Abstract
MicroRNAs (miRNAs) are small RNA molecules that regulate gene expression. They are characterized by specific maturation processes defined by canonical and non-canonical biogenic pathways. Analysis of ∼0.5 billion sequences from mouse data sets derived from different tissues, developmental stages and cell types, partly characterized by either ablation or mutation of the main proteins belonging to miRNA processor complexes, reveals 66 high-confidence new genomic loci coding for miRNAs that could be processed in a canonical or non-canonical manner. A proportion of the newly discovered miRNAs comprises mirtrons, for which we define a new sub-class. Notably, some of these newly discovered miRNAs are generated from untranslated and open reading frames of coding genes, and we experimentally validate these. We also show that many annotated miRNAs do not present miRNA-like features, as they are neither processed by known processing complexes nor loaded on AGO2; this indicates that the current miRNA miRBase database list should be refined and re-defined. Accordingly, a group of them map on ribosomal RNA molecules, whereas others cannot undergo genuine miRNA biogenesis. Notably, a group of annotated miRNAs are Dgcr8 independent and DICER dependent endogenous small interfering RNAs that derive from a unique hairpin formed from a short interspersed nuclear element.
INTRODUCTION
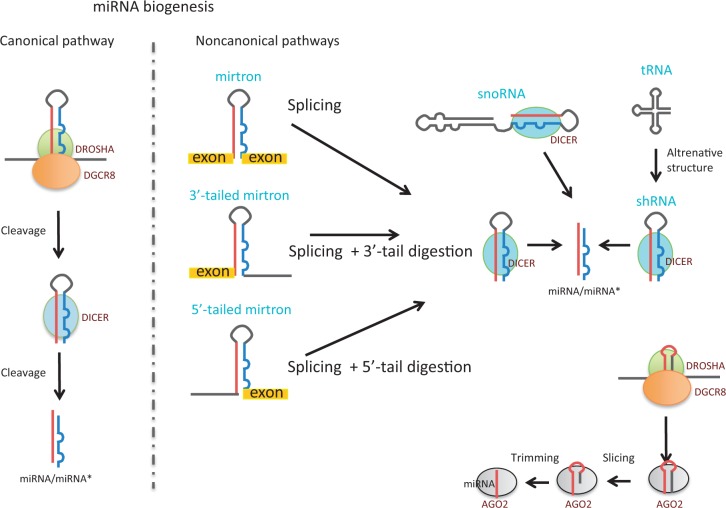
MicroRNAs (miRNAs) are small RNA molecules that range from 21 to 25 nucleotides (nt) in length, capable of negatively regulating gene expression. They usually mediate their action by base pairing with the 3′ untranslated region (3′-UTR) of messenger RNA (mRNA) targets (1). The majority of miRNAs are transcribed as a long primary transcript (pri-miRNA) that undergoes a canonical pathway of biogenesis characterized by a dual processing event (Figure 1). The first cleavage is carried out by the RNAse III, DROSHA and its partner Dgcr8 (called microprocessor complex) in the nucleus (2–4). This cut converts the pri-miRNA into a ∼70-nt hairpin-loop precursor miRNA (pre-miRNA), leaving a 5′ phosphate and 2-nt 3′ overhang (2,3). The second cleavage occurs in the cytoplasm and is carried out by the RNAse III enzyme DICER, which cuts out the loop converting the pre-miRNA into miRNA/miRNA* duplex, ∼22 nt in length (Figure 1). This cleavage again leaves a 2-nt 3′ overhang (4). After maturation, one of the two strands from the duplex is predominantly loaded onto an miRNA-induced silencing complex (miRISC), composed of Argonaute (Ago) proteins, producing the effector complex. Recently, a number of alternative mechanisms of miRNA biogenesis, so-called non-canonical pathways, have been characterized and include both DROSHA-independent and DICER independent processes (Figure 1) (5).
Figure 1.
Schematic representation for the known biogenic pathway of miRNA processing and maturation.
Mirtrons are short hairpin introns that are spliced and debranched from mRNA transcripts directly forming a pre-mirna DICER substrate, escaping DROSHA–Dgcr8 processing (Figure 1) (6–8). Furthermore a sub-class of ‘tailed mirtrons’ contains only one end of the pre-mirna formed directly by splicing, but its maturation is still DROSHA independent and DICER dependent (Figure 1). It is thought that whereas Drosophila expresses only 3′-tailed mirtrons (the tail is removed by the exosome) (9), vertebrates produce only 5′-tailed mirtrons, but the nuclease involved in the removal of their tail has not been defined yet (5). However, two articles recently reported the expression of 3′-tailed mirtrons in mammals (10,11). In addition, miRNAs can be directly transcribed as endogenous short hairpin RNAs (shRNAs) (12) or derived from both C/D and H/ACA C/D box and H/ACA box small nucleolar RNA (snoRNA) types that comprise additional DROSHA independent DICER dependent sub-classes (Figure 1) (5,13).
Another class of small RNAs generated by DICER, independently of DROSHA, is the endogenous small interfering RNAs (endo-siRNAs). They are generated by a sequential DICER cleavage of long double-stranded RNA molecules. Although they have been described in various organisms and also in mouse oocytes and mouse embryonic stem cells (mESCs) (12,14,15), they remain uncharacterized in other mammalian tissues, with many doubting they exist in these cell types.
It has been recently shown that miRNA processing could also be independent of DICER but mediated by Argonaute 2 (AGO2). miR-451 is processed by DROSHA in the nucleus, producing an unusually small pre-mir-451, which is then loaded and directly matured by AGO2 (Figure 1) (16–18).
In this study, we performed an examination of deep sequencing of small RNA sequences from published data sets, partly derived from mouse cells, in which the main effectors of miRNA biogenesis pathways have been mutated or completely removed. We mapped reads derived from these data sets on miRNAs elucidated after the examined studies had been published. Using this approach, we determined that a group of annotated canonical miRNAs is processed through a non-canonical pathway. We also showed that many currently annotated miRNAs do not present miRNA-like features, as they are neither processed by any of the known processing complexes, nor are they loaded onto AGO2, indicating that the current miRNA list in the miRBase database (www.miRBase.org) (19) should be refined and re-defined.
Using newly developed highly efficient algorithms of miRNA predictions (20), plus genomic inspection of the small sequence reads, we discovered novel canonical and non-canonical miRNAs, comprising miRNAs derived from coding and untranslated regions of protein-coding genes in mammals [previously only described in Drosophila (21)]. Finally, we found that previously annotated miRNAs located on short interspersed nuclear elements (SINEs) originated from a unique long hairpin RNA structure processed by DICER to produce endo-siRNAs in somatic tissues (as per our flowchart in Supplementary Figure S1).
