Abstract
Nuclear (nDNA) and mitochondrial DNA (mtDNA) communication is essential for cell function, but it remains unclear whether the replication of these genomes is linked. We inspected human cells with a novel fluorescence in situ hybridization protocol (mitochondrial Transcription and Replication Imaging Protocol) that identifies mitochondrial structures engaged in initiation of mtDNA replication and unique transcript profiles, and reconstruct the temporal series of mitochondrial and nuclear events in single cells during the cell cycle. We show that mtDNA transcription and initiation of replication are prevalently coordinated with the cell cycle, preceding nuclear DNA synthesis, and being reactivated towards the end of S-phase. This coordination is achieved by modulating the fraction of mitochondrial structures that intiate mtDNA synthesis and/or contain transcript at a given time. Thus, although replication of the mitochondrial genome is active through the entire cell cycle, but in a limited fraction of mitochondrial structures, peaks of these activities are synchronized with nDNA synthesis. After release from blockage of mtDNA replication with either nocodazole or double thymidine treatment, prevalent mtDNA and nDNA synthesis occurred simultaneously, indicating that mitochondrial coordination with the nuclear phase can be adjusted in response to physiological alterations. These findings will help redefine other nuclear–mitochondrial links in cell function.
INTRODUCTION
To function, mitochondria rely on nuclear-encoded genes as well as on a few genes coded by their own genome [mitochondrial DNA (mtDNA)], a 16.5 bp circular molecule that in humans codes for 13 proteins essential for oxidative phosphorylation, 22 tRNAs and 2 rRNAs. mtDNA molecules are packaged in a few copies into nucleoprotein complexes, nucleoids, that include the mitochondrial replicative DNA polymerase γ (Polγ), and transcription factor A, mitochondrial (TFAM), a transcription and DNA-packaging factor (1). Several hundreds nucleoids are present in cells, resulting in 103–104 copies of mtDNA in most human somatic cells, but this number changes during development and depends on tissue-specific factors (2). The elevated number of mtDNA copies is essential for cell function, and depletion of mtDNA may cause disease (3). Mutations of mtDNA have been also associated with diseases, including when only a fraction of the molecules is mutated (4). Mitochondria exist as isolated entities as well as interconnected mitochondrial network, where dynamics are regulated by fusion and fission processes (5). Fusion mixes individual compartments and complements possible defects, as in the presence of mutated mtDNAs (6). Release of mtDNA from the organelle, detected for instance in some cancers, is considered an indicator of cell damage (7). mtDNA fragments are occasionally integrated in the nuclear genome of eukaryotes including humans (8,9), with activity linked to DNA replication in yeast (10).
The mitochondrial genome is replicated and transcribed autonomously in the organelle, but the proteins necessary for these processes are coded in the nucleus. Strong nuclear–mitochondrial communication is therefore necessary to ensure the mitochondrial function. A large number of evidences indicate that unidirectional mtDNA replication starts at the OH origin, located in the regulatory D-loop region, and displaces the parental heavy strain until a second replication origin OL on the light strain is exposed, which initiates DNA synthesis in the opposite direction [reviewed in (11)]. However, alternative replication strategies have also been reported (12,13). Replication at origin OH frequently terminates ∼700 bp downstream resulting in 7S DNA, which forms a triple-stranded structure with the parental DNA, the ‘displacement loop’ or D-loop (14). The frequency of D-loop structures in mtDNA molecules is variable depending on the cell and growth conditions, but the function of the D-loop is still unknown (11).
Differently from hundreds or thousands of mtDNA copies per cell, two copies of the nuclear genome are present in somatic cells, where replication is strictly regulated by a complex series of events and checkpoints (15). The cell cycle comprises a long growth phase (G1), DNA replication (S), a short growth phase (G2) and cell division or mitosis (M). Whether the synthesis of mtDNA is coordinated with the cell cycle is still debated, and controversial observation on this topic has been reported in the past 40 years. Indeed, mtDNA replication is believed to occur continuously (11,16,17), although mtDNA synthesis at specific phases of the cell cycle, in particular S and G2, has also been reported (18–20). Moreover, the use of different synchronization procedures resulted in either mtDNA synthesis restricted to phases S and G2 (selective cell detachment) or continuous synthesis during the cell cycle (double thymidine) (21). Interestingly, high levels of Polγ activity were observed in phases S and G2 (22). Furthermore, mitochondrial biogenesis (18) appears to be regulated during the cell cycle, although a previous report indicated no link between mitochondrial growth and the cell cycle (23). Coordination between mitochondrial energetics and cell cycle progression has also been shown (24), and the molecular mechanisms linking cell proliferation and the mitochondrial metabolic machinery start to be elucidated (25). Importantly, the formation of a giant mitochondrial tubular network has been shown during the G1-S transition phase that triggers expression of cyclin E and therefore S-phase progression (26), thereby linking mitochondria and cell cycle. Moreover, it has been found that 7S DNA synthesis is synchronized with nuclear DNA (nDNA) synthesis (27). Thus, an increasing number of evidences support a link between mitochondrial dynamics and cellular events, in agreement with the notion that dividing cells must be metabolically prepared to deal with the energy demand of proliferation. However, it remains unresolved whether mtDNA synthesis and transcription are correlated with the cell cycle.
The distinct analysis of mitochondrial and nuclear markers at the single-cell level may help elucidating this point. We used a novel imaging protocol (28), which identifies mitochondrial subpopulations engaged in the initiation of mtDNA replication and/or rich in transcripts, and at the same time inspected nuclear events. We thus assessed the dynamics of mtDNA processing in single cells and show that mtDNA replication and transcription are prevalently coordinated with the cell cycle.
MATERIALS AND METHODS
Culture conditions and synchronization
Human HeLa cells were grown in minimal essential medium (Gibco) with 10% foetal bovine serum (FBS) at 37°C and in the presence of 5% CO2. Cells were synchronized at 25–30% confluence. Cells synchronized by serum starvation were cultured in 0.1% FBS for 3 days to accumulate in G0. Cells were treated with nocodazole (200 ng/ml) for 16 h to induce mitotic arrest (G2/M). Synchronization in G1/S with double thymidine was done with 2 mM thymidine for 16 h followed by 10 h in fresh medium, then 14 h with 2 mM thymidine. Following the synchronization procedure, cells were washed three times in phosphate buffered saline buffer (PBS) and released in 10% FBS containing medium for the indicated times.
Bromodeoxyuridine incorporation
Cells were incubated for 10 min in the presence of 100 µM bromodeoxyuridine (BrdU), fixed in 2% paraformaldehyde (PFA) (10 min), treated for 10 min with 4 N HCl and 0.5% Triton X-100, and neutralized for 30 min with 100 mM sodium borate (26). Cells were then immunolabelled with anti-BrdU antibody.
