Abstract
The URE2 gene of Saccharomyces cerevisiae has been cloned and sequenced. It encodes a predicted polypeptide of 354 amino acids with a molecular weight of 40,226. Deletion of the first 63 amino acids does not have any effect on the function of the protein. Studies with disruption alleles of the URE2 and GLN3 genes showed that both genes regulate GLN1 and GDH2, the structural genes for glutamine synthetase and NAD-linked glutamate dehydrogenase, respectively, at the transcriptional level, but expression of the regulatory genes does not appear to be regulated. Active URE2 gene product was required for the inactivation of glutamine synthetase upon addition of glutamine to cells growing with glutamate as the source of nitrogen. The predicted URE2 gene product has homology to glutathione S-transferases. The gene has been mapped to chromosome XIV, 5.9 map units from petX and 3.4 map units from kex2.
Full text
PDF


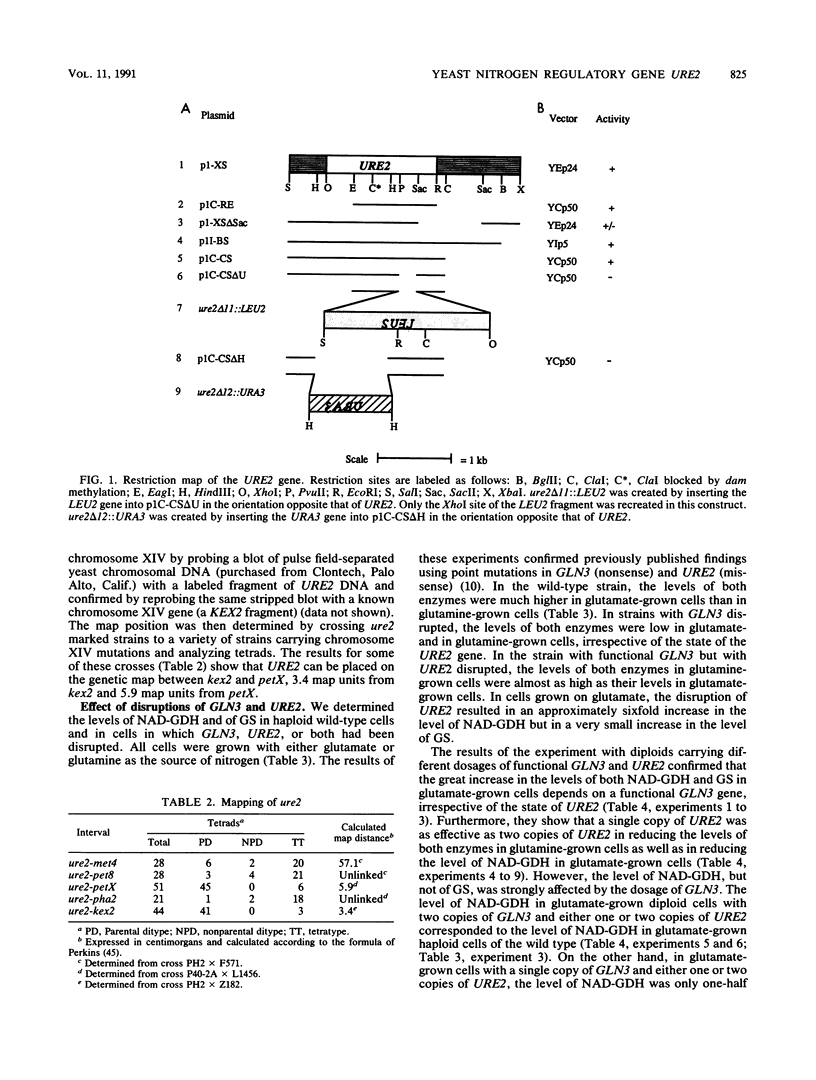







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benjamin P. M., Wu J. I., Mitchell A. P., Magasanik B. Three regulatory systems control expression of glutamine synthetase in Saccharomyces cerevisiae at the level of transcription. Mol Gen Genet. 1989 Jun;217(2-3):370–377. doi: 10.1007/BF02464906. [DOI] [PubMed] [Google Scholar]
- Board P. G., Webb G. C. Isolation of a cDNA clone and localization of human glutathione S-transferase 2 genes to chromosome band 6p12. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2377–2381. doi: 10.1073/pnas.84.8.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T. G., Ferguson D., Rai R., Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol. 1990 Feb;172(2):1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne W. E., Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988 Feb;170(2):708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillien R., Lacroute F. Ureidosuccinic acid uptake in yeast and some aspects of its regulation. J Bacteriol. 1972 Jan;109(1):203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol. 1988 Feb;8(2):647–654. doi: 10.1128/mcb.8.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 May;7(5):1691–1696. doi: 10.1128/mcb.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson M., Dubois E., Piotrowska M., Drillien R., Aigle M. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Evidence for the gdhA locus being a structural gene for the NADP-dependent glutamate dehydrogenase. Mol Gen Genet. 1974;128(1):73–85. doi: 10.1007/BF00267295. [DOI] [PubMed] [Google Scholar]
- Grenson M. The utilization of exogenous pyrimidines and the recycling of uridine-5'-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur J Biochem. 1969 Dec;11(2):249–260. doi: 10.1111/j.1432-1033.1969.tb00767.x. [DOI] [PubMed] [Google Scholar]