MATERIALS AND METHODS
Data sets and pre-processing
Small RNA reads used in this study were downloaded from the NCBI Gene Expression Omnibus (GEO) and the Sequence Read Archive (SRA). The accession numbers of all data sets analyzed are summarized in the Supplementary Table S1. When raw sequences were available, the 3′ adaptors were clipped out with the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) before further analysis. GEO format files were transformed to multi-Fasta files using the auxiliary scripts available from the miRDeep2 package (20).
Read mapping and small RNA quantification
Pre-processed reads were mapped to the University of California at Santa Cruz mm9 genome assembly using Bowtie version 0.12.8, allowing for zero mismatches. All reads that mapped perfectly to the genome at <500 loci were considered for further analysis. Genome inspection was performed visualizing the mapped reads on the University of California at Santa Cruz genome browser (22). Reads were mapped on known miRNAs using the quantifier script from the miRDeep2 package (20). miRNA sequences were downloaded from miRBase release 18, November 2011. Quantification of known miRNAs between various mutant backgrounds was performed as described (12).
miRNA discovery and miRNA expression profiles
To discover novel miRNAs, the miRDeep2 algorithm was used (20) maintaining default settings and filtering reads by size ≤17 nt. Among the new miRNAs discovered using this approach, high-confidence miRNAs containing both mature and star sequences complementary with 2-nt 3′ overhang detected in multiple samples were considered. Genomic inspection of the novel miRNAs was performed directly using Blat links located on the html output files, obtained at the end of the mirRDeep2 runs. Newly discovered miRNAs were then quantified among the various tissue samples using the quantifier module (20). Obtained normalized reads were used to build an intensity plot with Partek Genomic Suite (Partek Incorporated, USA). Secondary RNA structures of the precursors were either directly obtained using miRDeep2, which uses RNAfold by default (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), or manually assembled using mfold (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
Evaluation of miRNAs located on genomic repeats
To evaluate whether annotated miRNAs are located on genomic repeats and to discover the nature of these repeats, downloaded miRNA precursors from the miRBase release 18 were analyzed using the RepeatMasker script, version 3.2.8 (http://www.repeatmasker.org/).
miRNA overexpression, luciferase reporter assays and small RNA deep sequencing
Genomic regions containing predicted miRNA hairpins were amplified by PCR and cloned into pcDNA 3.3 TOPO TA vectors (Life Technologies, Paisley, UK). A pool of eight hairpins, verified to be mutation free by sequencing, or the same amount of pcDNA empty vector was transfected in HEK293T cells with lipofectamine 2000 (Life Technologies). Deep sequencing of small RNAs then verified that they were processed to miRNAs. Libraries of small RNAs were prepared using the True-seq small RNA preparation kit (Illumina, Essex, UK), according to the manufacturer’s instructions. Pre-processing, quantification and normalization of the reads were performed using auxiliary scripts from the miRDeep2 package (n = 3). Sequencing was performed using a Hiseq 2000 instrument (Illumina). To verify that one of the novel miRNAs we describe induces gene silencing, six complementary sites of the putative miRNA were cloned in the 3′-UTR of the firefly luciferase gene of the pMIR-REPORT vector (Life Technologies). This construct was then co-expressed with either the hairpin containing the putative miRNA and the pRLTK Renilla luciferase vector (Promega, Southampton, UK) or the pcDNA vector control and the pRLTK Renilla vector, and luciferase expression was measured. This experiment was performed in triplicate on three independent occasions.
RESULTS
Re-analysis of known miRNAs to identify additional Dgcr8 independent or DICER independent miRNAs
Babiarz et al. (12,23) sequenced small RNAs derived from wild-type (WT), Dgcr8−/− and DICER−/− mESCs, as well as mouse hippocampus and cortex tissues. Using this approach, they identified mouse mirtrons, endogenous miRNA producing shRNAs and new snoRNAs with miRNA-like features (12,23). The newly discovered miRNAs from mESCs were annotated on miRBase release 13. Considering that in miRBase release 13, there were 547 annotated miRNAs, whereas the new miRBase release 18 contains 741 miRNAs, the biogenesis of 193 miRNAs could potentially be re-evaluated using sequencing reads derived from these experiments and miRNAs downloaded from the current version of the miRBase as a substrate for analysis with mESCs.
To discover new canonical and non-canonical miRNAs, we re-analyzed these sequencing data, mapping the reads onto miRNAs derived from the miRBase annotation, release 18, and on the mm9 mouse genome assembly, considering only perfect mapping for further analysis. As previously described (12), normalized reads that decreased <2-fold in either mutant background were considered as Dgcr8 or DICER independent. We also included reads derived from photo–cross-linking immunoprecipitation (CLIP) of AGO2 followed by deep sequencing (CLIP-seq) in WT and DICER−/− mESCs to evaluate possible AGO2 loading (24). This approach indicated that 23 miRNAs were Dgcr8 independent, but DICER dependent (Supplementary Table S2 and Figure 2A), 16 more than the ones described in the original study (12), confirming our hypothesis that using these methods, we can discover non-canonical miRNAs. Nevertheless, some of those have already been re-classified in an experimental evaluation of novel and annotated miRNAs (25). In Table 1, we listed the Dgcr8 independent and DICER dependent miRNAs that we retrieved with this analysis and summarized a previous classification of some of the Dgcr8 independent miRNAs that we recovered (8,12,13,25,26).
Figure 2.
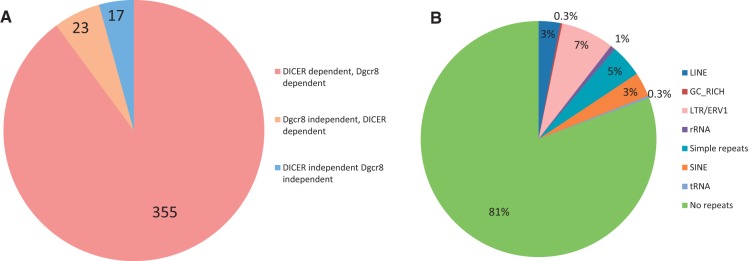
A fraction of the annotated miRNAs is Dgcr8 independent DICER dependent or Dgcr8 independent DICER independent. (A) Number of canonical miRNAs (Dgcr8 dependent, DICER dependent), Dgcr8 independent miRNAs and DICER independent miRNAs from the miRBase release 18, expressed in mESCs. (B) Classification of genomic repeats containing miRNAs using RepeatMasker on precursors downloaded from the miRBase release 18.
Table 1.