Immunofluorescence, reagents and antibodies
Cells plated on slides were fixed with 2% PFA and permeabilized with 0.5% Triton X-100. Slides were incubated in blocking buffer (bovine serum albumin 5% in PBS) for 1 h and then with the primary antibody for 1 h. DNA was stained with 10 µg/ml of Hoechst. BrdU, anti-TOM22 Atto488, Hoechst 33342, nocodazole and thymidine were purchased from Sigma; anti-BrdU antibody from BD Biosciences; proliferating cell nuclear antigen (PCNA), TFAM, Polγ, and glyceraldheyde 3-phosphate dehydrogenase (GAPDH) antibodies from SantaCruz Biotechnology; goat anti-mouse and goat anti-rabbit Alexa® Fluor 555 or Alexa® Fluor 488 conjugated secondary antibodies (Invitrogen).
Probe labelling and denaturation
DNA probes for fluorescence in situ hybridization (FISH) were labelled by nick translation of PCR products, incorporating Atto425-dUTP, or Atto488-dUTP, or Atto550-dUTP (Atto425/Atto488/Atto550 NT Labeling kit, Jena Bioscience). Primers used for PCR amplification of DNA probes are listed in Supplementary Table S2. In all, 40 ng of labelled probes were mixed with 400 ng of sonicated salmon sperm DNA (Sigma) in the hybridization buffer [50% formamide, 10% dextran sulfate, in 2 × saline-sodium citrate buffer (SSC) pH 7.0], denatured at 80°C for 10 min and then kept at 37°C for 30 min.
Mitochondrial transcription and replication imaging protocol
Cells plated on slides were fixed with 2% PFA and permeabilized with 0.5% Triton X100, incubated in 50% formamide/2 × SSC pH 7.0 for 30 min at room temperature, and denaturated in 70% formamide/2 × SSC for 4 min at 75°C. Hybridization was done with 40 ng of probe (single probe or mix) for 16 h at 37°C. Slides were washed in SSC, and DNA was stained with 10 µg/ml of Hoechst.
Confocal acquisition, 3D reconstruction and quantification
Confocal acquisitions were performed using a spinning-disk Perkin-Elmer Ultraview RS Nipkow Disk, an inverted laser-scanning confocal microscope Zeiss Axiovert 200 M with an Apochromat 63×/1.4 oil objective and a Hamamatsu ORCA-II-ER camera (Imagopole, Institut Pasteur). Optical slices were taken every 200-nm interval along the z-axis covering the whole depth of the cell, at resolution of 1.024/1.024 pixels. 3D reconstruction was achieved using the IMARIS software (Bitplane). Fluorescence quantification was done using a single-imaging frame collection and ImageJ 1.34-s software (post-acquisition analysis). For each sample, 50 cells were analysed from three independent experiments.
Statistical analysis
The significance of differences between data was determined using Student’s t-test for unpaired observations. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Real-time quantitative polymerase chain reaction
Total RNA was isolated from HeLa cells using the RNAeasy Mini kit (Qiagen), treated with DNaseI (Qiagen), then reverse transcribed using Superscript®III Reverse transcriptase (Invitrogen). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using Power Sybr Green PCR Master Mix (Applied Biosystems), and the rate of dye incorporation was monitored using the StepOne™ Plus RealTime PCR system (Applied Biosystems). Three biological replicates were used for each condition. Data were analysed by StepOne Plus RT PCR software v2.1 and Microsoft Excel. TATA box binding protein transcript levels were used for normalization of each target (=ΔCT). RT-PCR CT values were analysed using the 2−ΔΔCt method to calculate the fold expression (29). Primers used for amplifications are listed in Supplementary Table S2, with indicates the relative reference.
mtDNA content analysis by quantitative polymerase chain reaction
The quantification of mtDNA was performed by quantitative polymerase chain reaction (qPCR) amplification on 200 pg of total DNA using the StepOne™ Plus RealTime PCR system (Applied Biosystems) and Power Sybr Green PCR Master mix (ABI) following the manufacturer’s instruction. Human mtDNA accession number: NC012920.1. The region tested on mtDNA was included in the 12S gene. When indicated, a region within 7S (primers A, B1) and a larger downstream region that also contain 7S (primers A, B2) were analysed, as described (27). The nuclear encoded 18S rRNA gene was used as an endogenous reference. The level of mtDNA was calculated using the ΔCT of average CT of mtDNA and nDNA (ΔCT = CT nDNA − CT mtDNA) as 2ΔCT (30). Primers used for amplification are listed in Supplementary Table S2.
Protein extraction and western blot
Cells were lysed by lysis solution [20 mM Tris, 18 mM NaCl, 0.5 % Lauryl β Maltoside, 1 mM MgCl2, 200 mM Na4P2O7, 1 mM ethylene glycol tetraacetic acid (EGTA), 20 mM NaF, 2 mM NaVO4, 1 mM Pefabloc (Sigma), 1 mM Aprotinin (Sigma), 1 mM Leupeptin (Sigma)]. Protein content was determined with the Bradford reagent (Sigma), and 30 µg of protein were loaded for sodium dodecyl sulphate-polyacrylamide gel electrophoresis. After blotting, Hybond ECL nitrocellulose filters were probed with anti-Polγ and anti-TFAM antibodies. Detection was performed using Odyssey Infrared Imaging system scanner and Odyssey application software v3.0 (LI-COR Biosciences).
RESULTS
Identification and quantification of mtDNA processing markers in single cells
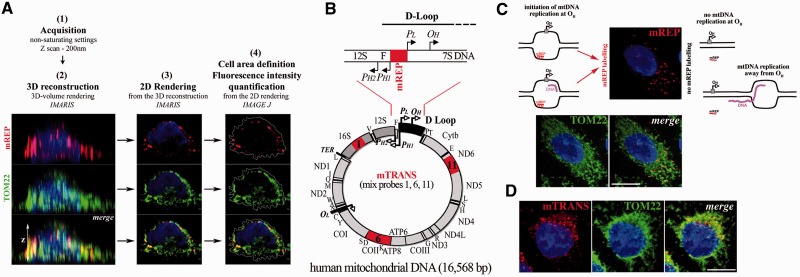
We recently developed an imaging approach, mitochondrial Transcription and Replication Imaging Protocol (mTRIP) that is a combination of DNA FISH and RNA FISH, and which can be also combined to immunofluorescence, as the treatment does not destroy proteins (28). With mTRIP, we performed 3D confocal acquisitions of the fluorescent signal of either DNA probes or protein immunomarkers, or both in colabelling experiments, and measured the intensity of fluorescence of each marker per cell. The measurement was performed in the entire cell area, defined in the 2D rendering file, which results from 3D reconstruction of z-stacks (Figure 1A). 3D reconstruction and 2D rendering were obtained with the software Imaris and fluorescence intensity quantification with the software ImageJ. 3D analysis allows detecting signal in the entire volume of the cell, rather than in a single section, thereby providing comprehensive data collection for quantification. Probes called mREP and mTRANS are used here as in situ mtDNA processing markers (coordinates in the mtDNA are in Supplementary Table S1). Probe mREP, located upstream of the main replication origin OH, specifically labels DNA engaged in initiation of mtDNA replication, see Figure 1B and C [and ref (28); in this article, we show that mREP labelling is specific to DNA, as the signal is fully DNAse1 sensitive, and RNAseA and RNAseH resistant; moreover, mREP labelling is associated with limited incorporation of BrdU, an indicator of DNA replication, compatibly with the synthesis of short DNA chains and with an open DNA bubble at the initiation site]. Probe mTRANS, a mix of three probes that label several rRNAs, mRNA and tRNAs is used as marker of mitochondrial transcripts [Figure 1B and D; in ref. (28), we show that mTRANS labelling is specific to RNA, as the signal is fully RNaseA sensitive, and DNaseI and RNaseH resistant]. The localization of the FISH probes within the mitochondrial network is revealed in colabelling experiments with anti-TOM22, which recognizes a protein of the outer mitochondrial membrane, member of the Translocase of the Outer Membrane complex [Figure 1C and D, and (28)]. More in-depth, we previously showed that mTRIP signal colabels to the largest extent with nucleoid markers (28), but given the limits of confocal fluorescence microscopy [200 nm versus ∼70 nm average size of nucleoids (31,32)] in this study, we refer to mitochondria or mitochondrial structures rather than nucleoids labelled by mTRIP.
