. [DOI] [PubMed] [Google Scholar]
- Leffers H., Gropp F., Lottspeich F., Zillig W., Garrett R. A. Sequence, organization, transcription and evolution of RNA polymerase subunit genes from the archaebacterial extreme halophiles Halobacterium halobium and Halococcus morrhuae. J Mol Biol. 1989 Mar 5;206(1):1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- Martin C., Okamura S., Young R. Genetic exploration of interactive domains in RNA polymerase II subunits. Mol Cell Biol. 1990 May;10(5):1908–1914. doi: 10.1128/mcb.10.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990 May 25;18(10):2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker J. H., Yamagishi M., Kolb J. M., Nomura M. Suppressor analysis of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae: suppression of mutations in a zinc-binding motif by transposed mutant genes. Mol Cell Biol. 1991 Feb;11(2):746–753. doi: 10.1128/mcb.11.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortin M. A. Use of second-site suppressor mutations in Drosophila to identify components of the transcriptional machinery. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4864–4868. doi: 10.1073/pnas.87.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Mémet S., Saurin W., Sentenac A. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J Biol Chem. 1988 Jul 25;263(21):10048–10051. [PubMed] [Google Scholar]
- Nogi Y., Fukasawa T. Functional domains of a negative regulatory protein, GAL80, of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jul;9(7):3009–3017. doi: 10.1128/mcb.9.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Young R. A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989 Dec;123(4):715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M., Schäffner A. R., Sentenac A., Hartmann G. R., Mustaev A. A., Zaychikov E. F., Grachev M. A. Active site labeling of the RNA polymerases A, B, and C from yeast. J Biol Chem. 1987 Oct 25;262(30):14377–14380. [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Levin J. R., Agabian N. Molecular characterization of the Trypanosoma brucei RNA polymerase I and III largest subunit genes. J Biol Chem. 1989 Oct 25;264(30):18091–18099. [PubMed] [Google Scholar]
- Smith J. L., Levin J. R., Ingles C. J., Agabian N. In trypanosomes the homolog of the largest subunit of RNA polymerase II is encoded by two genes and has a highly unusual C-terminal domain structure. Cell. 1989 Mar 10;56(5):815–827. doi: 10.1016/0092-8674(89)90686-7. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. W., Meacock P. A. Extranuclear gene expression in yeast: evidence for a plasmid-encoded RNA polymerase of unique structure. Nucleic Acids Res. 1988 Aug 25;16(16):8097–8112. [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Dodd J., Vu L., Kolb J. M., Buhler J. M., Sentenac A., Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol Cell Biol. 1988 Oct;8(10):3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M., Kolb J. M., Dodd J., Yamagishi M., Mémet S., Buhler J. M., Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol Cell Biol. 1990 May;10(5):2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. RNA polymerase II: subunit structure and function. Trends Biochem Sci. 1990 Sep;15(9):347–351. doi: 10.1016/0968-0004(90)90074-l. [DOI] [PubMed] [Google Scholar]
- Yamagishi M., Nomura M. Cloning and sequence determination of the gene encoding the largest subunit of the fission yeast Schizosaccharomyces pombe RNA polymerase I. Gene. 1988 Dec 30;74(2):503–515. doi: 10.1016/0378-1119(88)90183-7. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]