- Grove G., Zarlengo R. P., Timmerman K. P., Li N. Q., Tam M. F., Tu C. P. Characterization and heterospecific expression of cDNA clones of genes in the maize GSH S-transferase multigene family. Nucleic Acids Res. 1988 Jan 25;16(2):425–438. doi: 10.1093/nar/16.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Roche S. D., Leisinger T. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J Bacteriol. 1990 Jan;172(1):164–171. doi: 10.1128/jb.172.1.164-171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai H. C., Li N., Weiss M. J., Reddy C. C., Tu C. P. The nucleotide sequence of a rat liver glutathione S-transferase subunit cDNA clone. J Biol Chem. 1984 May 10;259(9):5536–5542. [PubMed] [Google Scholar]
- Legrain C., Vissers S., Dubois E., Legrain M., Wiame J. M. Regulation of glutamine synthetase from Saccharomyces cerevisiae by repression, inactivation and proteolysis. Eur J Biochem. 1982 Apr;123(3):611–616. doi: 10.1111/j.1432-1033.1982.tb06576.x. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Magasanik B. Reversible phosphorylation of an enhancer binding protein regulates the transcription of bacterial nitrogen utilization genes. Trends Biochem Sci. 1988 Dec;13(12):475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- Martin C., Young R. A. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989 Jun;9(6):2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990 Sep;172(9):4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Ludmerer S. W. Identification of a glutaminyl-tRNA synthetase mutation Saccharomyces cerevisiae. J Bacteriol. 1984 May;158(2):530–534. doi: 10.1128/jb.158.2.530-534.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Biochemical and physiological aspects of glutamine synthetase inactivation in Saccharomyces cerevisiae. J Biol Chem. 1984 Oct 10;259(19):12054–12062. [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Purification and properties of glutamine synthetase from Saccharomyces cerevisiae. J Biol Chem. 1983 Jan 10;258(1):119–124. [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2758–2766. doi: 10.1128/mcb.4.12.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P., Magasanik B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2767–2773. doi: 10.1128/mcb.4.12.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. P. The GLN1 locus of Saccharomyces cerevisiae encodes glutamine synthetase. Genetics. 1985 Oct;111(2):243–258. doi: 10.1093/genetics/111.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949 Sep;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toung Y. P., Hsieh T. S., Tu C. P. Drosophila glutathione S-transferase 1-1 shares a region of sequence homology with the maize glutathione S-transferase III. Proc Natl Acad Sci U S A. 1990 Jan;87(1):31–35. doi: 10.1073/pnas.87.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. L., Jarai G., Fu Y. H., Marzluf G. A. Nucleotide sequence and analysis of NMR, a negative-acting regulatory gene in the nitrogen circuit of Neurospora crassa. Mol Gen Genet. 1990 Jun;222(1):120–128. doi: 10.1007/BF00283032. [DOI] [PubMed] [Google Scholar]