Figure 1.
Experimental paradigm of FISH labelling with the mTRIP protocol. (A) Scheme of the 3D analysis of cells labelled with the mTRIP protocol (in this case probe mREP, upper panels, red) and also immunolabelled with TOM22 (middle panels, green). Merge is shown in lower panels. Steps 1–4 of data acquisition and processing are indicated. Briefly, optical z-slices of confocal acquisitions were 3D reconstructed, and then 2D volume rendered for fluorescence quantification, using the entire cell surface. (B) Schematic representation of the human mitochondrial genome (circular, below), and detail of the D-loop region at the level of the main replication origin (linear, above). The D-loop extends shortly beyond the 7S locus. In red are indicated the regions recognized by FISH probes used in this study (mREP and mTRANS). Single genes are indicated (tRNA genes with the corresponding letter). Ribosomal RNAs (16S and 12S) are in dark grey. The D-loop region is shown in black in the circular genome. PL and PH1–PH2, promoters of the L and H strand, respectively; OH, H-strand origin of replication. Probe mTRANS (mix of probes 1, 6 and 11, in red on the mt genome) labels only RNA, and includes 16S rRNA as well as mRNAs and tRNAs (see corresponding genes in the circular map). (C) Probe mREP (in red) recognizes only DNA in open structure (28). Scheme on the left represents structures that are recognized by probe mREP within an open replication bubble either with or without DNA synthesis (the new DNA chain is in purple). Position of the origin OH is indicated with a grey square, and the direction of replication with an arrow. Scheme on the right indicates structures that are not recognized by probe mREP because the replication bubble is either not formed at the level of OH (no DNA replication) or it is present in a region far from OH (elongating mtDNA). The mREP probe is indicated either as bound to the DNA (hatches represent hydrogen bonds) or unbound (line). Images show mTRIP labelling with probe mREP (see red foci) and immunostaining of the mitochondrial protein TOM22 (green) in a HeLa cell. Merge shows the distribution of mREP-labelled mitochondrial structures (initiation of mtDNA replication) within the mitochondrial network. (D) HeLa cell labelled with probe mTRANS and with anti-TOM22. Merge shows the distribution of transcript-rich mitochondrial structures (mTRANS) within the mitochondrial network. Scale bars = 10 µm.
With the mTRIP protocol and various combinations of single, double and triple labelling with probes mREP, mTRANS and the mitochondrial marker TOM22, we identified for the first time mitochondria engaged in initiation of mtDNA replication and distinct mitochondrial transcription profiles in single human cells. We detected a mosaic of mtDNA processing (replication and transcription) activities, namely (i) replication initiation active and transcript positive; (ii) replication initiation silent and transcript positive; and (iii) replication initiation silent and transcript negative, indicating that mitochondria may not be functionally alike inside a single cell.
Initiation of mtDNA replication precedes nDNA synthesis and is promoted again at the end of the S-phase in most mitochondrial structures
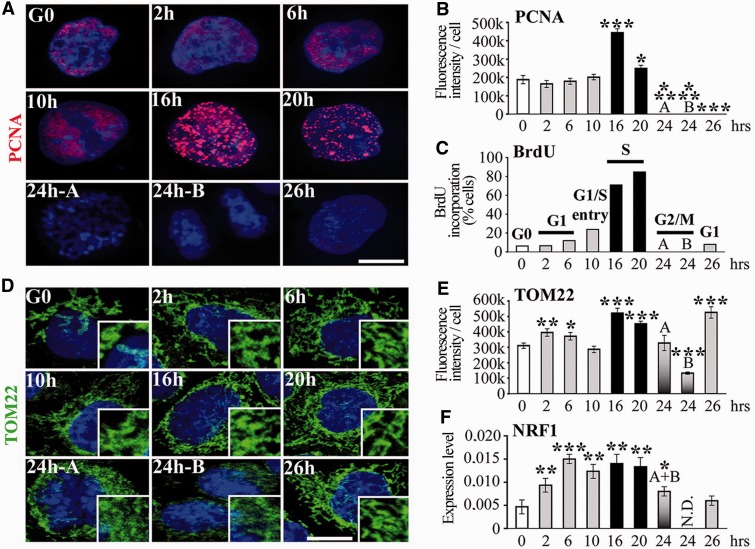
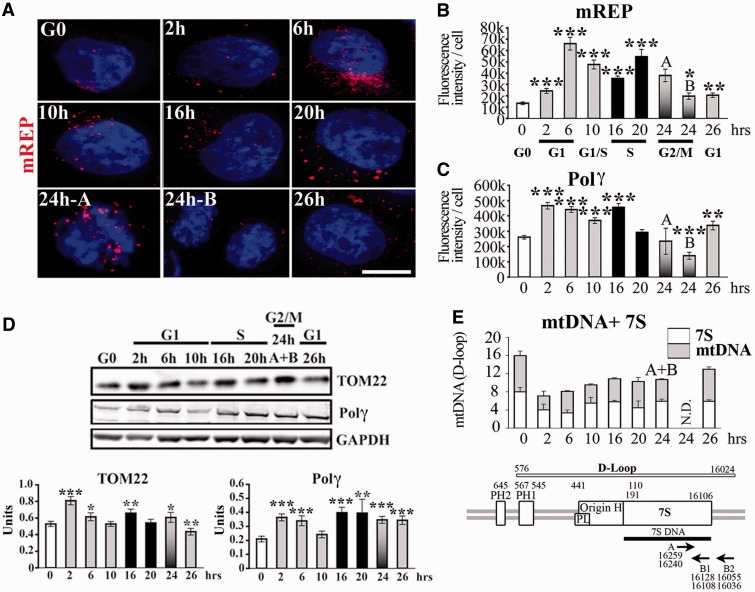
We used the mTRIP assay to assess the dynamics of DNA processing in mitochondria during the cell cycle in single cells. Mitochondria were analysed in synchronized HeLa cells released from G0 by relief from serum starvation, as described (26). Cell cycle progression monitored by BrdU incorporation and PCNA (a protein involved in DNA replication) immunolabelling defined the S-phase at 16–20 h after release from serum starvation (Figure 2A–C). We first observed an increase in the mitochondrial mass [measured by immunostaining of TOM22, as in (33) and (28), Figure 2D and E] in G1 and in the S-phase, compatible with increased mitochondrial mass detected in proliferating cells with a mitochondrial ligand (25). In the present experiments, we refer to the mitochondrial mass without specifying whether mitochondria are present as individual units or structured in the interconnected mitochondrial network. The increase in mitochondrial mass was confirmed by western blot of TOM22, and by high levels of transcripts of the mitochondrial biogenesis marker NRF1, from G1 to S (Figures 2F and 3D). Variations in the mitochondrial mass were not directly correlated with the cell size (Supplementary Figure S1A and D). Mitochondrial biogenesis was associated with a sharp increase in mREP labelling (initiation of mtDNA replication) in G1 and at the end of S-phase (Figure 3A and B). This finding was confirmed by increased levels of Polγ in early G1 and in the middle of S-phase by immunostaining (Figure 3C and Supplementary Figure S2A) and western blot (Figure 3D). The increase in mREP signal during the cell cycle was associated with DNA synthesis of the H-strand and with the transient triple stranded 7S structure at about equivalent amounts (Figure 3E), as measured by qPCR using two sets of primers, as described (27).