Classification according to previous literature of Dgcr8 independent DICER dependent miRNAs retrieved by this study in mESCs
| miRNA | miRNA biogenesis mechanism | Description by Berezikov et al. 2007 | Description by Babiarz et al. 2008 | Description by Chiang et al. 2010 | Description by Ender et al. 2008 | Description by Westholm et al. 2012 |
|---|---|---|---|---|---|---|
| miR-1186 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1195 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1196 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1839 | DICER dependent | ACA45 snoRNA | ||||
| Dgcr8 independent | ||||||
| miR-1843 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1843b | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1935 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1965 | DICER dependent | |||||
| Dgcr8 independent | ||||||
| miR-1981 | DICER dependent | Mirtron | ||||
| Dgcr8 independent | ||||||
| miR-1982 | DICER dependent | 5′-tailed mirtron | ||||
| Dgcr8 independent | ||||||
| miR-1983 | DICER dependent | tRNA-shRNA | ||||
| Dgcr8 independent | ||||||
| miR-3062 | DICER dependent | Canonical | ||||
| Dgcr8 independent | ||||||
| miR-3068 | DICER dependent | Canonical | ||||
| Dgcr8 independent | ||||||
| miR-3084 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-3102 | DICER dependent | Sequentially diced mirtron | ||||
| Dgcr8 independent | ||||||
| miR-320 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-344 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-484 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-5132 | DICER dependent | Mirtron | ||||
| Dgcr8 independent | ||||||
| miR-664 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-668 | DICER dependent | shRNA | ||||
| Dgcr8 independent | ||||||
| miR-702 | DICER dependent | Mirtron | ||||
| Dgcr8 independent | ||||||
| miR-877 | DICER dependent | Mirtron | ||||
| Dgcr8 independent |
Interestingly, we could also appreciate that at least 17 of the annotated miRNAs appeared independent of DICER activity (Figure 2A and Supplementary Table S2). Considering thus far that only miR-451 has been described as a DICER independent miRNA, we proceeded to investigate the possibility that other miRNAs could show similar characteristics.
Re-evaluation of the current miRBase list of miRNAs
To investigate the possibility that the annotated miRNAs we found to be independent of DICER processing are also dependent on AGO2 catalytic activity, we analyzed the expression of these miRNAs using small RNA reads from embryonic liver tissue of mice that have been engineered to have a catalytically inactive AGO2 protein (17) (Supplementary Table S2). We again included in this analysis reads derived from AGO2-CLIP-seq, to evaluate whether these miRNAs were loaded onto AGO2. As expected, reads corresponding to the genuine DICER independent miR-451 were 70-fold less in the setting of an AGO2 mutant background compared with WT. Furthermore, AGO2-CLIP–derived reads contained the DICER independent miR-451 (Supplementary Table S2). Notably, all the other DICER independent miRNAs were not predominantly present in WT compared with AGO2 mutants, as the ratio between the number of normalized reads corresponding to these miRNAs in WT and the number of reads from AGO2 mutant approached 1, indicating that they cannot be derived from AGO2 cleavage (Supplementary Table S2). Interestingly, these miRNAs appeared to be also Dgcr8 independent. Because they do not appear to be processed by any known effector(s) of miRNA biogenesis and, in addition, were not loaded onto AGO2 (Supplementary Table S2), it emerged that these reads could arise from erroneous annotations of other kind of small RNA molecules or RNA degradation products.
To elucidate the nature of these miRNAs, we first ran RepeatMasker (http://www.repeatmasker.org) on the entire set of mouse pre-mirnas that we had downloaded from miRBase 18. This indicated that although 81% of the annotated miRNAs map onto specific localized regions of the genome, they can also be located on genomic repeats (Figure 2B and Supplementary Table S3). The possibility that genuine miRNAs can map onto genomic repeats that diverged from the consensus sequence has also been demonstrated by others (20,27,28).
Remarkably, 1% of annotated miRNAs mapped onto ribosomal RNAs (rRNAs), whereas just two of them onto transfer RNAs (tRNAs) (Figure 2B). miRNAs that map onto rRNA sequences corresponded to miR-2182, miR-5102, miR-5105, miR-5109 and miR-5115. Although we could not retrieve any reads derived from miR-2182 in mESCs, the other four were among the miRNAs that we found to be unprocessed by any known miRNA processing factor and not even loaded onto AGO2 (Supplementary Table S2), indicating an erroneous annotation of these sequences derived from rRNAs. Regarding the two miRNAs that map onto tRNAs, one corresponded to miR-1983 that has been demonstrated to be an RNA molecule that can assume two different structures, functioning either as a tRNA or as an endogenous shRNA processed by DICER (12). The second, miR-5097 was again among the miRNAs that are neither processed by DICER nor loaded onto AGO2 (Supplementary Table S2), indicating again an erroneous annotation of a tRNA as miRNA. Among other miRNAs that do not fall into these two categories, miR-3096, miR-3096b and miR-5117 contained reads corresponding to miRNAs that map inconsistently with DICER processing. miR-720 is conserved between primates and mice, but it is too short to be an miRNA because it is only 16–18 nt long, and in addition, the miRNA derived from the predicted hairpin structure is produced from the 3′ end of the putative stem in mouse and from the 5′ end in primates. miR-18a, an authentic miRNA, is instead perfectly conserved across species, including its position within the stem and its structure.
The remaining DICER independent miRNAs (Supplementary Table S2) had reads that map on the predicted precursor with a highly heterogeneous 5′ end, suggesting that they could derive from non-specific degradation of a non–pre-mirna transcript.
Analysis of deep small RNA sequencing data identifies a new sub-class of mammalian mirtrons previously annotated as canonical miRNAs
miR-3062 is among the miRNAs that we found to be independent of Dgcr8, but dependent on DICER processing (Table 1 and Supplementary Table S2). It is expressed at low levels in all the tissues analyzed, with a mean number of 1.06 reads per million (rpm) (www.miRBase.org), and it has been described as a canonical miRNA (25). We could determine reads from both WT and Dgcr8 backgrounds, but never from the DICER backgrounds in both mESCs and cortex (Supplementary Table S2 and Figure 3), indicating that its maturation bypasses Dgcr8 activity. Genomic inspection indicates that it derives from a short intron capable to fold into a hairpin loop. In contrast to other described mirtrons, neither of its ends appears to be formed by splicing, and we have thus proposed it as a two-tailed mirtron (Figure 3).
Figure 3.
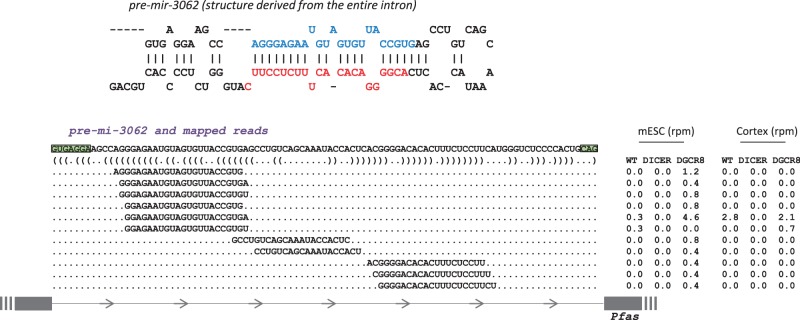
miR-3062 belongs to a new class of mirtrons. Top: the predicted RNA secondary structure of miR-3062; bottom: the distribution of reads across the precursor. Comparison of rpm between WT, DICER KO and Dgcr8 KO mESCs and cortex indicates that their biogenesis is Dgcr8 independent and DICER dependent. Green boxes indicate acceptor and donor splicing sites.