Figure 2.
Mitochondrial biogenesis during the cell cycle. (A) 3D-reconstructed HeLa cells synchronized in G0 by serum starvation. Anti-PCNA labelling (red) identifies phase of cell cycle. (B) Fluorescence intensity quantification of PCNA labelling. (C) Percentage of cells that incorporate BrdU (S-phase). Phases of cell cycle are indicated on top of columns. Code colours are constant for the histograms. (D) 3D-reconstructed HeLa cells immunostained with anti-TOM22 (green) to label the mitochondrial network. Cells in G2/M: pre-mitotic ‘A’ and post-mitotic ‘B’, according to the size of the nuclei. Magnification of the mitochondrial network is shown on the right low corner of each time point. (E) Fluorescence intensity quantification of TOM22 labelling. (F) RT-qPCR of mitochondrial biogenesis marker NRF1. n = 3. G2/M ‘A’ and ‘B’ cells cannot be separated in RT-qPCR experiments (single A + B column). t-test, each time point was compared with G0. Scale bars = 10 μm.
Figure 3.
Initiation of mtDNA replication during the cell cycle. (A) 3D-reconstructed HeLa cells synchronized in G0 by serum starvation and labelled with the mREP probe (red). Scale bar = 10 μm. Fluorescence intensity quantification of (B) mREP, and (C) Polγ (images shown in Supplementary Figure S2A). (D) Western blot of TOM22, Polγ and loading control GAPDH protein (upper panel). Quantification of signal for TOM22 and Polγ is expressed in arbitrary units normalized to GAPDH (lower panels). (E) Estimation of 7S DNA and mtDNA content by qPCR using primers described (27); see scheme below for position; n = 3. t-test, each time point was compared with G0.
Thus, mtDNA initiation of replication is elevated in G1 and during S-phase after release from serum starvation, and it precedes the biogenesis of the organelle. Interestingly, the distribution of mREP labelling (Figure 3A), combined to mREP and TOM22 signal intensity indicates that the higher levels of mREP are owing to the labelling of a larger fraction of mitochondria, rather than to an increased signal in a fixed fraction of mitochondria. Therefore, although mREP signal accounts for only ∼4% of the labelling of the mitochondrial mass (ratio of mREP/TOM22; Supplementary Figure S3A) in G0, this percentage increases to ∼18% in G1 and 12% at the end of S-phase. Interestingly, at each phase of the cell cycle, a large fraction of mitochondria did not appear to be involved in initiation of replication. Taken together, these data indicate a temporally consequent series of events linked to activation of mtDNA replication and organelle biogenesis during the cell cycle, which is modulated by varying the fraction of organelles implicated in this process.
Mitochondrial transcription precedes mitochondrial and nDNA synthesis in most mitochondrial structures
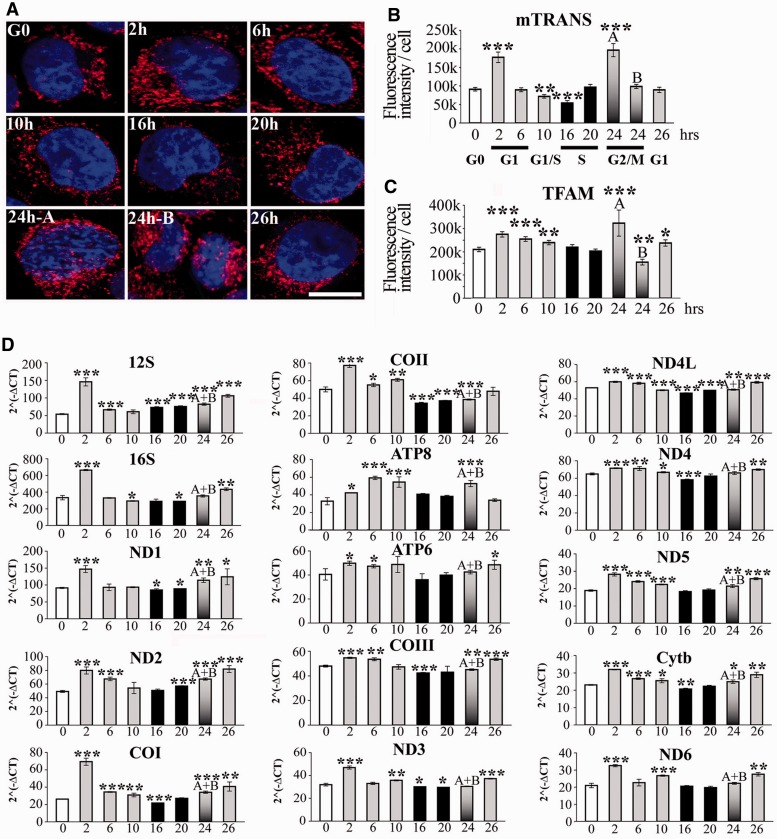
We then assessed whether transcription of mtDNA was affected during the cell cycle. HeLa cells labelled with mTRANS showed a 2-fold increase in signal intensity in G1 and in G2 (events ‘A’, but not in events ‘B’, defined as pre-mitotic and post-mitotic cells, respectively, based the size of the nuclei, in Figure 4A and B; for cell size measurements, see Supplementary Figure S1A) compared with G0. Interestingly, the lowest value of mTRANS signal was observed in the middle of S-phase (16 h after release from starvation). The increase in mTRANS signal in early G1 and in pre-mitotic cells (events ‘A’ at 24 h) was accompanied by a corresponding increase of immunolabelling of the mitochondrial transcription and DNA binding factor TFAM (Figure 4C and Supplementary Figure S2B).
Figure 4.
Transcription of the mitochondrial genome during the cell cycle. (A) 3D-reconstructed synchronized HeLa cells labelled with mTRANS probe (red), and Hoechst (blue). Scale bar = 10 μm. Fluorescence intensity quantification of (B) mTRANS, and (C) TFAM. (D) RT-qPCR analysis of the individual mitochondrial genes during the cell cycle (x-axis, h). n = 3. t-test, each time point was compared with G0.
We also performed RT-qPCR of each mitochondrial gene at the different phases of the cell cycle, with the exception of tRNAs, using RNA preparations from experiments in Figures 2 and 3. Transcript levels during the cell cycle were more heterogeneous for certain genes (e.g. ND2, COI) than for others (e.g. ND4, COIII). Nevertheless, for each gene, we observed the highest levels of transcripts in early G1 (2 h), with exception of ATP8 (Figure 4D), confirming the FISH results obtained with probe mTRANS. Moreover, the lowest values of transcripts for each gene were found in the middle of S-phase (16 h), with the exception of 12S, again confirming the FISH data. Therefore, mTRIP results were validated by RT-qPCR data, and they showed similar trends for the transcription of mitochondrial genes, with the highest levels in G1 and in G2/M, and the lowest at the peak of S-phase. In cells synchronized by detachment, high incorporation of labelled ribonucleotides in mitochondria was observed in G2 (34), in agreement with the present study, but also to some extent in the S-phase, in contrast to present FISH and RT-qPCR data.
Importantly, the distribution of mTRANS labelling (Figure 4A) combined to mTRANS and TOM22 signal intensity indicate that variations in the level of mitochondrial transcripts during the cell cycle depend on the fraction of transcript-carrying mitochondria rather than on the amount of transcripts per mitochondria, and this was also the case for the initiation of mtDNA replication shown earlier in the text. Indeed, mTRANS signal accounts for ∼10% of the labelling of the mitochondrial mass (ratio of mTRANS/TOM22; Supplementary Figure S3B) in the middle of the S-phase, and 60–74% in G2/M. In each phase of the cell cycle tested here, a fraction of mitochondria within single cells did not contain detectable levels of transcripts, at least those identified by mTRANS.
mtDNA initiation of replication and transcription are prevalently coordinated with nDNA synthesis after treatment with double thymidine or nocodazole
Previous reports regarding the coordination of mtDNA replication with the cell cycle, have been controversial, and we wondered whether different synchronization procedures used in those studies might account for some of these differences (16,17,19–23,27). To investigate this possibility, we synchronized cells with two additional treatments: double thymidine, which blocks the cell cycle in G1/S by generating and imbalanced nucleotide pool, and nocodazole, which interferes with the polymerization of microtubules, and arrests cells in the G2/M phase (35,36).
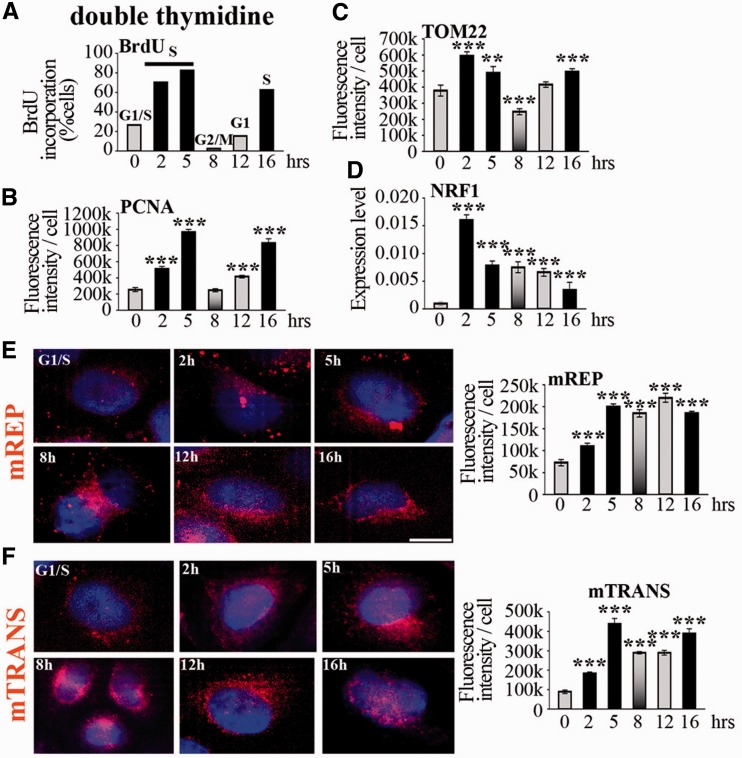
After release from treatment with double thymidine, cells resumed S-phase as shown by BrdU incorporation and PCNA immunostaining, completed the cycle and re-entered the next S-phase ∼11 h after the end of the first one (Figure 5A and B and Supplementary Figure S4A). The release from G1/S blockage resulted in increase of the mitochondrial mass (measured by TOM22 immunostaining) in S-phase (Figure 5C and Supplementary Figure S4B), confirmed by high expression of the biogenesis factor NRF1 (Figure 5D), as it was also the case with serum starvation. The levels of mREP were highest in S-phase, following the profile of nDNA synthesis, and remained high until the next S-phase (Figure 5E), whereas mTRANS levels, which were also the highest in the S-phase, decreased afterwards, and subsequently increased in the next S-phase (Figure 5F). These data are in agreement with a previous study that reported high levels of mtDNA synthesis in the S, G2 and in the next G1 phases after double thymidine treatment, leading to the notion that mtDNA synthesis is continuous during the cell cycle (21). Importantly, we observed also low levels of mtDNA initiation of replication (by mREP labelling) in early S-phase and in G1/S, and this was not examined in the previous study. Taken together, these results define a profile of mtDNA synthesis that appears to be coordinated with nDNA synthesis.
Figure 5.
Initiation of mtDNA replication and transcription in cells synchronized with double thymidine. (A) BrdU incorporation and (B) anti-PCNA immunostaining identify phases of cell cycle, indicated on top of columns in (A). The code colour is constant for histograms. (C) Quantification of TOM22 immunofluorescence during cell cycle. For cell size measurement, see Supplementary Figure S1. (D) Expression of NRF1 during cell cycle analysed by RT-qPCR; n = 3. t-test, each time point was compared with G1/S. (E) mREP and (F) mTRANS labelling (red) in 3D-reconstructed cells during the cell cycle; scale bar = 10 µm. Fluorescence intensity quantification is indicated on the right.