Endo-siRNAs derived from SINE elements are expressed in various mammalian tissues
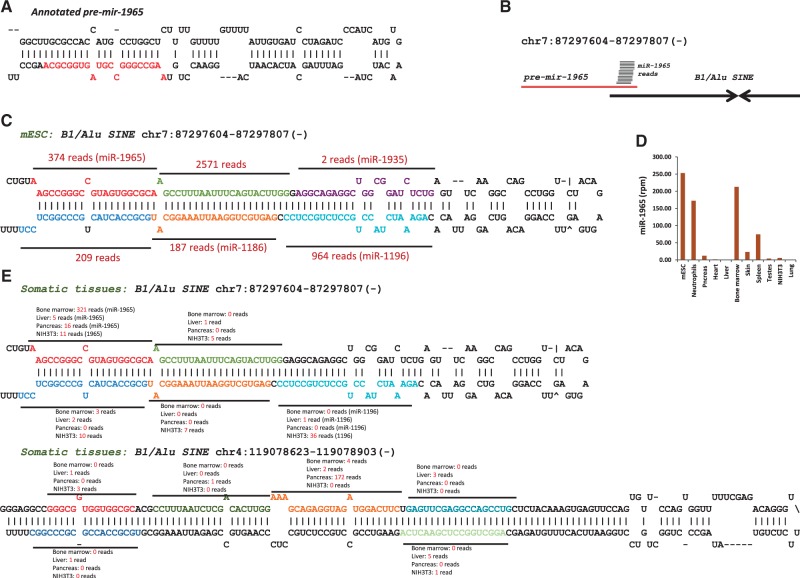
miR-1965 was an additional DICER dependent and Dgcr8 independent miRNA that we have re-classified using our approach (Table 1 and Supplementary Table S2). The annotated miRNA is located in a hairpin structure for which the corresponding miRNA* species has never been identified (Figure 4A). Genomic inspection of the reads derived from this locus indicated that the arm producing the small RNA sequences overlaps with the described B1/Alu sub-class of SINE elements in mESCs (Figure 4B). This short repeat is predicted to form a long hairpin loop, an RNA structure that DICER processes serially, producing endo-siRNAs in mESC cells (12). We next mapped again the most abundant reads derived from the WT mESCs on the B1/Alu hairpin, as performed by Babiarz et al. in their original study (Figure 4C and Supplementary Figure S2). This indicates that miR-1965 represents an endo-siRNA formed from the DICER processing of this element, as in this context, it contains a complementary small RNA* on the opposite arm, which shared a 2-nt 3′ overhang (Figure 4C), demonstrating that the annotated pre-mir-1965 is not a real miRNA precursor. Furthermore, other Dgcr8 independent and DICER dependent small RNAs (Supplementary Table S2), annotated as miR-1186, miR-1196 and miR-1935, do not show any miRNA* counterparts in the annotated precursors; they also derive from this Alu element, where instead they pair with their star counterparts with 2-nt 3′ overhang (Figures 4C and Supplementary Figure S2). Again, this indicates an erroneous annotation of these small RNAs as miRNAs in the miRBase.
Figure 4.
Somatic tissues express endogenous siRNAs derived from sequential DICER cleavage (A) Annotated predicted RNA secondary structure of mmu-miR-1965 retrieved from miRBase database. Highlighted in red is the miR-1965 mapped on the predicted annotated structure. (B) Chromosomal localization of the predicted precursor of miR-1965. It overlaps with two convergent SINE elements that after transcription fold in a long double-stranded RNA structure that gets processed by DICER. (C) Predicted RNA secondary structure of the SINE elements that are sequentially processed by DICER. Number of reads derived from WT mESCs and corresponding annotated miRNAs are mapped on the RNA structure. The number represents exclusively the most represented reads. (D) Reads per million of miR-1965 in different mouse tissues. (E) Predicted secondary structure of the transcript derived from SINE elements located on two different chromosomal locations, with the most abundant small RNAs derived from bone marrow, pancreas, liver and NIH3T3 embryonic fibroblasts from mouse.
Because miR-1965 was first identified from leukemia cells (23), we hypothesized that various tissues, not only mESCs, can also produce endo-siRNAs from the long hairpin loop derived from repeated elements, through sequential DICER processing. We first quantified the amount of miR-1965 in various mouse tissues and cells as an index of the amount of endo-siRNAs produced in these contexts (Figure 4D), indicating a substantial amount of endo-siRNAs production in various tissues, comparable in some cases with the ones produced from mESCs (Figure 4D). We then mapped reads derived from these mouse somatic tissues and cells on both Alu elements that have already been described by Babiarz et al. (12) to produce endo-siRNAs in mESCs (Figures 4E and Supplementary Figure S2), thereby showing that reads mapped on these repeats did not correspond exclusively to miR-1965 and the star counterpart, but instead are derived from different parts of the stems, phased at ∼21-nt intervals. Importantly, the reads from the 5′ arms of the hairpins shared 2-nt overhangs with those from the 3′ arms. We also demonstrated that endo-siRNAs derived from these elements are extensively detected in AGO2-CLIP and AGO2-immunoprecipitation (IP) libraries from mESC and NIH3T3 cells, respectively, indicating that after dicing, they are actively loaded on AGO2 complexes (Supplementary Figure S2). Interestingly, in testes, endo-siRNAs spanning the entire stem are mainly produced from the Alu elements located on chromosome 4, whereas only two-phased reads were detected from the one located on chromosome 7, of which one exactly corresponds to miR-1965 (Supplementary Figure S2). We then inspected genomic clusters from a chromatin immunoprecipitation experiment followed by deep sequencing (ChIP-seq) that used an antibody that recognizes the heterochromatin marker histone H3 lysine 9 trimethylation, a marker of transcriptional repression, from WT mESCs (29). There were no clusters associated with these loci, indicating that these two Alu/B1 producing endo-siRNAs are not heterochromatic; therefore, this is likely to exclude any possibility of silencing mediated by AGO2–siRNA interaction with these loci (data not shown).