Several parameters linked to DNA synthesis and transcription were dramatically elevated after double thymidine treatment compared with untreated cells, or cells blocked by serum starvation, namely PCNA labelling, mREP and mTRANS levels, as well as mtDNA and 7S content, despite comparable levels of the mitochondrial mass (Figure 5 and Supplementary Figure S5).
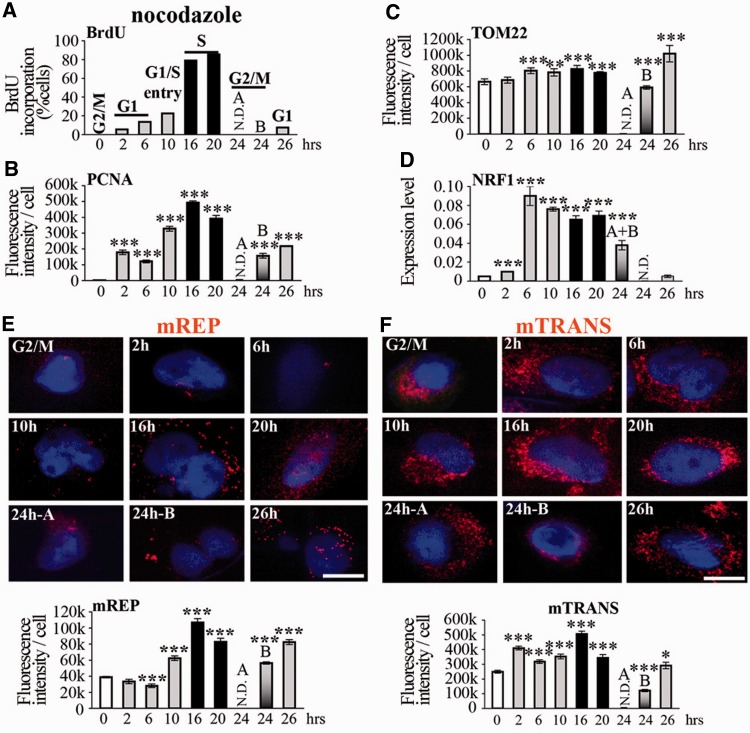
Release from G2/M cell cycle blocking with nocodazole resulted in an increase in mREP label until S-phase, with a profile similar to that of nDNA synthesis (cf. PCNA and mREP fluorescence intensity histograms, Figure 6B and E, Supplementary Figure S4C), and this was also the case for double thymidine treatment. Interestingly, during the cell cycle, 7S levels were consistently higher than mtDNA values, in particular in G1/S (4.6-fold), indicating that a large part of initiation of replication actually results in production of 7S. It is thus possible that variations in the levels of 7S (but not of mtDNA) during the cell cycle that were previously reported for cells treated with double thymidine and nocodazole (27) were largely owing to the synchronization procedures. Moreover, synchronization with nocodazole severely altered the mtDNA content during the cell cycle (with the highest values at the end of the S-phase, Supplementary Figure S5A), differently from serum starvation treatment, which maintained relatively constant levels of mtDNA probably by balanced DNA synthesis and loss.
Figure 6.
Initiation of mtDNA replication and transcription in cells synchronized with nocodazole. Treatment with nocodazole. Legend as in Figure 5. t-test, each time point was compared with G2/M.
After release from nocodazole treatment, the mitochondrial mass remained high, with a slight increase in S-phase, a decrease in G2/M and again higher values in the next G1 (Figure 6C, Supplementary Figure S4D), a similar but less accentuated profile than with the other cell cycle blockage procedures. We note that the mild increase in the mitochondrial mass in G1 was associated with 9-fold higher transcript levels of the mitochondrial biogenesis factor NRF1 (Figure 6D). Finally, mitochondrial transcription, evaluated by mTRANS labelling, was highest in G1, as it was after serum starvation, but also during S-phase (Figure 6F), in coordination with the nuclear phase.
Removal of cell blockage by nocodazole globally increased several mitochondrial parameters throughout the entire cell cycle, namely mitochondrial transcription (2.5-fold higher levels of mTRANS and 15–30-fold higher levels of transcripts by RT-qPCR), mtDNA content and, to a lower extent mitochondrial mass compared with untreated cells, or cells synchronized by serum starvation (Figure 6 and Supplementary Figure S5).
DISCUSSION
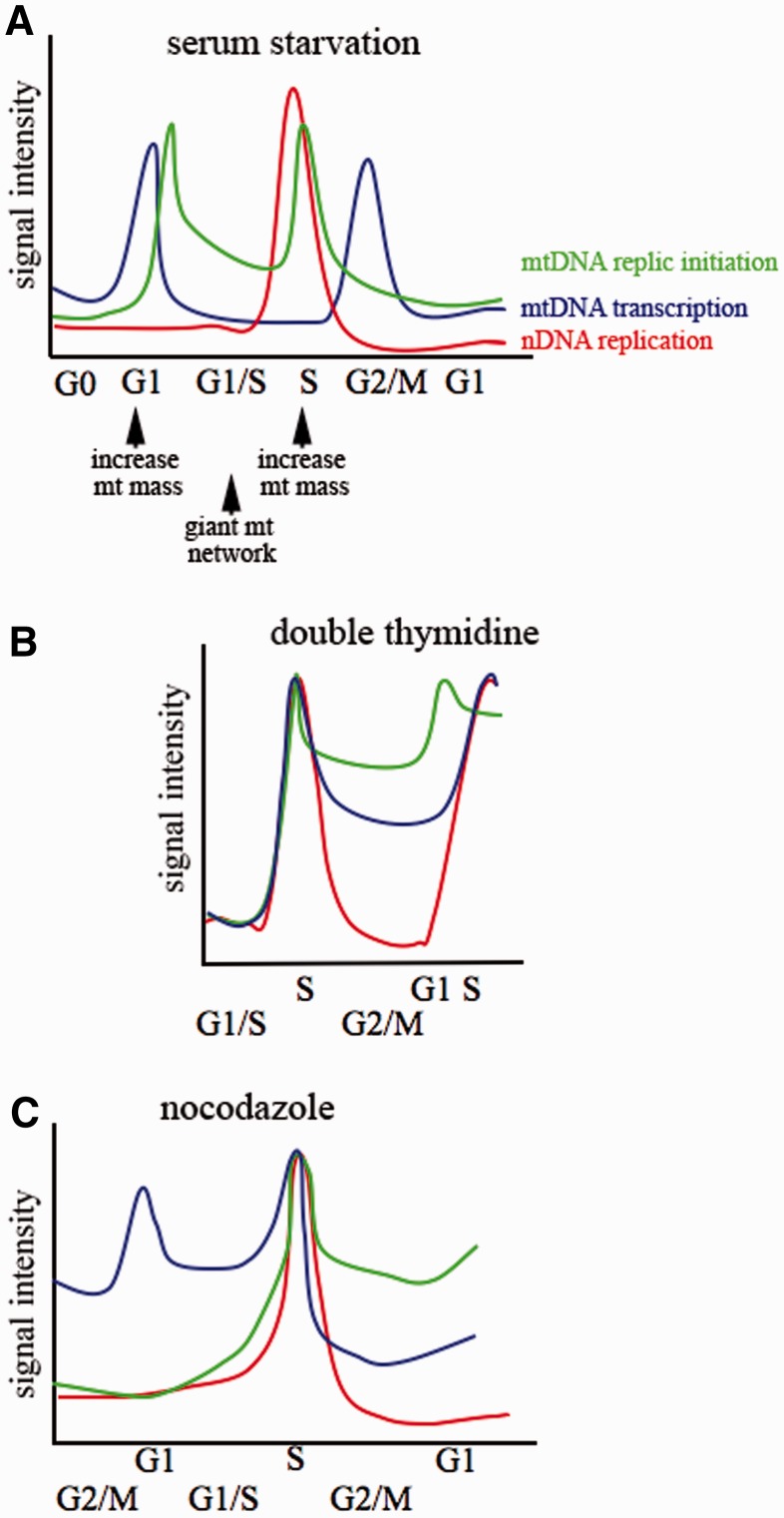
The analysis of single cells allows detecting distinct mitochondrial and nuclear events, thereby overcoming the risk of contamination among subcellular components that might have occurred in other studies, and provides a unique tool for reconstructing the temporal consequent series of mtDNA processing events during the cell cycle. Taken together, we report that mitochondrial transcription increases in G1 just after release from serum depletion, and it is followed, a few hours later again in G1, by a sharp increase in the initiation of mtDNA replication (Figure 7A). Accordingly, mitochondria were reported to promote the G1 to S progression (26). mtDNA initiation of replication and transcription are lowest during nDNA replication. Initiation of mtDNA replication increases again at the end of the S-phase, and it declines subsequently. This last event is followed by high levels of mitochondrial transcription in G2 before cells undergo mitosis. Thus, in cells synchronized by serum depletion mitochondria initiate DNA replication and produce transcripts at the highest levels twice during the cell cycle, the first event before nDNA synthesis, and the second before mitosis.
Figure 7.
Schematic representation of the temporal consequent series of mitochondrial and nuclear events during the cell cycle. Cell cycle synchronized by (A) serum starvation, (B) double thymidine and (C) nocodazole. nDNA replication as from BrdU incorporation, mtDNA initiation of replication as from mREP labelling, mitochondrial transcription from mTRANS labelling and RT-qPCR of mitochondrial transcripts. x-axis and y-axis coordinates are arbitrarily represented. X-axis is shifted to place S-phases at the same point for the three synchronization procedures. Arrows below panel (A) indicate the increase of the mitochondrial mass, as from TOM22 immunolabelling, and formation of the giant mitochondrial network promoting G1 to S transition as from reference (26). The same colour code is used for the three panels.
Notably, the coordination of the levels of mtDNA processing during the cell cycle is achieved by modulating the fraction of mitochondria or of mitochondrial structures that initiate mtDNA replication or contain transcripts at a given time. Thus, the processing of the mitochondrial genome, which is present in hundreds/thousands of copies per cell, is active throughout the entire cell cycle but in a limited fraction of mitochondria or mitochondrial structures, whereas peaks of these activities are synchronized with nDNA synthesis, resulting in a prevalent coordination with the cell cycle. Importantly, at each tested phase of the cell cycle, a relevant fraction of mitochondria is silent for initiation of replication and/or transcript accumulation.
Interestingly, blocking the cell cycle in G1/S with double thymidine or in G2/M with nocodazole reveals that mtDNA initiation of replication and transcription are prevalently coordinated with the cell cycle, and that they are synchronized with nDNA synthesis (Figure 7B and C). However, both drugs not only affect the cell cycle but also block mtDNA synthesis (37,38). Thus, restoring physiological concentrations of deoxynucleotide triphosphates (dNTPs) (after release from double thymidine treatment) and re-establishing a functional microtubule network (after release from nocodazole treatment) might simultaneously promote mtDNA and nDNA synthesis, explaining why these processes appear coordinated.
These results also show that the method of synchronization affects the response of mitochondria, in addition to perturbing the cell cycle (39), and that several key mitochondrial parameters are greatly altered after treatment with double thymidine and nocodazole, compared with serum starvation or untreated cells. Serum starvation also metabolically perturbs cells, as they must perform additional metabolic reactions to return to the cycle (40). However, these alterations appear to be less drastic than drug-induced cell cycle blockage in directly affecting the processing of mtDNA. Despite these differences, we note that all treatments showed that mtDNA synthesis is coordinated with nDNA synthesis in at least one distinct period of the cell cycle, the S-phase (serum starvation additionally showed an earlier coordination between mtDNA replication and transcription with the cell cycle in G1).
The large availability of nucleotides during the nuclear S-phase [∼20-fold more than in resting cells and several fold more than in G1 (41)] might lead to the prevalent synchronization of mtDNA with nDNA synthesis. However, reported levels of dNTPs do not necessarily correlate with the profile of initiation of mtDNA replication and related parameters that we examined, in particular for synchronization by serum starvation. Moreover, dNTPs especially supplied for mtDNA synthesis by the ribonucleotide reductase p53R3 are produced at low but constant levels throughout the cell cycle (42). Thus, it seem unlikely that mtDNA replication bursts are solely driven by the concentration of dNTPs, and the mechanism of prevalent coordination of mtDNA replication and transcription with the cell cycle remains to be elucidated.
By analysis of nuclear and mitochondrial events in single cells largely based on a novel approach, we reconcile apparently contradictory reports that appeared in the past four decades. We provide evidences that transcription and replication of mtDNA are prevalently coordinated with nDNA synthesis events and also show that this coordination can be adapted in response to alterations in cell physiology. These findings have implications in redefining functional nuclear–mitochondrial links and may help redirect investigations on these interactions in diseased cell states.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–5, Supplementary Results and Supplementary Reference [43].
FUNDING
Association Nationale contre le Cancer (ARC 4022 and SFI20111204038), PTR-Institut Pasteur (PTR217), DARRI-Institut Pasteur (project p790319), Agence Nationale pour la Recherche (ANR 11BSV202502), and Centre National de la Recherche Scientifique (CNRS, UMR 3525, Team Stability of Nuclear and Mitochondrial DNA). L.C. was recipient of a Bourse Roux, and was supported by F. Lacoste. Funding for open access charge: ARC, Institut Pasteur, ANR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank F. Brandizzi, B. Dujon, S. Tajbakhsh and C. Zurzolo, for comments on the manuscript, and the Imagopole (PFID) of Institut Pasteur for advice.