Deep sequencing data from various tissues identify novel canonical and non-canonical miRNAs and reveal miRNA production from coding regions in mammals
To identify a substantial amount of novel miRNAs from mouse sequences, we downloaded additional samples containing deep sequences of small RNAs from the GEO and the SRA databases. For this study, we used 474 439 169 reads that were >16 nt in length, derived from several different adult mouse tissues, developmental stages and cell types in addition to three different mutant backgrounds, and reads derived from IP and CLIP of AGO2. To discover novel miRNAs, we used miRDeep2, a new algorithm capable of discovering canonical and non-canonical miRNAs with high efficiency from various animal clades (20). For miRNA selection, we used strict criteria that were previously successfully undertaken to annotate miRNAs from small RNA deep sequencing with high confidence (20,21,25). In doing so, we considered as confident for annotation only miRNAs that (i) paired with 2-nt overhang, with the miRNA star on the stem of the predicted precursor, and (ii) also contained a uniform 5′ terminus compared with the 3′ terminus that was expressed in multiple tissues. Using this approach, we selected 66 genomic locations that can produce miRNAs (Supplementary Figure S3, Table 2 and Supplementary Table S4). The miRNAs identified were predominantly derived from the sense strand of introns or were from intergenic loci (61 and 23%, respectively) as expected (30) (Figure 5A). The minority of these derived from exonic regions (16%) (Figure 5A). Surprisingly, only one of the new discovered miRNAs were produced from the exon of a non-coding RNA, whereas nine of these were produced from coding (CDS) and untranslated regions of protein-coding genes. To the best of our knowledge. miRNAs derived from coding regions were discovered only in Drosophila recently (21). We found one miRNA is produced from the 3′-UTR of Fnbp1l, one from the 5′-UTR of Alg10b, four from CDSs and, interestingly, three from a region that spans the 5′-UTR with the coding region, indicating that DICER could compete with the translational machinery of those mRNAs (Table 2 and Supplementary Table S4). We also found two miRNAs derived from genomic repeats, one from a SINE element and one for a long interspersed nuclear element (LINE) (Figure 5B, Table 2 and Supplementary Table S4). Furthermore, 12% of the discovered miRNAs derived from the antisense strand of previously annotated ones (Figure 5B, Table 2 and Supplementary Table S4). We also discovered four novel 5′-tailed mirtrons, and one that contains a long tail at 5′ end and a short one (11 nt) at the 3′, which we propose as a second two-tailed mirtron (chr6_15427) other than miR-3062 (Figure 5B, Table 2 and Supplementary Table S4).
Table 2.
Description of the novel miRNAs identified in this study
| Provisional ID | Total read count | Precursor coordinate | Genomic location | Gene | Type |
|---|---|---|---|---|---|
| chr1_2655 | 853 | chr1:172994721..172994783:− | Intron | Fcgr3 | shRNA (DGCR8−/−) |
| chr15_37558 | 788 | chr15:25877982..25878042:+ | Intron | Fam134b | Canonical |
| chrX_47068 | 292 | chrX:132281255..132281309:+ | Intron | Grasp1 | shRNA (DGCR8−/−) |
| chr5_14110 | 254 | chr5:123972645..123972704:− | Intron | Diablo | shRNA (DGCR8−/−) |
| chr15_38333 | 227 | chr15:90054791..90054844:+ | 5′-UTR | Alg10b | DGCR8−/− |
| chr6_17117 | 156 | chr6:128924857..128924913:− | Exon | AK040061 | Canonical |
| chr9_23196 | 146 | chr9:15117055..15117114:+ | Intron | Taf1d | shRNA |
| chr11_28442 | 136 | chr11:6301633..6301697:+ | Intron | Zmiz2 | Canonical |
| chr2_3181 | 117 | chr2:29701167..29701232:+ | Intergenic | Canonical | |
| chr5_11864 | 108 | chr5:34916800..34916863:+ | Intron | Add1 | Canonical |
| chr11_28874 | 101 | chr11:55314728..55314784:+ | Intron | G3bp1 | Canonical |
| chr11_29306 | 106 | chr11:77992329..77992390:+ | Span 5′-UTR-CDS | Tlcd1 | Canonical |
| chr8_21975 | 96 | chr8:8690092..8690162:− | CDS | Arglu1 | Canonical |
| chr2_3037 | 88 | chr2:17979473..17979531:+ | Intron | Mllt10 | shRNA (DGCR8−/−) |
| chr9_23976 | 76 | chr9:94652248..94652310:+ | Intron | Slc9a9 | Canonical |
| chr3_8489 | 93 | chr3:122244542..122244599:− | 3′-UTR | Fnbp1l | DGCR8−/− |
| chr4_11627 | 78 | chr4:155604167..155604224:− | CDS | Klhl17 | Canonical |
| chr15_39359 | 73 | chr15:79829399..79829456:− | Intron | Pdgfb | Canonical |
| chr10_27859 | 70 | chr10:77802894..77802952:− | Intron | Agpat3 | Canonical |
| chr6_15465 | 54 | chr6:115725675..115725738:+ | Intron | Cand2 | Canonical |
| chr6_15427 | 62 | chr6:108439737..108439798:+ | Intron | Itpr1 | Two-tailed mirtron |
| chr10_27686 | 49 | chr10:61656914..61656975:− | Intron | Tspan15 | Canonical |
| chr10_27394 | 41 | chr10:22449177..22449238:− | Intron | Tbpl1 | Canonical |
| chr2_3125 | 46 | chr2:24915331..24915394:+ | Intron | Nelf | Canonical |
| chr2_4218 | 45 | chr2:119843253..119843316:+ | Intron | Mapkbp1 | 5′-tailed mirtron |
| chr8_22282 | 700 | chr8:41393228..41393283:− | Intron | Fgf20 | shRNA (DGCR8−/−) |
| chr17_33821 | 390 | chr17:29732483..29732546:+ | Intron | Tbc1d22b | Canonical |
| chr7_14441 | 301 | chr7:30691528..30691587:+ | Intron | Zfp420 | shRNA (DGCR8−/−) |
| chr3_6937 | 176 | chr3:89815074..89815132:− | Intron | Ubap2l | Canonical |