REFERENCES
- 1.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 2.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 3.Rotig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim. Biophys. Acta. 2009;1792:1103–1108. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Liu D, Lu J, Bai Y. Physiology and pathophysiology of mitochondrial DNA. Adv. Exp. Med. Biol. 2012;942:39–51. doi: 10.1007/978-94-007-2869-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 6.Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- 7.Kohler C, Radpour R, Barekati Z, Asadollahi R, Bitzer J, Wight E, Burki N, Diesch C, Holzgreve W, Zhong XY. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol. Cancer. 2009;8:105. doi: 10.1186/1476-4598-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 9.Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatre L, Ricchetti M. Nuclear mitochondrial DNA activates replication in saccharomyces cerevisiae. PLoS One. 2011;6:e17235. doi: 10.1371/journal.pone.0017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 12.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 14.Chang DD, Hauswirth WW, Clayton DA. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985;4:1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 16.Bogenhagen D, Clayton DA. Mouse L cell mitochondrial DNA molecules are selected randomly for replication throughout the cell cycle. Cell. 1977;11:719–727. doi: 10.1016/0092-8674(77)90286-0. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson J, Orth M, Lestienne P, Taanman JW. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp. Cell Res. 2003;289:133–142. doi: 10.1016/s0014-4827(03)00249-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Kim S, Sun X, Lee JH, Cho H. Cell cycle-dependent mitochondrial biogenesis and dynamics in mammalian cells. Biochem. Biophys. Res. Commun. 2007;357:111–117. doi: 10.1016/j.bbrc.2007.03.091. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Diez M, Santamaria G, Ortega AD, Cuezva JM. Biogenesis and dynamics of mitochondria during the cell cycle: significance of 3'UTRs. PLoS One. 2006;1:e107. doi: 10.1371/journal.pone.0000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volpe P, Eremenko T. Nuclear and cytoplasmic DNA synthesis during the mitotic cycle of HeLa cells. Eur. J. Biochem. 1973;32:227–232. doi: 10.1111/j.1432-1033.1973.tb02600.x. [DOI] [PubMed] [Google Scholar]
- 21.Pica-Mattoccia L, Attardi G. Expression of the mitochondrial genome in HeLa cells. IX. Replication of mitochondrial DNA in relationship to cell cycle in HeLa cells. J. Mol. Biol. 1972;64:465–484. doi: 10.1016/0022-2836(72)90511-6. [DOI] [PubMed] [Google Scholar]
- 22.Radsak K, Schutz E. Changes of mitochondrial DNA polymerase-gamma activity in synchronized mouse cell cultures. Eur. J. Biochem. 1978;89:3–9. doi: 10.1111/j.1432-1033.1978.tb20889.x. [DOI] [PubMed] [Google Scholar]
- 23.Posakony JW, England JM, Attardi G. Mitochondrial growth and division during the cell cycle in HeLa cells. J. Cell Biol. 1977;74:468–491. doi: 10.1083/jcb.74.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schieke SM, McCoy JP, Jr, Finkel T. Coordination of mitochondrial bioenergetics with G1 phase cell cycle progression. Cell Cycle. 2008;7:1782–1787. doi: 10.4161/cc.7.12.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garedew A, Andreassi C, Moncada S. Mitochondrial dynamics, biogenesis, and function are coordinated with the cell cycle by APC/C(CDH1) Cell Metab. 2012;15:466–479. doi: 10.1016/j.cmet.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl Acad. Sci. USA. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antes A, Tappin I, Chung S, Lim R, Lu B, Parrott AM, Hill HZ, Suzuki CK, Lee CG. Differential regulation of full-length genome and a single-stranded 7S DNA along the cell cycle in human mitochondria. Nucleic Acids Res. 2010;38:6466–6476. doi: 10.1093/nar/gkq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatre L, Ricchetti M. Large heterogenietyof mitochondrial DNA transcription and initiation of replication exposed by single-cell imaging. J. Cell Sci. 2012 doi: 10.1242/jcs.114322. December 13 (doi: 10.1242/jcs.114322; epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 30.Fan AX, Radpour R, Haghighi MM, Kohler C, Xia P, Hahn S, Holzgreve W, Zhong XY. Mitochondrial DNA content in paired normal and cancerous breast tissue samples from patients with breast cancer. J. Cancer Res. Clin. Oncol. 2009;135:983–989. doi: 10.1007/s00432-008-0533-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell Biol. 2011;31:4994–5010. doi: 10.1128/MCB.05694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Latil M, Rocheteau P, Chatre L, Sanulli S, Memet S, Ricchetti M, Tajbakhsh S, Chretien F. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nat. Commun. 2012;3:903. doi: 10.1038/ncomms1890. [DOI] [PubMed] [Google Scholar]
- 34.Pica-Mattoccia L, Attardi G. Expression of the mitochondrial genome in HeLa cells. V. Transcription of mitochondrial DNA in relationship to the cell cycle. J. Mol. Biol. 1971;57:615–621. doi: 10.1016/0022-2836(71)90113-6. [DOI] [PubMed] [Google Scholar]
- 35.Bjursell G, Reichard P. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 1973;248:3904–3909. [PubMed] [Google Scholar]
- 36.Harper JV. Synchronization of cell populations in G1/S and G2/M phases of the cell cycle. Methods Mol. Biol. 2005;296:157–166. doi: 10.1385/1-59259-857-9:157. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez-Vioque E, Torres-Torronteras J, Andreu AL, Marti R. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) PLoS Genet. 2011;7:e1002035. doi: 10.1371/journal.pgen.1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karbowski M, Spodnik JH, Teranishi M, Wozniak M, Nishizawa Y, Usukura J, Wakabayashi T. Opposite effects of microtubule-stabilizing and microtubule-destabilizing drugs on biogenesis of mitochondria in mammalian cells. J. Cell Sci. 2001;114:281–291. doi: 10.1242/jcs.114.2.281. [DOI] [PubMed] [Google Scholar]
- 39.Urbani L, Sherwood SW, Schimke RT. Dissociation of nuclear and cytoplasmic cell cycle progression by drugs employed in cell synchronization. Exp. Cell Res. 1995;219:159–168. doi: 10.1006/excr.1995.1216. [DOI] [PubMed] [Google Scholar]
- 40.Pardee AB, Keyomarsi K. Modification of cell proliferation with inhibitors. Curr. Opin. Cell Biol. 1992;4:186–191. doi: 10.1016/0955-0674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 41.Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 42.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, et al. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat. Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 43.Xiong W, Jiao Y, Huang W, Ma M, Yu M, Cui Q, Tan D. Regulation of the cell cycle via mitochondrial gene expression and energy metabolism in HeLa cells. Acta. Biochim. Biophys. Sin. (Shanghai) 2012;44:347–358. doi: 10.1093/abbs/gms006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.