| chr6_12274 | 109 | chr6:38457325..38457394:+ | Intron | Ubn2 | Canonical |
| chr11_24328 | 76 | chr11:53577098..53577156:− | Intergenic | Canonical | |
| chr14_30051 | 71 | chr14:27489032..27489094:− | Span 5′-UTR-CDS | Pde12 | Canonical |
| chrX_38038 | 46 | chrX:91519913..91519977:+ | Intron | Klhl15 | Canonical |
| chr7_14452 | 53 | chr7:31374399..31374457:+ | AS intron | Gapdhs | shRNA (DGCR8−/−) |
| chr2_3012 | 50 | chr2:31906466..31906529:+ | Intron | Nup214 | 5′-tailed mirtron |
| chr12_25218 | 42 | chr12:11306693..11306751:+ | Intron | Smc6 | Canonical |
| chr15_38048 | 519 | chr15:78179029..78179091:+ | CDS | Csf2rb2 | Canonical |
| chr15_39273 | 519 | chr15:78115217..78115279:− | CDS | Csf2rb2 | Canonical |
| chr2_4044 | 178 | chr2:94081535..94081600:+ | Intergenic | Canonical | |
| chr7_18793 | 171 | chr7:3219198..3219260:− | Intergenic | Canonical | |
| chr10_27945 | 179 | chr10:80878984..80879042:− | Intron | Nfic | Canonical |
| chr18_44110 | 44 | chr18:61113527..61113587:+ | Intron | Camk2a | Canonical |
| chr17_41247 | 24 | chr17:27228190..27228257:+ | Intron | Itpr3 | 5′-tailed mirtron |
| chr2_6270 | 36 | chr2:164180048..164180119:− | Intron | Slpi | 5′-tailed mirtron |
| chr11_30735 | 286 | chr11:82798504..82798567:− | Intron | Slfn9 | Canonical |
| chr16_40566 | 301 | chr16:21256085..21256139:− | Intron | AK043964 | Canonical |
| chr19_45771 | 77 | chr19:55267168..55267225:+ | Intron | Tectb | Canonical |
| chrX_47993 | 293 | chrX:146981927..146981988:− | Intron | Apex2 | Canonical |
| chrX_47598 | 369 | chrX:64075830..64075888:− | Intergenic | Canonical | |
| chr1_19965 | 56 | chr1:53906268..53906341:+ | AS intron | Hecw2 | Canonical |
| chr1_20693 | 305 | chr1:153289523..153289581:+ | Intron | Trm1l | Canonical |
| chr1_8658 | 1116 | chr1:173571968..173572033:− | Intron | Slamf7 | Canonical |
| chr11_4307 | 48 | chr11:28646198.28646265:− | Intergenic | Canonical | |
| chr15_6934_1 | 29 | chr15:82224348..82224409:− | Span 5′-UTR-CDS | Cyp2d11 | Canonical |
| chr15_6934_2 | 29 | chr15:82443316..82443377:− | Intergenic | Canonical | |
| chr15_6934_3 | 29 | chr15:82612504..82612572:+ | Span 5′-UTR-CDS | Cyp2d41 | Canonical |
| chr17_15196 | 347 | chr17:34161957..34162019:+ | Intron | M73959 | Canonical |
| chr2_13190 | 80 | chr2:26446881..26446942:− | AS intron | Egfl7 | Canonical |
| chr2_23582 | 185 | chr6:28972622..28972685:− | Intergenic | Canonical | |
| chr2_25696 | 52 | chr2:158069240..158069301:− | Intergenic | Canonical | |
| chr7_32398 | 1608 | chr7:147143465..147143521:− | Intergenic | Canonical | |
| chr7_6169 | 71 | chr7:3218640..3218696:− | Intergenic | Canonical | |
| chr8_40622 | 649 | chr8:123062596..123062654:+ | Intron | Gse1 | Canonical |
| chr18_17432 | 642 | chr18:56697793..56697853:− | Intron | Aldh7a1 | shRNA (DGCR8−/−) |
| chr10_1158 | 925 | chr10:92895852..92895909:+ | Intergenic | LTR ERV1 (DGCR8−/−) | |
| chr5_30975 | 581 | chr5:36644219..36644278:+ | AS intron | Sorcs2 | SINE Mirb (DGCR8−/−) |
Figure 5.
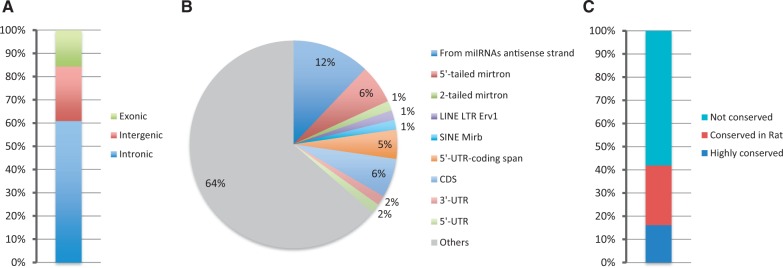
Classification of the novel miRNAs. (A) Genomic location of novel miRNAs. (B) Classification of the transcripts that produce novel miRNAs. (C) Level of conservation of the newly identified miRNAs.
Accordingly and in conjunction with previous findings (26), 97% of the mirtrons that we identified in this study contained miRNAs derived from the 3′ part of the precursor either in mono-uridylated or in mono-adenylated forms (Supplementary Figure S3).
The majority of the novel miRNAs identified in this study (58%) were not conserved; 26% were conserved in the rat, whereas 16% were conserved in several species (Figure 5C, Table 2 and Supplementary Table S4).
Experimental validation demonstrates that hairpins derived from coding regions are processed to miRNAs
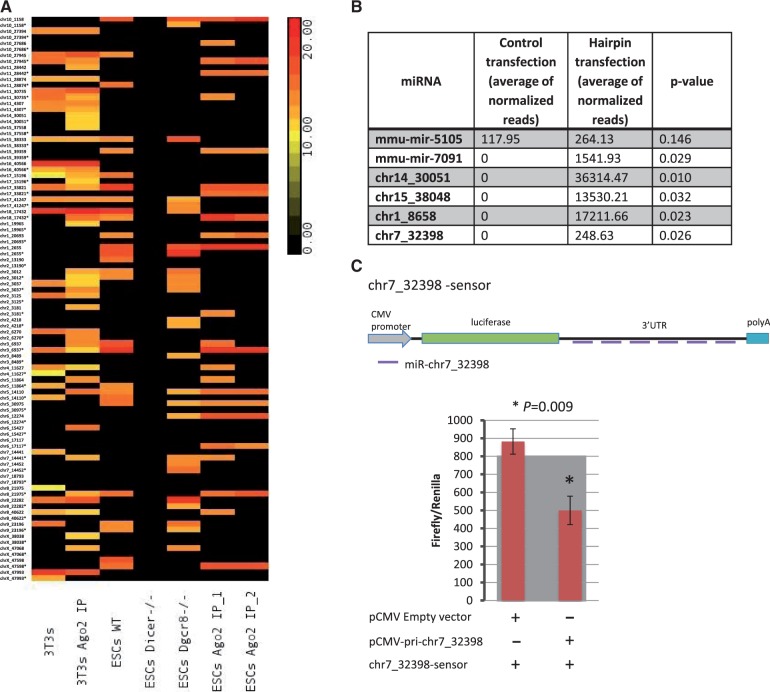
To validate our findings, we first mapped reads derived from WT, DICER knockout (KO) and Dgcr8 KO mESCs, from CLIP of AGO2 from mESCs and from cells derived from AGO2-IP (NIH3T3 and mESC cells) onto our newly discovered miRNAs (Figure 6A and Supplementary Table S5). In doing so, we demonstrated that 70% of these novel miRNAs are loaded on AGO2 and/or processed by DICER (they are absent from DICER KO), indicating that our methods elucidate authentic miRNAs (Figure 6A and Supplementary Table S5). Importantly, we found that a group of miRNAs was independent of Dgcr8 processing (Figure 6A, Supplementary Tables S4 and S5). Among these, there were mirtrons and miRNAs derived from repeats as expected. Chr15_38333, derived from the 5′-UTR of Alg10b, was the only miRNA encoded by a protein-coding sequence expressed in both NIH3T3 and mESCs and could be verified to be loaded on AGO2 and dependent on DICER processing (Figure 6A and Supplementary Table S5). Surprisingly, it appears to be independent of Dgcr8 processing (Figure 6A and Supplementary Table S5). In addition, chr3_8489, a second novel miRNAs derived from the 3′-UTR of Fnb1l, contains reads just in Dgcr8−/− mESCs, indicating microprocessor independency. A group of miRNAs that appeared dependent on DICER but independent of DGCR8 does not seem to belong to mirtrons or snoRNA sub-classes, and we refer to these as potential shRNA (Table 2, Supplementary Tables S4 and S5) as described (12,25).
Figure 6.
Validation of the novel candidate miRNAs. (A) Heat map showing the expression of the novel miRNAs in NIH3T3, AGO2 from NIH3T3, WT mESCs, DICER−/− mESCs, Dgcr8−/− mESCs and CLIP of AGO2 from mESCs from two repeats. The sum of the normalized reads is provided in the Supplementary Table S5. (B) Table showing normalized miRNA expression and P-values from deep sequencing of small RNAs after ectopic expression in cells of a group of miRNAs selected from this study. This experiment has been conducted in triplicate. (C) Top: the schematic representation of the cloning of six complementary sites to the novel miRNA chr7_32398 as a UTR of the firefly luciferase in the pMIR-REPORT vector, forming a chr7_32398-sensor construct; bottom: firefly luciferase relative levels from cells transfected with empty vector and sensor versus cells transfected with sequences coding for pri-chr7_32398 and chr7_32398-sensor construct. This experiment has been conducted in triplicate on three independent occasions.
We also selected and cloned into a CMV vector four novel hairpins of miRNAs [including two canonical miRNAs (chr1_8658 and chr7_32398) and two miRNAs derived from protein-coding regions: chr14_30051, derived from a region that spans the 5′-UTR and CDS of Pde12, and chr15_38048, from the CDS of Csf2rb2, which could not be validated using the previous approach (because they are not expressed in those cells)] and ectopically expressed these in HEK293T cell lines. Small RNA sequencing of these cells revealed that all the miRNAs tested were efficiently processed compared with cells expressing an empty vector control (Figure 6B). We also cloned and ectopically expressed miR-5105, which we discovered as an erroneous annotation because it was derived from rRNA, and in subsequent experiments observed no significant changes in reads from this region and miR-7091 as positive control (Figure 6B). We also cloned an miRNA interaction site six times in a row into a vector, incorporating it as the 3′-UTR of the luciferase gene, and co-expressed this with the CMV vector containing the hairpin of one of the newly discovered miRNAs (chr7_32398). We found that this miRNA was able to confer post-transcriptional gene regulation/silencing (Figure 6C).
Quantification of the newly identified miRNAs in multiple tissues
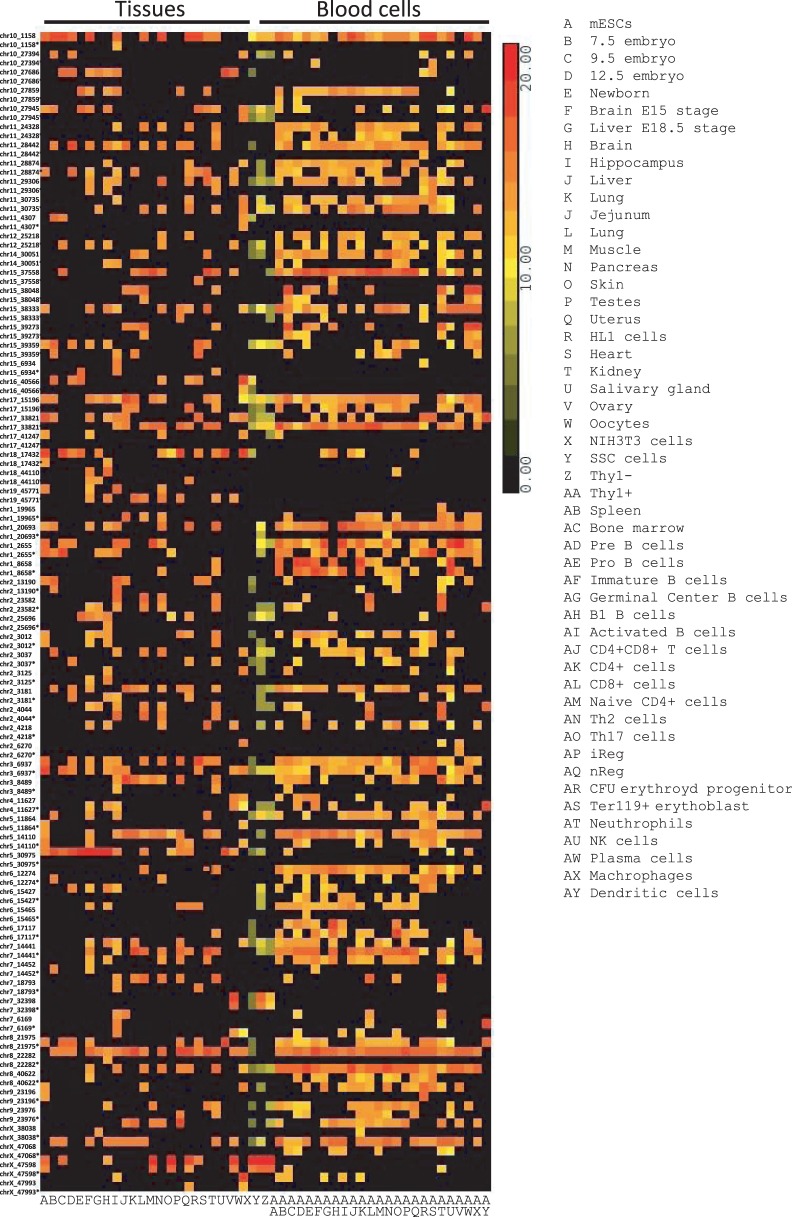
We then performed quantification of the newly discovered miRNAs among the tissues analyzed in this study (Figure 7 and Supplementary Table S5). This derives a map of the expression of the novel miRNAs in tissues and different developmental stages, and indicates their presence in multiple tissues (Figure 7 and Supplementary Table S6). Interestingly, the miRNA heat map clearly differentiates blood cells from the remainder (Figure 7).
Figure 7.
Novel candidate miRNA expression profiles. Expression of normalized reads of novel discovered miRNAs across different embryonic and adult mouse tissues. The sum of the normalized reads is provided in the Supplementary Table S6.
DISCUSSION
In this study, we re-analyzed small RNA sequences from multiple mouse tissues, partly characterized by mutation or KO of the main effectors of miRNA biogenesis, to provide a more reliable miRNA list and to discover and characterize these novel miRNAs. We also used small RNA sequences isolated from IP of AGO2. Such an analysis permitted the discovery of mammalian mirtrons, putative shRNAs and miRNAs derived from coding and untranslated region of protein-coding genes and antisense to annotated miRNAs, in addition to canonical miRNAs. We revealed 66-high confidence loci coding for miRNAs and experimentally validated some of these. Intriguingly, we could also demonstrate that at least two miRNAs derived from coding genes, one from the 3′-UTR and one from the 5′-UTR that are independent of Dgcr8 processing. It has been demonstrated that DROSHA–Dgcr8 processes and regulates mRNAs directly recognizing hairpin structures within these, but this processing is not destined to lead to miRNA production (31). It is possible that DICER directly targets messengers recognizing hairpin structures, directly regulating mRNAs levels and producing miRNAs at the same time, but this observation requires further validation.
We have used two methods to validate our findings: we mapped reads derived from WT, DICER KO and Dgcr8 KO mESCs; from CLIP of AGO2 from mESCs; and from NIH3T3 cells and AGO2 from NIH3T3 cells on our novel miRNAs, discovering that 70% of these miRNAs were loaded on AGO2 and/or processed by DICER. Next, we selected four novel miRNAs and one derived from rRNA (erroneous annotation), and ectopically expressed the corresponding hairpins in HEK293T cells. We observed that these were efficiently processed to miRNAs except the one discovered to be an erroneous annotation. We also experimentally demonstrated that one of the novel miRNAs is able to confer gene regulation/silencing.
Lai’s group recently published an article on the discovery of hundreds of mirtrons (10). Because of these data, we subsequently excluded 30 mirtrons from our study owing to overlap; in contrast, this indicated our ability to discover authentic miRNAs using our analysis. We also mapped reads derived from WT, DICER−/− and Dgcr8−/− mESCs on miRNAs downloaded from the current miRBase release 18, to evaluate whether part of the new annotated miRNAs, missed in the original analysis, belonged to a non-canonical pathway of biogenesis, and to verify that all the annotated miRNAs are processed by DICER. We found that many annotated small RNAs did not conform to the rules of miRNA biogenesis, indicating that the current miRBase list should be carefully re-defined. Mapping reads derived from mutated backgrounds showed us that a group of annotated miRNAs was not processed by DICER or Dgcr8, nor were they loaded on the AGO2 complex. We also found that some of these reads map on rRNA or tRNA, and that they have a heterogenous 5′ end (mapping on the precursor inconsistent with DICER processing). For this approach, we used miRNAs derived from the mirbase release 18, and accordingly, at least some of them (miR-2182, miR-5102 and miR-720) have been already removed from the newest release of the miRBase (miRBase 19).
We also characterized a new group of mirtrons in which both ends are not directly defined by splicing, which we alternatively define as two-tailed mirtrons. miR-3062, previously annotated as a canonical miRNA, is usually expressed at low levels (25). After splicing, the entire 97-nt-long intron, containing miR-3062, forms a hairpin that contains a 5′ tail of 21 nt and a 3′ tail of 13 nt from the mature 5p and 3p miRNA reads, respectively. The size of the two tails is incompatible with DROSHA processing, as it has been demonstrated that precursors should contain at least 40 nt on each side of the hairpin to be recognized as a DROSHA substrate (32). In addition, we could demonstrate that its processing is independent of Dgcr8, but dependent on DICER in both mESCs and mouse cortex. Because the entire intron forms a hairpin loop, including the two tails, it is likely that in this case, the two tails are not removed by any nucleases, which probably occurs for ‘canonical’ tailed mirtrons, but instead the entire hairpin is transported from the nucleus into the cytoplasm, where DICER recognizes it as a substrate, cleaving it directly under the loop, but above the double-stranded end. Similarly, snoRNAs, in which the hairpin structure does not contain a typical 2-nt overhang at the end of the stem, can be subjected to DICER processing (13).
Another important observation was that somatic tissues express endo-siRNAs derived from long hairpins. Although endo-siRNAs have been previously cloned from mice, not only fruit flies, plants and nematodes (14), it has been shown thus far that they are a characteristic of cells that do not possess an interferon response, such as oocytes and mESCs (12,33,34). We found here that some annotated miRNAs are endo-siRNAs that are produced from one convergent Alu/B1 SINE element located on chromosome 7 that, together with another one located on chromosome 4, is described as the source of endo-siRNAs in mESCs (12). Because one of these miRNAs, miR-1965, which we show to correspond to the end of the long stem formed by the transcription of one of the Alu elements, is widely expressed in tissues types, we postulated that endo-siRNAs derived from these two Alu elements are not restricted to mESCs. We could map reads derived from these stems on the tissues that we analyzed, but not in post-mitotic neurons. However, the amount of reads generated by sequential DICER processing and the region that is predominantly cleaved by it can be diverse in different tissues, indicating regulation of their processing in various tissue types. Nevertheless, Kaneco et al. demonstrated that macular degeneration is caused by DICER loss, but not by a loss of miRNAs (35). The lack of DICER expression causes an up-regulation of Alu elements that, in turn, causes cytotoxicity, indicating the possibility that DICER is directly implicated in the silencing of the Alu element. Moreover, the authors propose that DICER could directly process Alu elements to render them inert (35). Because we found that somatic tissues produce endo-siRNAs from Alu elements, this could indeed be possible. We also demonstrated that the produced endo-siRNAs are then loaded onto AGO2 complexes. We analyzed the genomic regions containing these Alu elements for the presence of genomic clusters derived from a chromatin immunoprecipitation experiment followed by deep sequencing (ChIP-seq) that used an antibody that recognizes the heterochromatin marker histone H3 lysine 9 trimethylation, performed in WT mESCs (29). This revealed to us that these two Alu/B1 elements producing endo-siRNAs are not heterochromatic, which indicates that the mechanisms of action of Alu element–derived endo-siRNAs are not yet clear and merit further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–3.
FUNDING
Association for International Cancer Research; the Imperial BRC and ECMC. Funding for open access charge: The Imperial BRC and ECMC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all the people who deposited their sequences data in openly available databases permitting us to perform this study, and Giannis Dzegoutanis for his critical help and advice.
REFERENCES
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III DROSHA initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass DROSHA processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 2011;18:1075–1082. doi: 10.1038/nsmb.2091. [DOI] [PubMed] [Google Scholar]
- 12.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, DICER dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poethig RS, Peragine A, Yoshikawa M, Hunter C, Willmann M, Wu G. The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 2006;71:165–170. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- 16.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of DICER requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for DICER independent, AGO2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. US A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus DICER1 knockouts and small RNA sequencing. RNA. 2011;17:1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of AGO2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westholm JO, Ladewig E, Okamura K, Robine N, Lai EC. Common and distinct patterns of terminal modifications to mirtrons and canonical microRNAs. RNA. 2012;18:177–192. doi: 10.1261/rna.030627.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between DROSHA and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 33.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.